Dependence of Fluorescence Quenching of CY3 Oligonucleotide Conjugates on the Oxidation Potential of the Stacking Base Pair
Abstract
:1. Introduction
2. Results
2.1. Optical Spectra
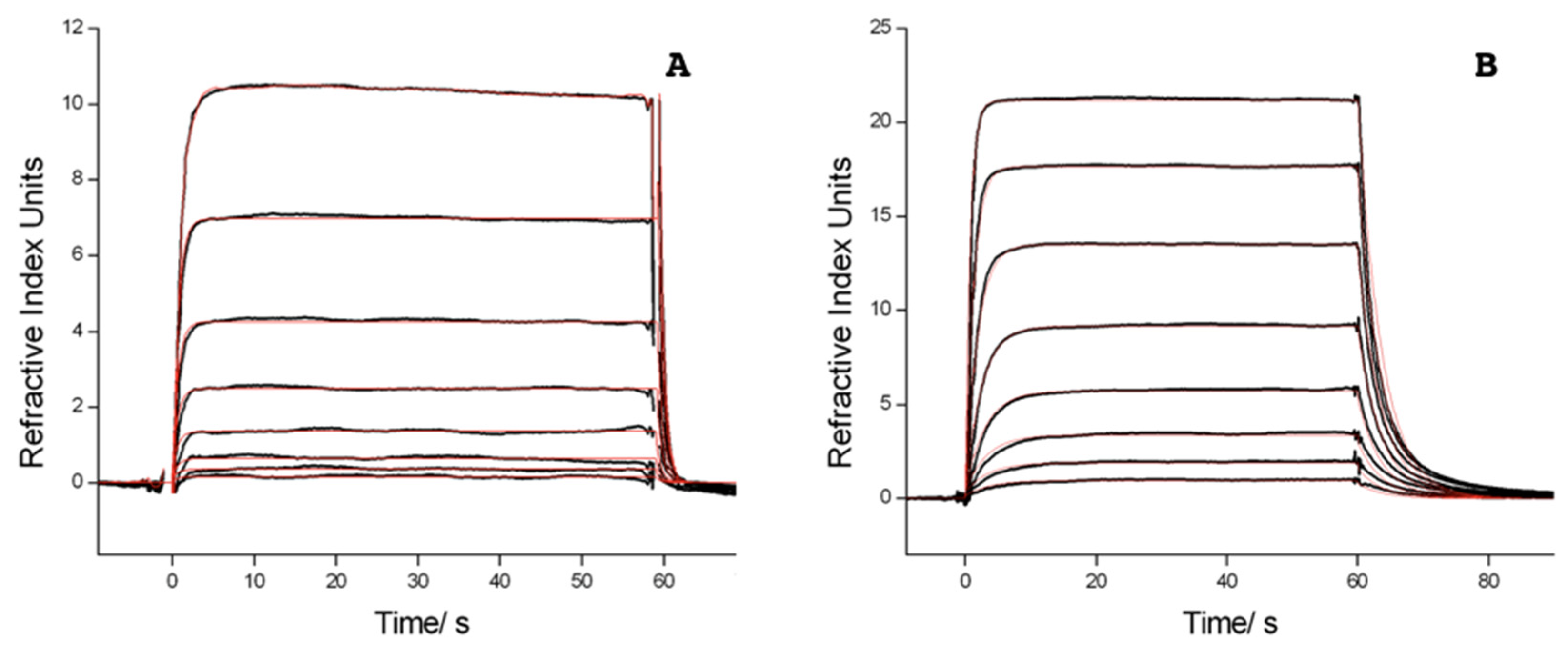
2.2. Surface Plasmon Resonance (SPR) Measurements
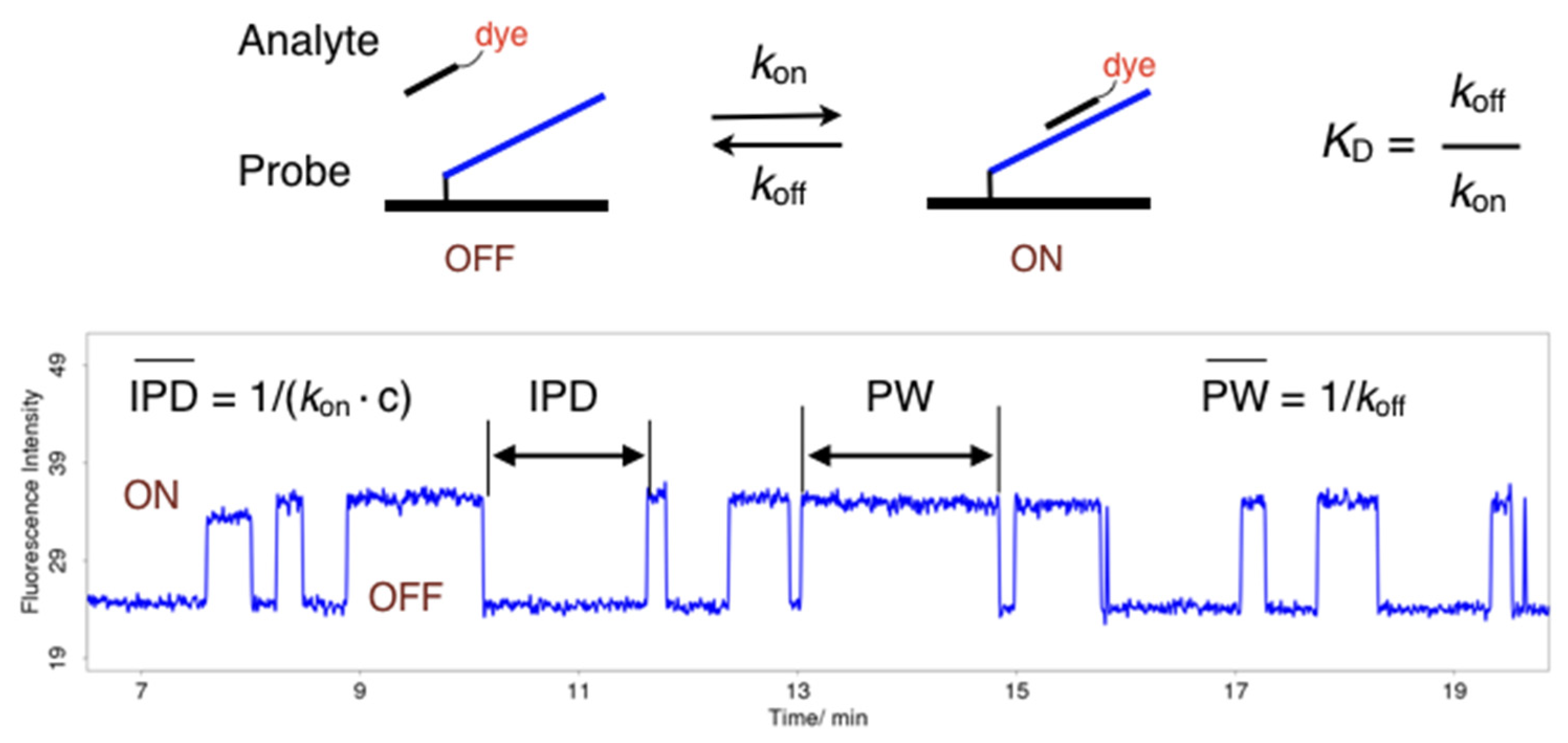
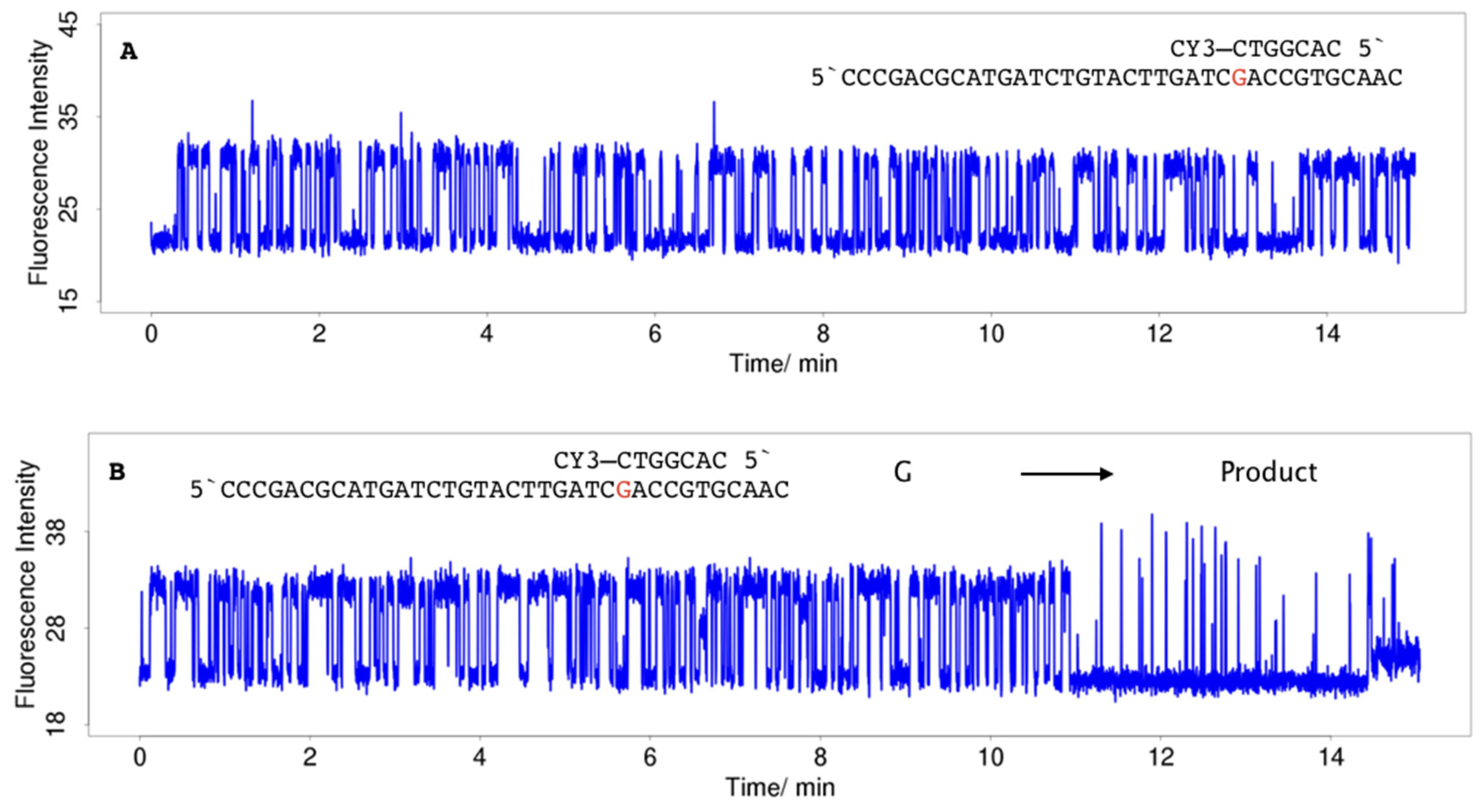
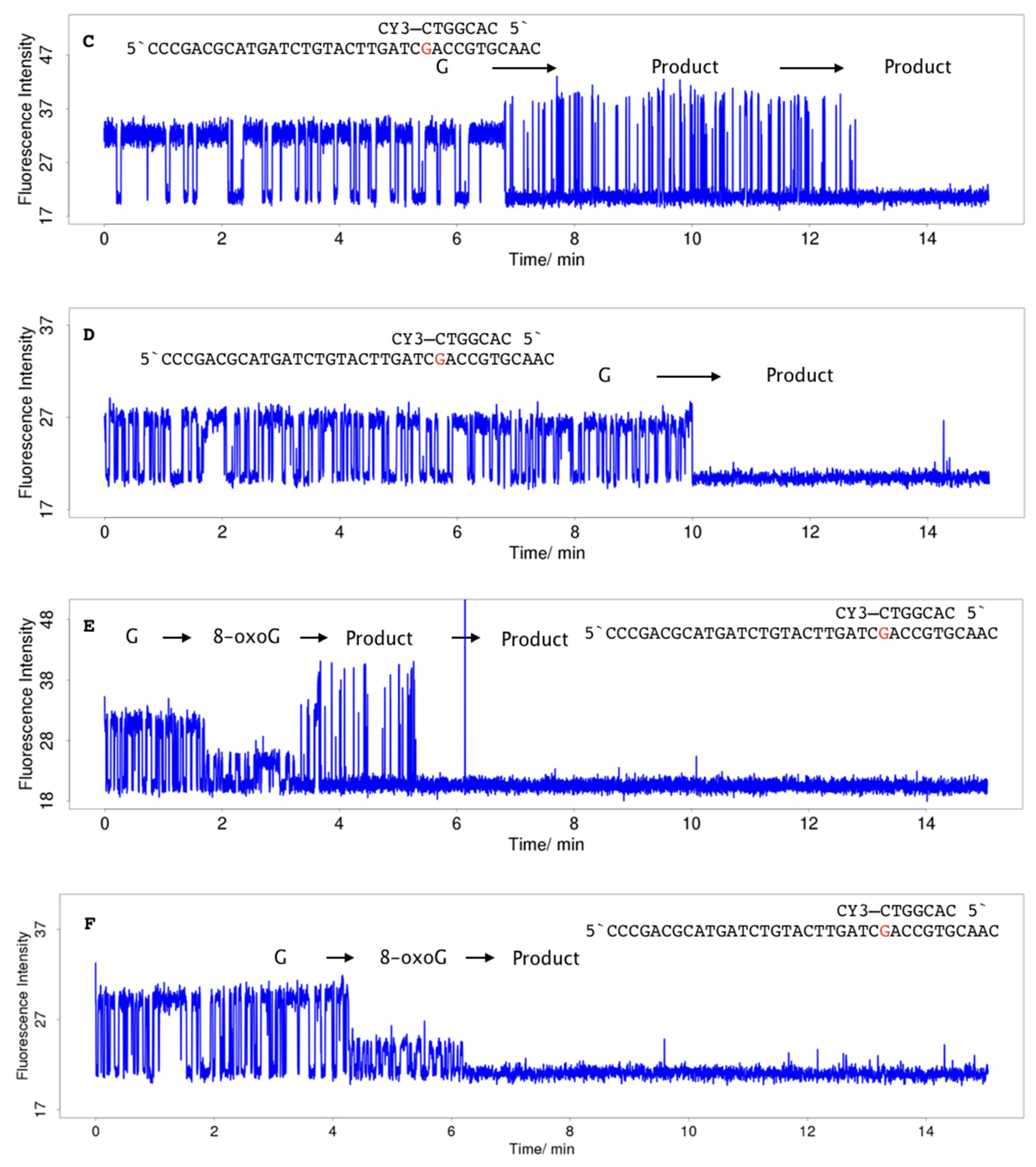
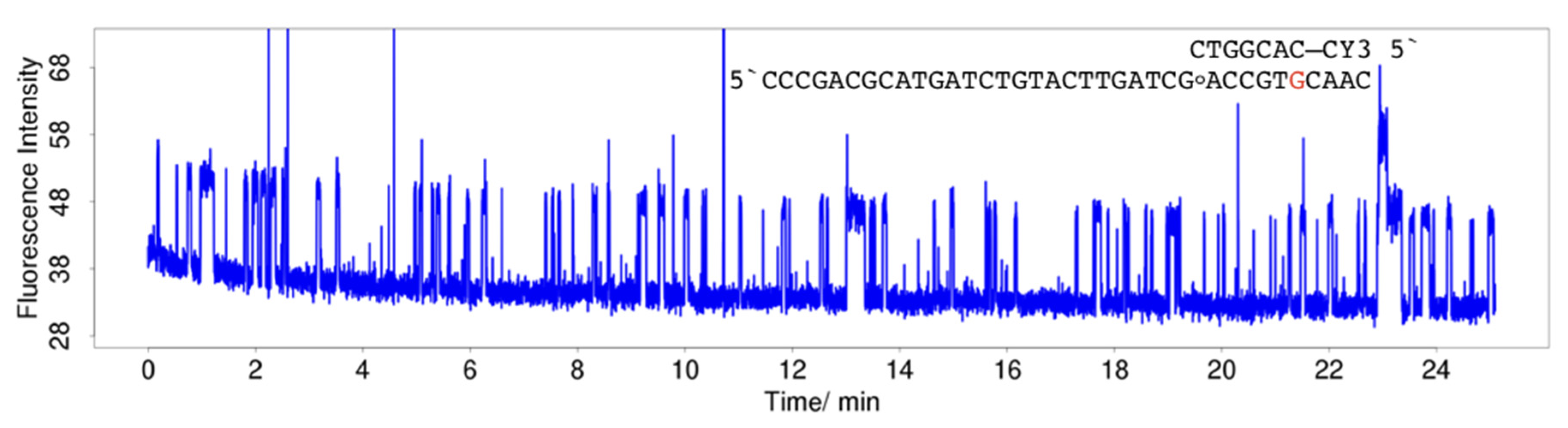
2.3. Single-Molecule Measurements
3. Discussion
4. Materials and Methods
4.1. Optical Measurements
4.2. Surface Plasmon Resonance Measurements
4.3. Single-Molecule Measurements
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- König, W. Über die Konstitution der Pinacyanole, ein Beitrag zur Chemie der Chinocyanine. Ber. Dtsch. Chem. Ges. 1922, 55, 3294–3313. [Google Scholar] [CrossRef] [Green Version]
- Randolph, J.B.; Waggoner, A.S. Stability, specificity and fluorescence brightness of multiply-labeled fluorescent DNA probes. Nucleic Acids Res. 1997, 25, 2923–2929. [Google Scholar] [CrossRef] [PubMed]
- Sanborn, M.E.; Connolly, B.K.; Gurunathan, K.; Levitus, M. Fluorescence properties and photophysics of the sulfoindocyanine Cy3 linked covalently to DNA. J. Phys. Chem. B 2007, 111, 11064–11074. [Google Scholar] [CrossRef] [PubMed]
- Harvey, B.J.; Perez, C.; Levitus, M. DNA sequence-dependent enhancement of Cy3 fluorescence. Photochem. Photobiol. Sci. 2009, 8, 1105. [Google Scholar] [CrossRef] [PubMed]
- Levitus, M.; Ranjit, S. Cyanine dyes in biophysical research: The photophysics of polymethine fluorescent dyes in biomolecular environments. Q. Rev. Biophys. 2011, 44, 123–151. [Google Scholar] [CrossRef]
- Stennett, E.M.S.; Ma, N.; van der Vaart, A.; Levitus, M. Photophysical and dynamical properties of doubly linked Cy3-DNA constructs. J. Phys. Chem. B 2014, 118, 152–163. [Google Scholar] [CrossRef]
- Jia, K.; Wan, Y.; Xia, A.; Li, S.; Gong, F.; Yang, G. Characterization of Photoinduced Isomerization and Intersystem Crossing of the Cyanine Dye Cy3. J. Phys. Chem. A 2007, 111, 1593–1597. [Google Scholar] [CrossRef]
- Ouellet, J.; Schorr, S.; Iqbal, A.; Wilson, T.J.; Lilley, D.M.J. Orientation of cyanine fluorophores terminally attached to DNA via long, flexible tethers. Biophys. J. 2011, 101, 1148–1154. [Google Scholar] [CrossRef] [Green Version]
- Linck, L.; Kapusta, P.; Resch-Genger, U. Spectroscopic and photophysical properties of dUTP and internally DNA bound fluorophores for optimized signal detection in biological formats. Photochem. Photobiol. 2012, 88, 867–875. [Google Scholar] [CrossRef]
- Linck, L.; Reiß, E.; Bier, F.; Resch-Genger, U. Direct labeling rolling circle amplification as a straightforward signal amplification technique for biodetection formats. Anal. Methods 2012, 4, 1215. [Google Scholar] [CrossRef]
- Resch-Genger, U.; Grabolle, M.; Cavaliere-Jaricot, S.; Nitschke, R.; Nann, T. Quantum dots versus organic dyes as fluorescent labels. Science 2008, 5, 763–775. [Google Scholar] [CrossRef] [PubMed]
- Kashida, H.; Morimoto, K.; Asanuma, H. A stem-less probe using spontaneous pairing between Cy3 and quencher for RNA detection. Sci. Technol. Adv. Mater. 2016, 17, 267–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sauer, M.; Heilemann, M. Single-molecule localization microscopy in Eukaryotes. Chem. Rev. 2017, 117, 7478–7509. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa-Ankerhold, H.C.H.; Ankerhold, R.R.; Drummen, G.P.C.G. Advanced fluorescence microscopy techniques—FRAP, FLIP, FLAP, FRET and FLIM. Molecules 2011, 17, 4047–4132. [Google Scholar] [CrossRef] [PubMed]
- Ha, T.; Tinnefeld, P. Photophysics of fluorescent probes for single-molecule biophysics and super-resolution imaging. Annu. Rev. Phys. Chem. 2012, 63, 595–617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morten, M.J.; Lopez, S.G.; Steinmark, I.E.; Rafferty, A.; Magennis, S.W. Stacking-induced fluorescence increase reveals allosteric interactions through DNA. Nucleic Acids Res. 2018, 46, 11618–11626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glembockyte, V.; Lincoln, R.; Cosa, G. Cy3 Photoprotection mediated by Ni2+ for extended single-molecule imaging: Old tricks for new techniques. J. Am. Chem. Soc. 2015, 137, 1116–1122. [Google Scholar] [CrossRef]
- Kawai, K.; Maruyama, A. Triple helix conformation-specific blinking of Cy3 in DNA. Chem. Commun. 2015, 51, 4861–4864. [Google Scholar] [CrossRef]
- Sobek, J.; Rehrauer, H.; Schauer, S.; Fischer, D.; Patrignani, A.; Landgraf, S.; Korlach, J.; Schlapbach, R. Single-molecule DNA hybridisation studied by using a modified DNA sequencer: A comparison with surface plasmon resonance data. Methods Appl. Fluoresc. 2016, 4, 015002. [Google Scholar] [CrossRef] [Green Version]
- Sobek, J.; Schmidt, M.; Grossmann, J.; Rehrauer, H.; Schmidt, L.; Schlapbach, R. Single-molecule chemistry. Part I: Monitoring oxidation of G in oligonucleotides using CY3 fluorescence. Methods Appl. Fluoresc. 2020, 8, 035010. [Google Scholar] [CrossRef]
- Harvey, B.; Levitus, M. Nucleobase-specific enhancement of Cy3 fluorescence. J. Fluoresc. 2009, 19, 443–448. [Google Scholar] [CrossRef] [PubMed]
- Lv, W.; Chen, X.; Aumiler, D.; Xia, A. Single molecule fluorescence fluctuations of the cyanine dyes linked covalently to DNA. Sci. China Ser. B Chem. 2009, 52, 1148–1153. [Google Scholar] [CrossRef]
- Mujumdar, R.B.; Ernst, L.A.; Mujumdar, S.R.; Lewis, C.J.; Waggoner, A.S. Cyanine dye labeling reagents: Sulfoindocyanine succinimidyl esters. Bioconjug. Chem. 1993, 4, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Hart, S.M.; Banal, J.L.; Bathe, M.; Schlau-Cohen, G.S. Identification of nonradiative decay pathways in Cy3. J. Phys. Chem. Lett. 2020, 11, 5000–5007. [Google Scholar] [CrossRef] [PubMed]
- Cooper, M.; Ebner, A.; Briggs, M.; Burrows, M.; Gardner, N.; Richardson, R.; West, R. Cy3B: Improving the performance of cyanine dyes. J. Fluoresc. 2004, 14, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Hall, L.M.; Gerowska, M.; Brown, T. A highly fluorescent DNA toolkit: Synthesis and properties of oligonucleotides containing new Cy3, Cy5 and Cy3B monomers. Nucleic Acids Res. 2012, 40, e108. [Google Scholar] [CrossRef]
- Stennett, E.M.S.; Ciuba, M.A.; Lin, S.; Levitus, M. Demystifying PIFE: The photophysics behind the protein-induced fluorescence enhancement phenomenon in Cy3. J. Phys. Chem. Lett. 2015, 6, 1819–1823. [Google Scholar] [CrossRef]
- Nazarenko, I.; Pires, R.; Lowe, B.; Obaidy, M.; Rashtchian, A. Effect of primary and secondary structure of oligodeoxyribonucleotides on the fluorescent properties of conjugated dyes. Nucleic Acids Res. 2002, 30, 2089–2195. [Google Scholar] [CrossRef] [Green Version]
- Seidel, C.; Schulz, A.; Sauer, M. Nucleobase-specific quenching of fluorescent dyes. 1. Nucleobase one-electron redox potentials and their correlation with static and dynamic quenching efficiencies. J. Phys. Chem. 1996, 100, 5541–5553. [Google Scholar] [CrossRef]
- Sauer, M.; Drexhage, K.; Lieberwirth, U.; Muller, R.; Nord, S.; Zander, C. Dynamics of the electron transfer reaction between an oxazine dye and DNA oligonucleotides monitored on the single-molecule level. Chem. Phys. Lett. 1998, 284, 153–163. [Google Scholar] [CrossRef]
- Widengren, J.; Dapprich, J.; Rigler, R. Fast interactions between Rh6G and dGTP in water studied by fluorescence correlation spectroscopy. Chem. Phys. 1997, 216, 417–426. [Google Scholar] [CrossRef]
- Unruh, J.R.; Gokulrangan, G.; Wilson, G.S.; Johnson, C.K. Fluorescence properties of fluorescein, tetramethylrhodamine and Texas Red linked to a DNA aptamer. Photochem. Photobiol. 2005, 81, 682–690. [Google Scholar] [CrossRef] [PubMed]
- Kroutil, O.; Romancová, I.; Šíp, M.; Chval, Z. Cy3 and Cy5 dyes terminally attached to 5′C end of DNA: Structure, dynamics, and energetics. J. Phys. Chem. B 2014, 118, 13564–13572. [Google Scholar] [CrossRef] [PubMed]
- Spiriti, J.; Binder, J.K.; Levitus, M.; van der Vaart, A. Cy3-DNA stacking interactions strongly depend on the identity of the terminal basepair. Biophys. J. 2011, 100, 1049–1057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Norman, D.G.; Grainger, R.J.; Uhrín, D.; Lilley, D.M.J. Location of Cyanine-3 on double-stranded DNA: Importance for fluorescence resonance energy transfer studies. Biochemistry 2000, 39, 6317–6324. [Google Scholar] [CrossRef] [Green Version]
- Moreira, B.G.; You, Y.; Owczarzy, R. Cy3 and Cy5 dyes attached to oligonucleotide terminus stabilize DNA duplexes: Predictive thermodynamic model. Biophys. Chem. 2015, 198, 36–44. [Google Scholar] [CrossRef] [Green Version]
- Guckian, K.M.; Schweitzer, B.A.; Ren, R.X.F.; Sheils, C.J.; Tahmassebi, D.C.; Kool, E.T. Factors contributing to aromatic stacking in water: Evaluation in the context of DNA. J. Am. Chem. Soc. 2000, 122, 2213–2222. [Google Scholar] [CrossRef] [Green Version]
- Lakowicz, J.R. Principles of Fluorescence Spectroscopy, 4th ed.; Springer Science & Business Media LLC: New York, NY, USA, 2006. [Google Scholar]
- Torimura, M.; Kurata, S.; Yamada, K.; Yokomaku, T.; Kamagata, Y.; Kanagawa, T.; Kurane, R. Fluorescence-quenching phenomenon by photoinduced electron transfer between a fluorescent dye and a nucleotide base. Anal. Sci. 2001, 17, 155–160. [Google Scholar] [CrossRef] [Green Version]
- Kawai, K.; Osakada, Y.; Fujitsuka, M.; Majima, T. Charge separation in acridine- and phenothiazine-modified DNA. J. Phys. Chem. B 2008, 112, 2144–2149. [Google Scholar] [CrossRef]
- Adhikary, A.; Khanduri, D.; Sevilla, M.D. Direct observation of the hole protonation state and hole localization site in DNA-oligomers. J. Am. Chem. Soc. 2009, 131, 8614–8619. [Google Scholar] [CrossRef] [Green Version]
- Steenken, S. Electron-transfer-induced acidity basicity and reactivity changes of purine and pyrimidine-bases—Consequences of redox processes for DNA-base pairs. Free Radic. Res. Commun. 1992, 16, 349–379. [Google Scholar] [CrossRef] [PubMed]
- Candeias, L.P.; Steenken, S. Structure and acid-base properties of one-electron-oxidized deoxyguanosine, guanosine, and 1-methylguanosine. J. Am. Chem. Soc. 1989, 111, 1094–1099. [Google Scholar] [CrossRef]
- Caruso, T.; Carotenuto, M.; Vasca, E.; Peluso, A. Direct experimental observation of the effect of the base pairing on the oxidation potential of guanine. J. Am. Chem. Soc. 2005, 127, 15040–15041. [Google Scholar] [CrossRef] [PubMed]
- Caruso, T.; Capobianco, A.; Peluso, A. The oxidation potential of adenosine and adenosine-thymidine base pair in chloroform solution. J. Am. Chem. Soc. 2007, 129, 15347–15353. [Google Scholar] [CrossRef]
- Crespo-Hernández, C.E.; Close, D.M.; Gorb, L.; Leszczynski, J. Determination of redox potentials for the watson−crick base pairs, DNA nucleosides, and relevant nucleoside analogues. J. Phys. Chem. B 1915, 111, 5386–5395. [Google Scholar] [CrossRef]
- Paukku, Y.; Hill, G. Theoretical determination of one-electron redox potentials for DNA bases, base pairs, and stacks. J. Phys. Chem. A 2011, 115, 4804–4810. [Google Scholar] [CrossRef]
- Colson, A.O.; Besler, B.; Sevilla, M.D. Ab initio molecular orbital calculations on DNA base pair radical ions: Effect of base pairing on proton-transfer energies, electron affinities, and ionization potentials. J. Phys. Chem. 1992, 96, 9787–9794. [Google Scholar] [CrossRef]
- Hutter, M.; Clark, T. On the enhanced stability of the guanine-cytosine base-pair radical cation. J. Am. Chem. Soc. 1996, 118, 7574–7577. [Google Scholar] [CrossRef]
- Kawai, K.; Wata, Y.; Hara, M.; Tojo, S.; Majima, T. Regulation of one-electron oxidation rate of guanine by base pairing with cytosine derivatives. J. Am. Chem. Soc. 2002, 124, 3586–3590. [Google Scholar] [CrossRef]
- Kan, Y.; Schuster, G.B. Long-range guanine damage in single-stranded DNA: Charge transport through a duplex bridge and in a single-stranded overhang. J. Am. Chem. Soc. 1999, 121, 10857–10864. [Google Scholar] [CrossRef]
- Burrows, C.J.; Muller, J.G. Oxidative nucleobase modifications leading to strand scission. Chem. Rev. 1998, 98, 1109–1152. [Google Scholar] [CrossRef] [PubMed]
- Kanvah, S.; Joseph, J.; Schuster, G.B.; Barnett, R.N.; Cleveland, C.L.; Landman, U. Oxidation of DNA: Damage to nucleobases. Acc. Chem. Res. 2010, 43, 280–287. [Google Scholar] [CrossRef] [PubMed]
- Muren, N.B.; Olmon, E.D.; Barton, J.K. Solution, surface, and single molecule platforms for the study of DNA-mediated charge transport. Phys. Chem. Chem. Phys. 2012, 14, 13754. [Google Scholar] [CrossRef] [Green Version]
- Genereux, J.C.; Barton, J.K. Mechanisms for DNA charge transport. Chem. Rev. 2010, 110, 1642–1662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giese, B. Hole injection and hole transfer through DNA: The hopping mechanism. In Longe-Range Charge Transfer in DNA I; Springer: Berlin/Heidelberg, Germany, 2004; pp. 151–164. [Google Scholar]
- Cadet, J.; Wagner, J.R. DNA base damage by reactive oxygen species, oxidizing agents, and UV radiation. Cold Spring Harb. Perspect. Biol. 2013, 5, a012559. [Google Scholar] [CrossRef] [PubMed]
- Nikolova, E.N.; Stull, F.; Al-Hashimi, H.M. Guanine to inosine substitution leads to large increases in the population of a transient G·C Hoogsteen base pair. Biochemistry 2014, 53, 7145–7147. [Google Scholar] [CrossRef]
- Modrich, P. DNA mismatch correction. Annu. Rev. Biochem. 1987, 56, 435–466. [Google Scholar] [CrossRef]
- Kingsland, A.; Maibaum, L. DNA base pair mismatches induce structural changes and alter the free-energy landscape of base flip. J. Phys. Chem. B 2018, 122, 12251–12259. [Google Scholar] [CrossRef] [Green Version]
- Wan, C.; Fiebig, T.; Schiemann, O.; Barton, J.K.; Zewail, A.H. Femtosecond direct observation of charge transfer between bases in DNA. Proc. Natl. Acad. Sci. USA 2000, 97, 14052–14055. [Google Scholar] [CrossRef] [Green Version]
- Sheu, C.; Foote, C.S. Photosensitized oxygenation of a 7,8-Dihydro-8-oxoguanosine derivative. Formation of dioxetane and hydroperoxide intermediates. J. Am. Chem. Soc. 1995, 117, 474–477. [Google Scholar] [CrossRef]
- Levene, M.J.; Korlach, J.; Turner, S.W.; Foquet, M.; Craighead, H.G.; Webb, W.W. Zero-mode waveguides for single-molecule analysis at high concentrations. Science 2003, 299, 682–686. [Google Scholar] [CrossRef] [PubMed]
- Eid, J.; Fehr, A.; Gray, J.; Luong, K.; Lyle, J.; Otto, G.; Peluso, P.; Rank, D.; Baybayan, P.; Bettman, B.; et al. Real-time DNA sequencing from single polymerase molecules. Science 2009, 323, 133–138. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Dalal, R.V.; Petrov, A.N.; Tsai, A.; O’Leary, S.E.; Chapin, K.; Cheng, J.; Ewan, M.; Hsiung, P.-L.; Lundquist, P.; et al. High-throughput platform for real-time monitoring of biological processes by multicolor single-molecule fluorescence. Proc. Natl. Acad. Sci. USA 2014, 111, 664–669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheu, C.; Foote, C.S. Reactivity toward singlet oxygen of a 7,8-Dihydro-8-oxoguanosine (“8-Hydroxyguanosine”) formed by photooxidation of a guanosine derivative. J. Am. Chem. Soc. 1995, 117, 6439–6442. [Google Scholar] [CrossRef]
- Ravanat, J.-L.; Saint-Pierre, C.; Cadet, J. One-electron oxidation of the guanine moiety of 2‘-deoxyguanosine: Influence of 8-Oxo-7,8-dihydro-2‘-deoxyguanosine. J. Am. Chem. Soc. 2003, 125, 2030–2031. [Google Scholar] [CrossRef]
- Fleming, A.M.; Muller, J.G.; Ji, I.; Burrows, C.J. Characterization of 2′-deoxyguanosine oxidation products observed in the Fenton-like system Cu(ii)/H2O2/reductant in nucleoside and oligodeoxynucleotide contexts. Org. Biomol. Chem. 2011, 9, 3338. [Google Scholar] [CrossRef] [Green Version]
- Fleming, A.M.; Burrows, C.J. 8-Oxo-7,8-dihydro-2′-deoxyguanosine and abasic site tandem lesions are oxidation prone yielding hydantoin products that strongly destabilize duplex DNA. Org. Biomol. Chem. 2017, 15, 8341–8353. [Google Scholar] [CrossRef]
- Jean Cadet, T.D.A.J.-L.R. Oxidatively generated damage to the guanine moiety of DNA: Mechanistic aspects and formation in cells. Acc. Chem. Res. 2008, 41, 1075–1083. [Google Scholar] [CrossRef]
- Kanvah, S.; Schuster, G.B. One-electron oxidation of DNA: Thymine versus guanine reactivity. Org. Biomol. Chem. 2010, 8, 1340. [Google Scholar] [CrossRef]
- Ghosh, A.; Joy, A.; Schuster, G.B.; Douki, T.; Cadet, J. Selective one-electron oxidation of duplex DNA oligomers: Reaction at thymines. Org. Biomol. Chem. 2008, 6, 916. [Google Scholar] [CrossRef]
- Li, X.; Yin, Y.; Yang, X.; Zhi, Z.; Zhao, X.S. Temperature dependence of interaction between double stranded DNA and Cy3 or Cy5. Chem. Phys. Lett. 2011, 513, 271–275. [Google Scholar] [CrossRef]
- Iqbal, A.; Arslan, S.; Okumus, B.; Wilson, T.J.; Giraud, G.; Norman, D.G.; Ha, T.; Lilley, D.M.J. Orientation dependence in fluorescent energy transfer between Cy3 and Cy5 terminally attached to double-stranded nucleic acids. Proc. Natl. Acad. Sci. USA 2008, 105, 11176–11181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Milas, P.; Gamari, B.D.; Parrot, L.; Krueger, B.P.; Rahmanseresht, S.; Moore, J.; Goldner, L.S. Indocyanine dyes approach free rotation at the 3′ terminus of A-RNA: A comparison with the 5′ terminus and consequences for fluorescence resonance energy transfer. J. Phys. Chem. B 2013, 117, 8649–8658. [Google Scholar] [CrossRef] [PubMed]
- Sjöback, R.; Nygren, J.; Kubista, M. Characterization of fluorescein-oligonucleotide conjugates and measurement of local electrostatic potential. Biopolymers 1998, 46, 445–453. [Google Scholar] [CrossRef]
- Neeley, W.L.; Essigmann, J.M. Mechanisms of formation, genotoxicity, and mutation of guanine oxidation products. Chem. Res. Toxicol. 2006, 19, 491–505. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, M.M. Reactivity of nucleic acid radicals. In Advances in Physical Organic Chemistry; Elsevier: Amsterdam, The Netherlands, 2016; Volume 50, pp. 119–202. [Google Scholar]
- Margolin, Y.; Shafirovich, V.; Geacintov, N.E.; DeMott, M.S.; Dedon, P.C. DNA sequence context as a determinant of the quantity and chemistry of guanine oxidation produced by hydroxyl radicals and one-electron oxidants. J. Biol. Chem. 2008, 283, 35569–35578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Douki, T.; Martini, R.; Ravanat, J.L.; Turesky, R.J.; Cadet, J. Measurement of 2,6-diamino-4-hydroxy-5-formamidopyrimidine and 8-oxo-7,8-dihydroguanine in isolated DNA exposed to gamma radiation in aqueous solution. Carcinogenesis 1997, 18, 2385–2391. [Google Scholar] [CrossRef] [Green Version]
- Bauer, N.C.; Corbett, A.H.; Doetsch, P.W. The current state of eukaryotic DNA base damage and repair. Nucleic Acids Res. 2015, 43, 10083–10101. [Google Scholar] [CrossRef] [Green Version]
- Cadet, J.; Davies, K.J.A.; Medeiros, M.H.; Di Mascio, P.; Wagner, J.R. Free radical biology and medicine. Free Radic. Biol. Med. 2017, 107, 13–34. [Google Scholar] [CrossRef]
- Fleming, A.M.; Burrows, C.J. Formation and processing of DNA damage substrates for the hNEIL enzymes. Free Radic. Biol. Med. 2017, 107, 35–52. [Google Scholar] [CrossRef]
- Kasai, H.; Yamaizumi, Z.; Berger, M.; Cadet, J. Photosensitized formation of 7,8-dihydro-8-oxo-2“-deoxyguanosine (8-hydroxy-2-”deoxyguanosine) in DNA by riboflavin: A nonsinglet oxygen-mediated reaction. J. Am. Chem. Soc. 1992, 114, 9692–9694. [Google Scholar] [CrossRef]
- Rokhlenko, Y.; Geacintov, N.E.; Shafirovich, V. Lifetimes and reaction pathways of guanine radical cations and neutral guanine radicals in an oligonucleotide in aqueous solutions. J. Am. Chem. Soc. 2012, 134, 4955–4962. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rokhlenko, Y.; Cadet, J.; Geacintov, N.E.; Shafirovich, V. Mechanistic aspects of hydration of guanine radical cations in DNA. J. Am. Chem. Soc. 2014, 136, 5956–5962. [Google Scholar] [CrossRef] [PubMed]
- Devasagayam, T.P.A.; Sundquist, A.R.; Di Mao, P.; Kaiser, S.; Sies, H. Activity of thiols as singlet molecular-oxygen quenchers. J. Photochem. Photobiol. B Biol. 1991, 9, 105–116. [Google Scholar] [CrossRef]
- Rehm, D.; Weller, A. Kinetics of fluorescence quenching by electron and H-atom transfer. Isr. J. Chem. 1970, 8, 259–271. [Google Scholar] [CrossRef]
- Stein, I.H.; Capone, S.; Smit, J.H.; Baumann, F.; Cordes, T.; Tinnefeld, P. Linking single-molecule blinking to chromophore structure and redox potentials. ChemPhysChem 2012, 13, 931–937. [Google Scholar] [CrossRef] [Green Version]
- Marcus, R.A. Electron-transfer reactions in chemistry—Theory and experiment (Nobel lecture). Angew. Chem. Int. Ed. Engl. 1993, 32, 1111–1121. [Google Scholar] [CrossRef]
- Batchelor-McAuley, C.; Li, Q.; Dapin, S.M.; Compton, R.G. Voltammetric characterization of DNA intercalators across the full pH range: Anthraquinone-2,6-disulfonate and anthraquinone-2-sulfonate. J. Phys. Chem. B 2010, 114, 4094–4100. [Google Scholar] [CrossRef]
- Grotz, K.K.; Nueesch, M.F.; Holmstrom, E.D.; Heinz, M.; Stelzl, L.S.; Schuler, B.; Hummer, G. Dispersion correction alleviates dye stacking of single-stranded DNA and RNA in simulations of single-molecule fluorescence experiments. J. Phys. Chem. B 2018, 122, 11626–11639. [Google Scholar] [CrossRef]
- Capobianco, A.; Caruso, T.; Celentano, M.; D’Ursi, A.M.; Scrima, M.; Peluso, A. Stacking interactions between adenines in oxidized oligonucleotides. J. Phys. Chem. B 2013, 117, 8947–8953. [Google Scholar] [CrossRef]
- Capobianco, A.; Caruso, T.; Peluso, A. Hole delocalization over adenine tracts in single stranded DNA oligonucleotides. Phys. Chem. Chem. Phys. 2015, 17, 4750–4756. [Google Scholar] [CrossRef] [PubMed]
- Capobianco, A.; Caruso, T.; D’Ursi, A.M.; Fusco, S.; Masi, A.; Scrima, M.; Chatgilialoglu, C.; Peluso, A. Delocalized hole domains in guanine-rich DNA oligonucleotides. J. Phys. Chem. B 2015, 119, 5462–5466. [Google Scholar] [CrossRef] [PubMed]
- Doose, S.; Neuweiler, H.; Sauer, M. Fluorescence quenching by photoinduced electron transfer: A reporter for conformational dynamics of macromolecules. ChemPhysChem 2009, 10, 1389–1398. [Google Scholar] [CrossRef]
- Rashid, F.; Raducanu, V.-S.; Zaher, M.S.; Tehseen, M.; Habuchi, S.; Hamdan, S.M. Initial state of DNA-Dye complex sets the stage for protein induced fluorescence modulation. Nat. Commun. 2019, 10, 2104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crockett, A.O.; Wittwer, C.T. Fluorescein-labeled oligonucleotides for real-time PCR: Using the inherent quenching of deoxyguanosine nucleotides. Anal. Biochem. 2001, 290, 89–97. [Google Scholar] [CrossRef]
- Noble, J.E.; Wang, L.; Cole, K.D.; Gaigalas, A.K. The effect of overhanging nucleotides on fluorescence properties of hybridising oligonucleotides labelled with Alexa-488 and FAM fluorophores. Biophys. Chem. 2005, 113, 255–263. [Google Scholar] [CrossRef]
- Clark, T.A.; Spittle, K.E.; Turner, S.W.; Korlach, J. Direct detection and sequencing of damaged DNA bases. Genome Integr. 2011, 2, 10. [Google Scholar] [CrossRef] [Green Version]



| Sample | λmax (abs) | λmax (fl) |
|---|---|---|
| CY3-26 | 550 | 566 |
| TLG | 549 | 564 |
| TLoG | 549 | 565 |
| TLA | 550 | 565 |
| TLI | 549 | 566 |
| ON34 | 550 | 565 |
| Probe | Analyte | KD/nM | KD Ratio | ∆G/kJ/mol |
|---|---|---|---|---|
| TLG | 7m | 2230(90) | ||
| 3CY3-7m | 107(3) | 20.8 | 7.4 | |
| 5CY3-7m | 77.4(23) | 28.8 | 8.2 | |
| TLoG | 7m | 4401(229) | ||
| 3CY3-7m | 253(6) | 17.4 | 7.0 | |
| 5CY3-7m | 155(3) | 28.4 | 8.2 | |
| TLA | 7m | 9475(1832) | 27.3 | 8.0 |
| 3CY3-7m | 347(12) | |||
| TLI | 7m | 8832(866) | 28.8 | 8.0 |
| 3CY3-7m | 307(5) | |||
| TLG | 8mG | 320(16) | 12.2 | 6.1 |
| 3CY3-8mG | 26.2(20) | |||
| TLoG | 8mG | 230(13) | 10.6 | 5.8 |
| 3CY3-8mG | 21.6(24) |
| Probe | Analyte | Dye Class | Dye Charge | KD/nM | KD Ratio | ∆G/kJ/mol |
|---|---|---|---|---|---|---|
| Bio34G | 7n | - | - | 978(25) | - | - |
| 5CY3-7n | sym. cyanine | +1 | 32.8(41) | 29.8 | 8.3 | |
| 3CY3-7n | sym. cyanine | +1 | 46.4(15) | 21.1 | 7.5 | |
| CY3-7nPEG | sym. cyanine | +1 | 84.6(56) | 11.6 | 6.0 | |
| CY3B-7n | sym. cyanine | 0 | 92.9(42) | 10.5 | 5.8 | |
| DY547-7n | sym. cyanine | −1 | 206(6) | 4.5 | 3.7 | |
| DY530-7n | rhodamine | −1 | 217(9) | 4.5 | 3.7 | |
| TAMRA-7n | rhodamine | 0 | 278(5) | 3.5 | 3.1 | |
| TexasRed-7n | rhodamine | 0 | 94.0(51) | 10.4 | 5.7 | |
| ATTO532-7n | rhodamine | −1 | 200(8) | 4.9 | 3.9 | |
| ATTO550-7n | rhodamine | +1 | 39.1(44) | 25.0 | 7.9 | |
| CY5-7n | sym. cyanine | +1 | 28.8(17) | 34.0 | 8.6 | |
| DY630-7n | asym. cyanine | 0 | 84.4(27) | 11.6 | 6.0 | |
| ATTO647N-7n | carbopyronine | 0 | 32.2(41) | 30.3 | 8.4 | |
| MB-7n | phenothiazine | +1 | 29.6(18) | 33.0 | 8.6 | |
| FAM-7n | fluorescein | −2 | 881(69) | 1.1 | 0.3 | |
| Bio34r | 7r | - | - | 506(210) | - | - |
| DY548-7r | sym. cyanine | −2 | 423(49) | 1.2 | 0.4 | |
| DY549-7r | sym. cyanine | −3 | 534(68) | 0.95 | −0.1 |
| Probe | Analyte | KD/nM | KD Ratio | ∆G/kJ/mol |
|---|---|---|---|---|
| Bio34G | 7T | 3025(68) | 19.7 | 7.3 |
| CY3-7T | 153(3) | |||
| Bio34I | 7n | 7419(61) | 28.8 | 8.2 |
| CY3-7n | 257(10) | |||
| Bio34U | 7n | 14,830(494) | 7.4 | 4.9 |
| CY3-7n | 1987(42) | |||
| Bio34ap | 7n | 45,111(24,127) | ||
| CY3-7n | 15,015(6605) | 3.0 | 2.7 | |
| 3CY3-7n | 1744(124) | 25.9 | 8.0 |
| Oligonucleotide | Sequence |
|---|---|
| TLG | 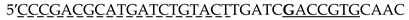 |
| TLoG |  |
| TLA |  |
| TLI | 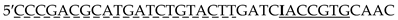 |
| 7m | 5′CACGGTC |
| 5CY3-7m | 5′CY3–CACGGTC |
| 3CY3-7m | 5′CACGGTC–CY3 |
| 8mG | 5′GGCACGGTG |
| 3CY3-8mG | 5′GCACGGTG–CY3 |
| Bio25LNA | 5′AGTACAGATCATGCGTCGGGTTTTT-Biotin |
| CY3-26 | 5′CY3–CGATCAAGTACAGATCATGCGTCGGG |
| CY3-27 | 5′CY3–TCGATCAAGTACAGATCATGCGTCGGG |
| ON34 | TTTTTGGAAACTGTATTGGCACTGAGTAGACTCC |
| Bio34G | 5′Biotin-TTTTTGGAAACTGTATTGGCACTGAGTAGACTCC |
| Bio34I | 5′Biotin-TTTTTGGAAACTGTATTGGCACTIAGTAGACTCC |
| Bio34U | 5′Biotin-TTTTTGGAAACTGTATTGGCACTUAGTAGACTCC |
| Bio34ap | 5′Biotin-TTTTTGGAAACTGTATTGGCACTapAGTAGACTCC |
| 7n | CAGTGCC |
| 5CY3-7n | 5′CY3-CAGTGCC |
| 3CY3-7n | 5′CAGTGCC-CY3 |
| DYE-7n | 5′DYE-CAGTGCC |
| 7T | 5′CY3-TCAGTGC |
| CY3-7T | 5′CY3-TCAGTGC |
| Bio34r | 5′Biotin-TTTTTGGAAACTGTATTGTCACGGAGTAGACTTT |
| 7r | 5′CCGTGAC |
| DY548-7r | 5′DY548-CCGTGAC |
| DY549-7r | 5′DY549-CCGTGAC |
Sample Availability: Samples of the compounds are not available from the authors. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sobek, J.; Schlapbach, R. Dependence of Fluorescence Quenching of CY3 Oligonucleotide Conjugates on the Oxidation Potential of the Stacking Base Pair. Molecules 2020, 25, 5369. https://doi.org/10.3390/molecules25225369
Sobek J, Schlapbach R. Dependence of Fluorescence Quenching of CY3 Oligonucleotide Conjugates on the Oxidation Potential of the Stacking Base Pair. Molecules. 2020; 25(22):5369. https://doi.org/10.3390/molecules25225369
Chicago/Turabian StyleSobek, Jens, and Ralph Schlapbach. 2020. "Dependence of Fluorescence Quenching of CY3 Oligonucleotide Conjugates on the Oxidation Potential of the Stacking Base Pair" Molecules 25, no. 22: 5369. https://doi.org/10.3390/molecules25225369






