Structure of a DNA G-Quadruplex Related to Osteoporosis with a G-A Bulge Forming a Pseudo-loop
Abstract
:1. Introduction
2. Results
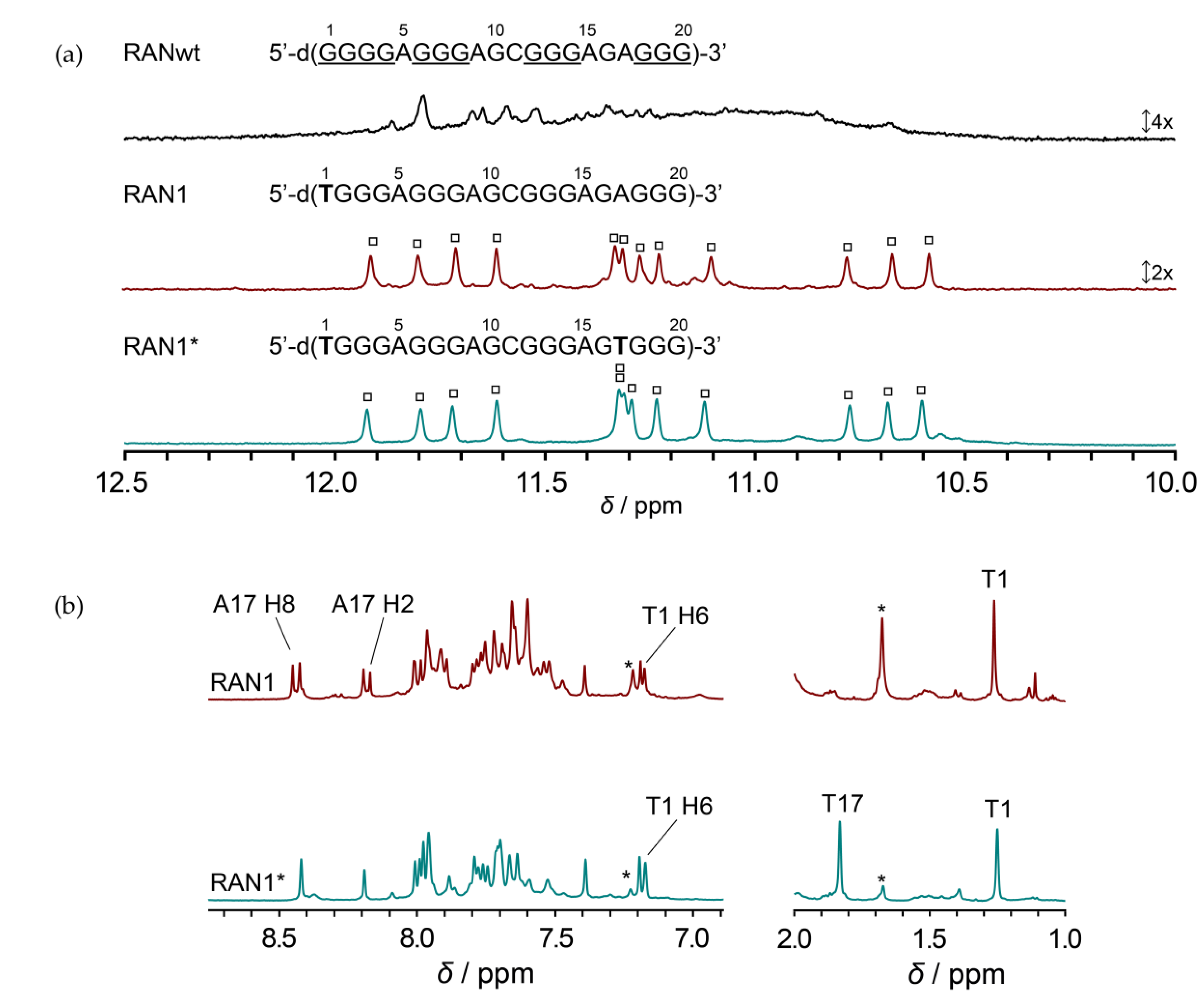
2.1. Favoring a Single Conformation by Sequence Modifications of RANwt
2.2. RAN1* Forms a Parallel G-Quadruplex with a G-A Bulge
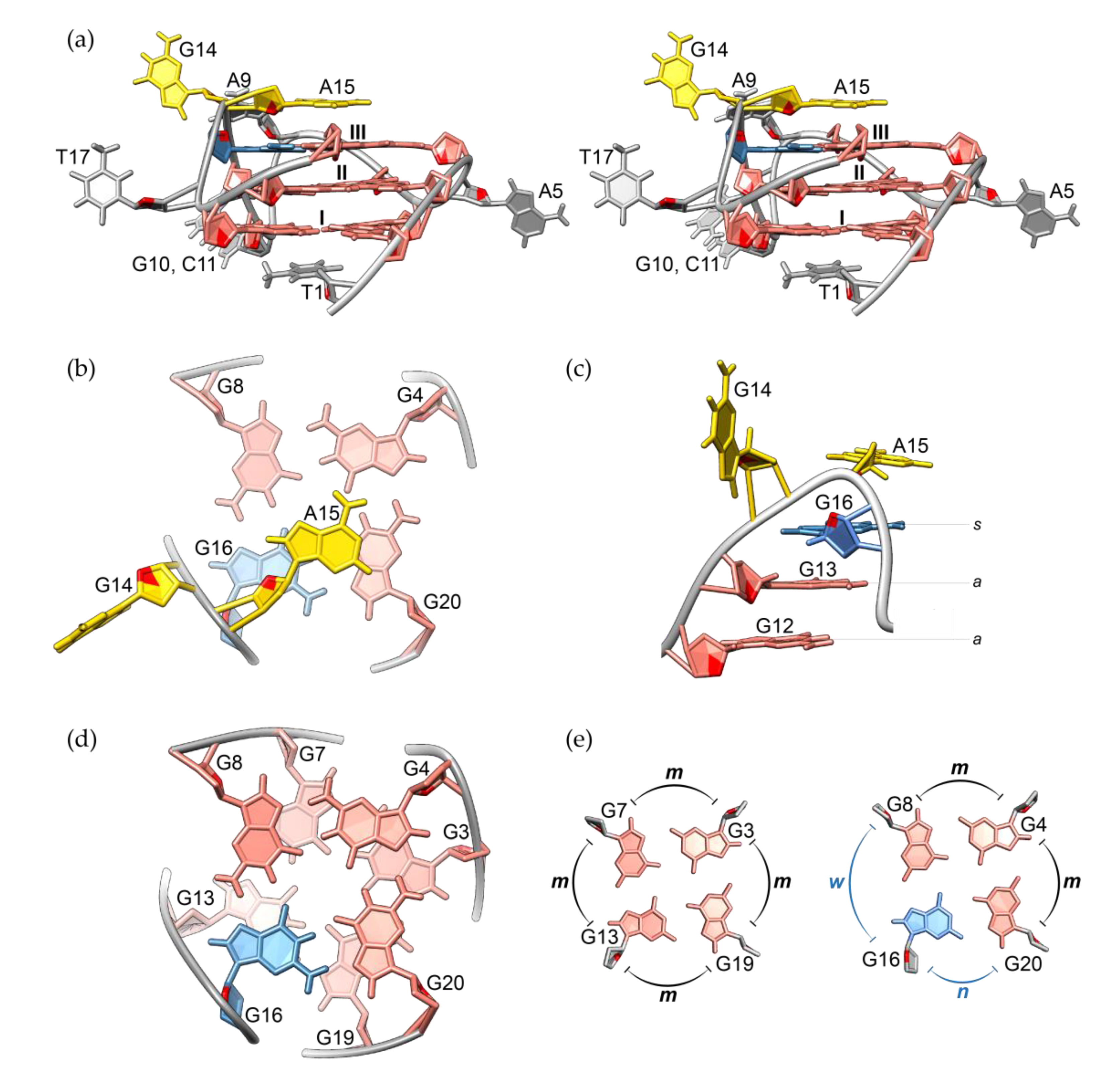
2.3. Solution Structure of the RAN1* G-Quadruplex
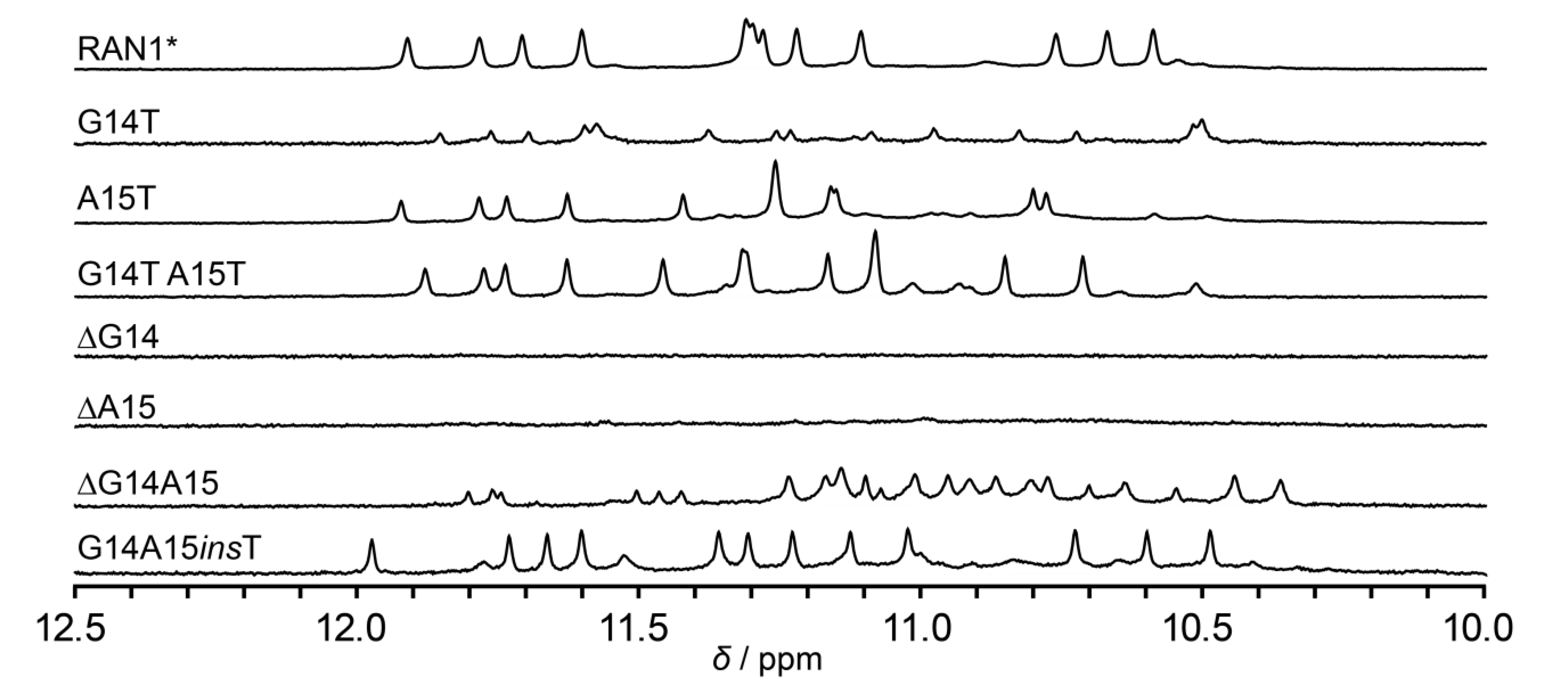
2.4. Effect of a Bulge Modifications on the RAN1* Structure
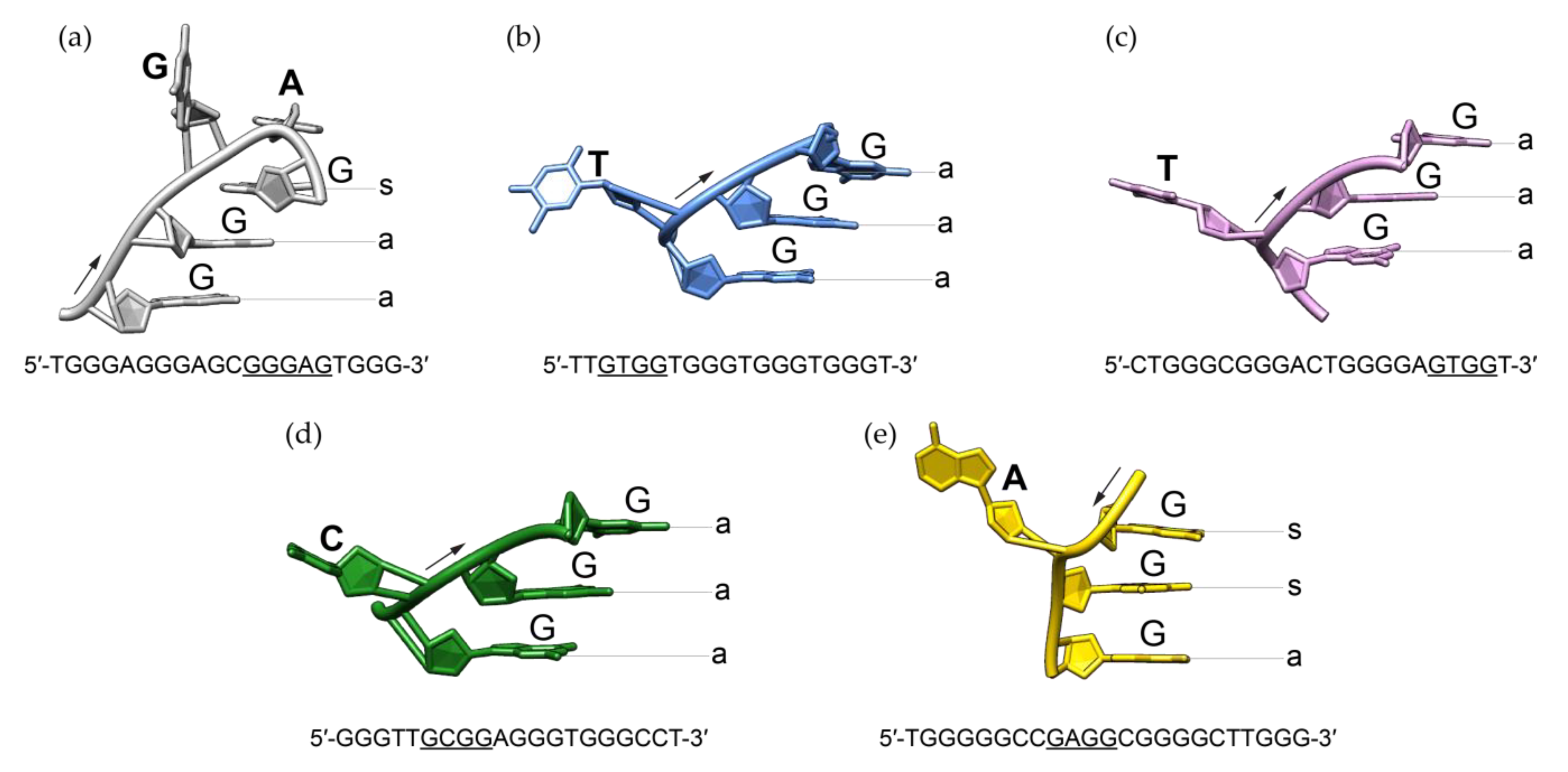
3. Discussion
4. Materials and Methods
4.1. Sample Preparation
4.2. CD and UV Experiments
4.3. NMR Experiments and Structure Calculations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Kaushik, M.; Kaushik, S.; Roy, K.; Singh, A.; Mahendru, S.; Kumar, M.; Chaudhary, S.; Ahmed, S.; Kukreti, S. A bouquet of DNA structures: Emerging diversity. Biochem. Biophys. Rep. 2016, 5, 388–395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwok, C.K.; Merrick, C.J. G-Quadruplexes: Prediction, Characterization, and Biological Application. Trends Biotechnol. 2017, 35, 997–1013. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Vasquez, K.M. Impact of alternative DNA structures on DNA damage, DNA repair, and genetic instability. DNA Repair 2014, 19, 143–151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rhodes, D.; Lipps, H.J. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015, 43, 8627–8637. [Google Scholar] [CrossRef] [Green Version]
- Saini, N.; Zhang, Y.; Usdin, K.; Lobachev, K.S. When secondary comes first--the importance of non-canonical DNA structures. Biochimie 2012, 95, 117–123. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.; Vasquez, K.M. Non-B DNA structure-induced genetic instability. Mutat. Res. Mol. Mech. Mutagen. 2006, 598, 103–119. [Google Scholar] [CrossRef]
- Tateishi-Karimata, H.; Kawauchi, K.; Sugimoto, N. Destabilization of DNA G-Quadruplexes by Chemical Environment Changes during Tumor Progression Facilitates Transcription. J. Am. Chem. Soc. 2017, 140, 642–651. [Google Scholar] [CrossRef]
- Kim, B.G.; Chalikian, T.V. Thermodynamic linkage analysis of pH-induced folding and unfolding transitions of i-motifs. Biophys. Chem. 2016, 216, 19–22. [Google Scholar] [CrossRef]
- Bielskutė, S.; Plavec, J.; Podbevšek, P. Impact of Oxidative Lesions on the Human Telomeric G-Quadruplex. J. Am. Chem. Soc. 2019, 141, 2594–2603. [Google Scholar] [CrossRef] [Green Version]
- Galer, P.; Wang, B.; Šket, P.; Plavec, J. Reversible pH Switch of Two-Quartet G-Quadruplexes Formed by Human Telomere. Angew. Chem. Int. Ed. 2016, 55, 1993–1997. [Google Scholar] [CrossRef]
- Nakano, S.-I.; Miyoshi, D.; Sugimoto, N. Effects of Molecular Crowding on the Structures, Interactions, and Functions of Nucleic Acids. Chem. Rev. 2013, 114, 2733–2758. [Google Scholar] [CrossRef] [PubMed]
- Trajkovski, M.; Endoh, T.; Tateishi-Karimata, H.; Ohyama, T.; Tanaka, S.; Plavec, J.; Sugimoto, N. Pursuing origins of (poly)ethylene glycol-induced G-quadruplex structural modulations. Nucleic Acids Res. 2018, 46, 4301–4315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huppert, J.L.; Balasubramanian, S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007, 35, 2105. [Google Scholar] [CrossRef] [Green Version]
- Biffi, G.; Tannahill, D.; McCafferty, J.; Balasubramanian, S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013, 5, 182–186. [Google Scholar] [CrossRef] [PubMed]
- Burge, S.; Parkinson, G.N.; Hazel, P.; Todd, A.K.; Neidle, S. Quadruplex DNA: Sequence, topology and structure. Nucleic Acids Res. 2006, 34, 5402–5415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Todd, A.K. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005, 33, 2901–2907. [Google Scholar] [CrossRef] [Green Version]
- Chambers, V.S.; Marsico, G.; Boutell, J.M.; Di Antonio, M.; Smith, G.P.; Balasubramanian, S. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat. Biotechnol. 2015, 33, 877–881. [Google Scholar] [CrossRef] [Green Version]
- Huppert, J.L. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005, 33, 2908–2916. [Google Scholar] [CrossRef] [Green Version]
- Lightfoot, H.L.; Hagen, T.; Tatum, N.J.; Hall, J. The diverse structural landscape of quadruplexes. FEBS Lett. 2019, 593, 2083–2102. [Google Scholar] [CrossRef]
- Murat, P.; Balasubramanian, S. Existence and consequences of G-quadruplex structures in DNA. Curr. Opin. Genet. Dev. 2014, 25, 22–29. [Google Scholar] [CrossRef] [Green Version]
- Jonchhe, S.; Pandey, S.; Karna, D.; Pokhrel, P.; Cui, Y.; Mishra, S.; Sugiyama, H.; Endo, M.; Mao, H. Duplex DNA Is Weakened in Nanoconfinement. J. Am. Chem. Soc. 2020, 142, 10042–10049. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, M.W. Geometric Formalism for DNA Quadruplex Folding. Chem. Eur. J. 2007, 13, 9738–9745. [Google Scholar] [CrossRef] [PubMed]
- Karsisiotis, A.I.; O’Kane, C.; Da Silva, M.W. DNA quadruplex folding formalism–A tutorial on quadruplex topologies. Methods 2013, 64, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Haase, L.; Dickerhoff, J.; Weisz, K. Sugar Puckering Drives G-Quadruplex Refolding: Implications for V-Shaped Loops. Chem. Eur. J. 2019, 26, 524–533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phan, A.T.; Kuryavyi, V.; Gaw, H.Y.; Patel, D.J. Small-molecule interaction with a five-guanine-tract G-quadruplex structure from the human MYC promoter. Nat. Chem. Biol. 2005, 1, 167–173. [Google Scholar] [CrossRef]
- Phan, A.T.; Kuryavyi, V.; Burge, S.; Neidle, S.; Patel, D.J. Structure of an unprecedented G-quadruplex scaffold in the human c-kit promoter. J. Am. Chem. Soc. 2007, 129, 4386–4392. [Google Scholar] [CrossRef] [Green Version]
- Mukundan, V.T.; Phan, A.T. Bulges in G-Quadruplexes: Broadening the Definition of G-Quadruplex-Forming Sequences. J. Am. Chem. Soc. 2013, 135, 5017–5028. [Google Scholar] [CrossRef]
- De Nicola, B.; Lech, C.J.; Heddi, B.; Regmi, S.; Frasson, I.; Perrone, R.; Richter, S.N.; Phan, A.T. Structure and possible function of a G-quadruplex in the long terminal repeat of the proviral HIV-1 genome. Nucleic Acids Res. 2016, 44, 6442–6451. [Google Scholar] [CrossRef]
- Meier, M.; Torres, A.M.; Krahn, N.J.; McDougall, M.D.; Orriss, G.L.; McRae, E.; Booy, E.P.; McEleney, K.; Patel, T.R.; A McKenna, S.; et al. Structure and hydrodynamics of a DNA G-quadruplex with a cytosine bulge. Nucleic Acids Res. 2018, 46, 5319–5331. [Google Scholar] [CrossRef]
- Sengar, A.; Vandana, J.J.; Chambers, V.S.; Di Antonio, M.; Winnerdy, F.R.; Balasubramanian, S.; Phan, A.T. Structure of a (3+1) hybrid G-quadruplex in the PARP1 promoter. Nucleic Acids Res. 2018, 47, 1564–1572. [Google Scholar] [CrossRef] [Green Version]
- Wada, T.; Nakashima, T.; Hiroshi, N.; Penninger, J.M. RANKL–RANK signaling in osteoclastogenesis and bone disease. Trends Mol. Med. 2006, 12, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Diao, L.; Sun, D.; Wang, D.; Zhu, J.; He, Y.; Liu, Y.; Xu, H.; Zhang, Y.; Liu, J.; et al. Osteoporos Atlas: A human osteoporosis-related gene database. PeerJ 2019, 7, e6778. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walsh, M.C.; Choi, Y. Biology of the RANKL-RANK-OPG system in immunity, bone, and beyond. Front. Immunol. 2014, 5, 511. [Google Scholar] [CrossRef] [Green Version]
- Sobacchi, C.; Menale, C.; Villa, A. The RANKL-RANK Axis: A Bone to Thymus Round Trip. Front. Immunol. 2019, 10, 629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McClung, M.R.; Lewiecki, E.M.; Cohen, S.B.; Bolognese, M.A.; Woodson, G.C.; Moffett, A.H.; Peacock, M.; Miller, P.D.; Lederman, S.N.; Chesnut, C.H.; et al. Denosumab in Postmenopausal Women with Low Bone Mineral Density. N. Engl. J. Med. 2006, 354, 821–831. [Google Scholar] [CrossRef] [PubMed]
- Cummings, S.R.; Martin, J.S.; McClung, M.R.; Siris, E.S.; Eastell, R.; Reid, I.R.; Delmas, P.; Zoog, H.B.; Austin, M.; Wang, A.; et al. Denosumab for Prevention of Fractures in Postmenopausal Women with Osteoporosis. N. Engl. J. Med. 2009, 361, 756–765. [Google Scholar] [CrossRef] [Green Version]
- Boonen, S.; Adachi, J.D.; Man, Z.; Cummings, S.R.; Lippuner, K.; Törring, O.; Gallagher, J.C.; Farrerons, J.; Wang, A.; Franchimont, N.; et al. Treatment with Denosumab Reduces the Incidence of New Vertebral and Hip Fractures in Postmenopausal Women at High Risk. J. Clin. Endocrinol. Metab. 2011, 96, 1727–1736. [Google Scholar] [CrossRef] [Green Version]
- Hoffmann, D.B.; Böker, K.O.; Schneider, S.; Eckermann-Felkl, E.; Schuder, A.; Komrakova, M.; Sehmisch, S.; Gruber, J. In Vivo siRNA Delivery Using JC Virus-like Particles Decreases the Expression of RANKL in Rats. Mol. Ther. Nucleic Acids 2016, 5, e298. [Google Scholar] [CrossRef]
- Hoffmann, D.B.; Gruber, J.; Böker, K.O.; Deppe, D.; Sehmisch, S.; Schilling, A.F.; Lemus-Diaz, N.; Komrakova, M.; Schneider, S. Effects of RANKL Knockdown by Virus-like Particle-Mediated RNAi in a Rat Model of Osteoporosis. Mol. Ther. Nucleic Acids 2018, 12, 443–452. [Google Scholar] [CrossRef] [Green Version]
- Živković, M.L.; Rozman, J.; Plavec, J. Adenine-Driven Structural Switch from a Two- to Three-Quartet DNA G-Quadruplex. Angew. Chem. Int. Ed. 2018, 57, 15395–15399. [Google Scholar] [CrossRef] [Green Version]
- Seenisamy, J.; Rezler, E.M.; Powell, T.J.; Tye, D.; Gokhale, V.; Joshi, C.S.; Siddiqui-Jain, A.; Hurley, L.H. The Dynamic Character of the G-Quadruplex Element in the c-MYC Promoter and Modification by TMPyP4. J. Am. Chem. Soc. 2004, 126, 8702–8709. [Google Scholar] [CrossRef]
- Chaires, J.B.; Jt, G.; C, H.; Dp, K.; Rw, H.; I, B.; A, H.; Ak, M.; H, S. Faculty Opinions recommendation of Conformational Dynamics of Strand Register Shifts in DNA G-Quadruplexes. Fac. Opin. Post-Publ. Peer Rev. Biomed. Lit. 2020, 142, 264–273. [Google Scholar] [CrossRef]
- Lech, C.J.; Heddi, B.; Phan, A.T. Guanine base stacking in G-quadruplex nucleic acids. Nucleic Acids Res. 2012, 41, 2034–2046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hazel, P.; Huppert, J.; Balasubramanian, S.; Neidle, S. Loop-Length-Dependent Folding of G-Quadruplexes. J. Am. Chem. Soc. 2004, 126, 16405–16415. [Google Scholar] [CrossRef]
- Bugaut, A.; Balasubramanian, S. A sequence-independent study of the influence of short loop lengths on the stability and topology of intramolecular DNA G-quadraplexes. Biochemistry 2008, 47, 689–697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takahama, K.; Sugimoto, C.; Arai, S.; Kurokawa, R.; Oyoshi, T. Loop Lengths of G-Quadruplex Structures Affect the G-Quadruplex DNA Binding Selectivity of the RGG Motif in Ewing’s Sarcoma. Biochemistry 2011, 50, 5369–5378. [Google Scholar] [CrossRef]
- Huang, Z.-L.; Dai, J.; Luo, W.-H.; Wang, X.-G.; Tan, J.-H.; Chen, S.-B.; Huang, Z.-S. Identification of G-Quadruplex-Binding Protein from the Exploration of RGG Motif/G-Quadruplex Interactions. J. Am. Chem. Soc. 2018, 140, 17945–17955. [Google Scholar] [CrossRef]
- Wu, G.; Xing, Z.; Tran, E.J.; Yang, D. DDX5 helicase resolves G-quadruplex and is involved in MYC gene transcriptional activation. Proc. Natl. Acad. Sci. USA 2019, 116, 20453–20461. [Google Scholar] [CrossRef] [Green Version]
- Tippana, R.; Hwang, H.; Opresko, P.L.; Bohr, V.A.; Myong, S. Single-molecule imaging reveals a common mechanism shared by G-quadruplex–resolving helicases. Proc. Natl. Acad. Sci. USA 2016, 113, 8448–8453. [Google Scholar] [CrossRef] [Green Version]
- Tippana, R.; Xiao, W.; Myong, S. G-quadruplex conformation and dynamics are determined by loop length and sequence. Nucleic Acids Res. 2014, 42, 8106–8114. [Google Scholar] [CrossRef] [Green Version]
- Tian, T.; Chen, Y.-Q.; Wang, S.-R.; Zhou, X. G-Quadruplex: A Regulator of Gene Expression and Its Chemical Targeting. Chem 2018, 4, 1314–1344. [Google Scholar] [CrossRef] [Green Version]
- Spiegel, J.; Adhikari, S.; Balasubramanian, S. The Structure and Function of DNA G-Quadruplexes. Trends Chem. 2020, 2, 123–136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kovačič, M.; Podbevšek, P.; Tateishi-Karimata, H.; Takahashi, S.; Sugimoto, N.; Plavec, J. Thrombin binding aptamer G-quadruplex stabilized by pyrene-modified nucleotides. Nucleic Acids Res. 2020, 48, 3975–3986. [Google Scholar] [CrossRef]
- Schnarr, L.; Jana, J.; Preckwinkel, P.; Weisz, K. Impact of a Snap-Back Loop on Stability and Ligand Binding to a Parallel G-Quadruplex. J. Phys. Chem. B 2020, 124, 2778–2787. [Google Scholar] [CrossRef] [PubMed]
- Jain, A.K.; Bhattacharya, S. Interaction of G-Quadruplexes with Nonintercalating Duplex-DNA Minor Groove Binding Ligands. Bioconjugate Chem. 2011, 22, 2355–2368. [Google Scholar] [CrossRef]
- Di Leva, F.S.; Novellino, E.; Cavalli, A.; Parrinello, M.; Limongelli, V. Mechanistic insight into ligand binding to G-quadruplex DNA. Nucleic Acids Res. 2014, 42, 5447–5455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, Q.; Li, Y.; Freisinger, E.; Qin, P.Z.; Sigel, R.K.O.; Mao, Z.-W. G-quadruplex DNA targeted metal complexes acting as potential anticancer drugs. Inorg. Chem. Front. 2017, 4, 10–32. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Zhong, Y.-F.; Liu, L.-Y.; Shen, C.-T.; Zeng, W.; Wang, F.; Yang, D.; Mao, Z.-W. Solution structures of multiple G-quadruplex complexes induced by a platinum(II)-based tripod reveal dynamic binding. Nat. Commun. 2018, 9, 1–11. [Google Scholar] [CrossRef]
- Nguyen, T.Q.N.; Lim, K.W.; Phan, A.T. Duplex formation in a G-quadruplex bulge. Nucleic Acids Res. 2020, 48, 10567–10575. [Google Scholar] [CrossRef]
- Goddard, T.D.; Kneller, D.G. Sparky 3; University of California: San Francisco, CA, USA, 2004. [Google Scholar]
- Case, D.A.; Babin, V.; Berryman, J.T.; Betz, R.M.; Cai, Q.; Cerutti, D.S.; Cheatham, T.E., III; Darden, T.A.; Duke, R.E.; Gohlke, H.; et al. AMBER 14; University of California: San Francisco, CA, USA, 2014. [Google Scholar]
- Pérez, A.; Marchán, I.; Svozil, D.; Sponer, J.; Cheatham, T.E.; Laughton, C.A.; Orozco, M. Refinement of the AMBER Force Field for Nucleic Acids: Improving the Description of α/γ Conformers. Biophys. J. 2007, 92, 3817–3829. [Google Scholar] [CrossRef] [Green Version]
- Krepl, M.; Zgarbová, M.; Stadlbauer, P.; Otyepka, M.; Banáš, P.; Koča, J.; Cheatham, I.T.E.; Jurečka, P.; Šponer, J. Reference Simulations of Noncanonical Nucleic Acids with Different χ Variants of the AMBER Force Field: Quadruplex DNA, Quadruplex RNA, and Z-DNA. J. Chem. Theory Comput. 2012, 8, 2506–2520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zgarbová, M.; Estarellas, C.; Šponer, J.; Otyepka, M.; Jurečka, P. A Novel Approach for Deriving Force Field Torsion Angle Parameters Accounting for Conformation-Dependent Solvation Effects. J. Chem. Theory Comput. 2012, 8, 3232–3242. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera: A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| NMR Statistics | |
|---|---|
| NOE-derived Distance Restraints | |
| Total | 630 |
| Intra-residual | 426 |
| Inter-residual | 204 |
| Sequential | 103 |
| Long-range | 101 |
| Hydrogen bond restraints | 24 |
| Torsion angle restraints | 37 |
| Planarity restraints | 36 |
| Structure Statistics | |
| Violations | |
| Mean NOE restraint violation (Å) | 0.058 ± 0.016 |
| Max. NOE restraint violation (Å) | 0.084 |
| Max. torsion angle restraint violation (°) | 2.50 |
| Deviations from idealized geometry | |
| Bonds (Å) | 0.0121 ± 0.0002 |
| Angles (°) | 2.33 ± 0.03 |
| Pairwise Heavy Atom RMSD (Å) | |
| Overall | 1.55 ± 0.49 |
| Overall without A9-G10-C11 and G14 | 0.51 ± 0.18 |
| G-quartets | 0.41 ± 0.17 |
Sample Availability: Samples of the compounds are not available from the authors. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lenarčič Živković, M.; Rozman, J.; Plavec, J. Structure of a DNA G-Quadruplex Related to Osteoporosis with a G-A Bulge Forming a Pseudo-loop. Molecules 2020, 25, 4867. https://doi.org/10.3390/molecules25204867
Lenarčič Živković M, Rozman J, Plavec J. Structure of a DNA G-Quadruplex Related to Osteoporosis with a G-A Bulge Forming a Pseudo-loop. Molecules. 2020; 25(20):4867. https://doi.org/10.3390/molecules25204867
Chicago/Turabian StyleLenarčič Živković, Martina, Jan Rozman, and Janez Plavec. 2020. "Structure of a DNA G-Quadruplex Related to Osteoporosis with a G-A Bulge Forming a Pseudo-loop" Molecules 25, no. 20: 4867. https://doi.org/10.3390/molecules25204867






