New Strategy to Preserve Phosphate by Ionic Liquid Matrices in Matrix-Assisted Laser Desorption/Ionization: A Case of Adenosine Nucleotides
Abstract
:1. Introduction
2. Results
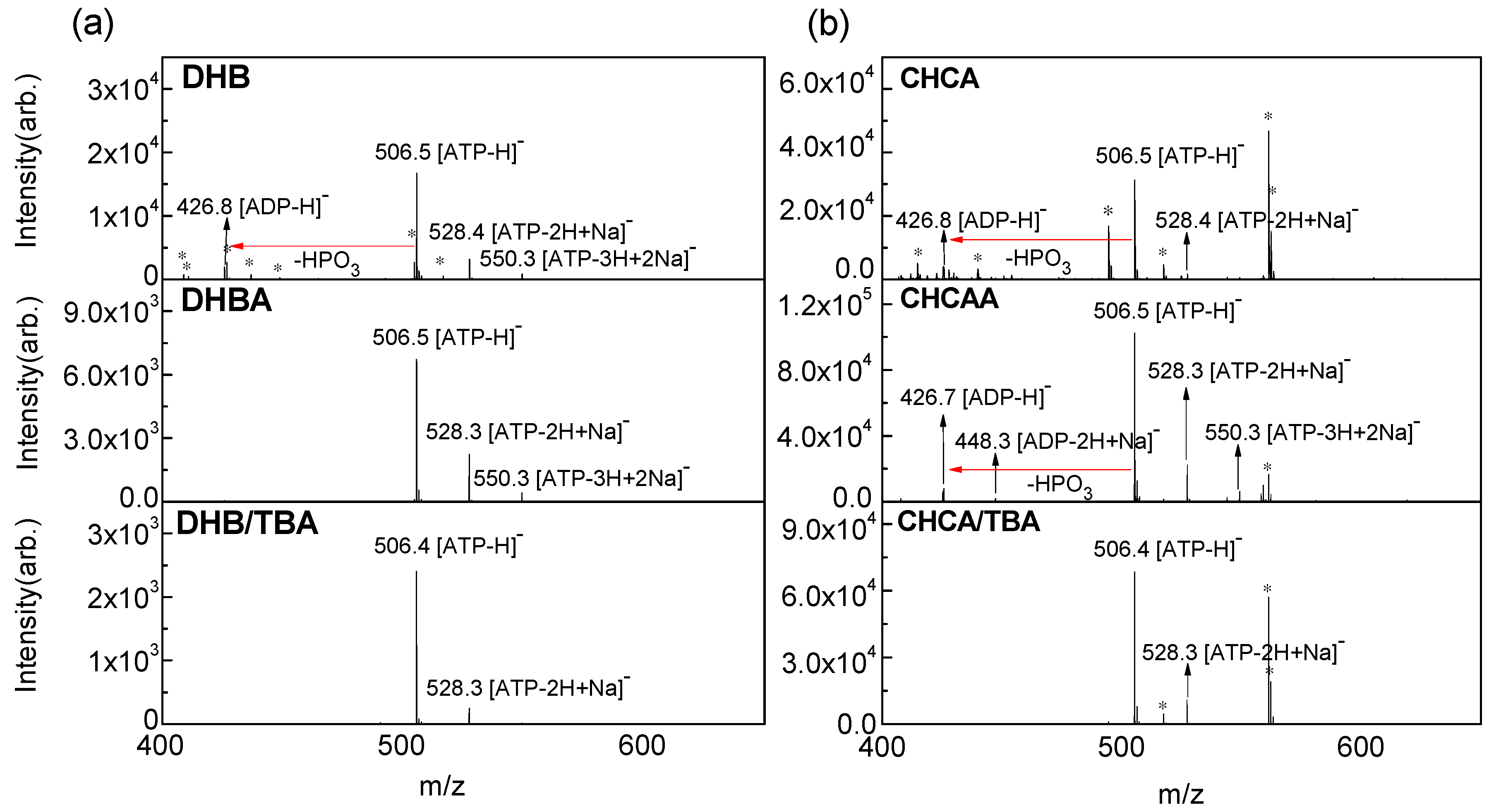
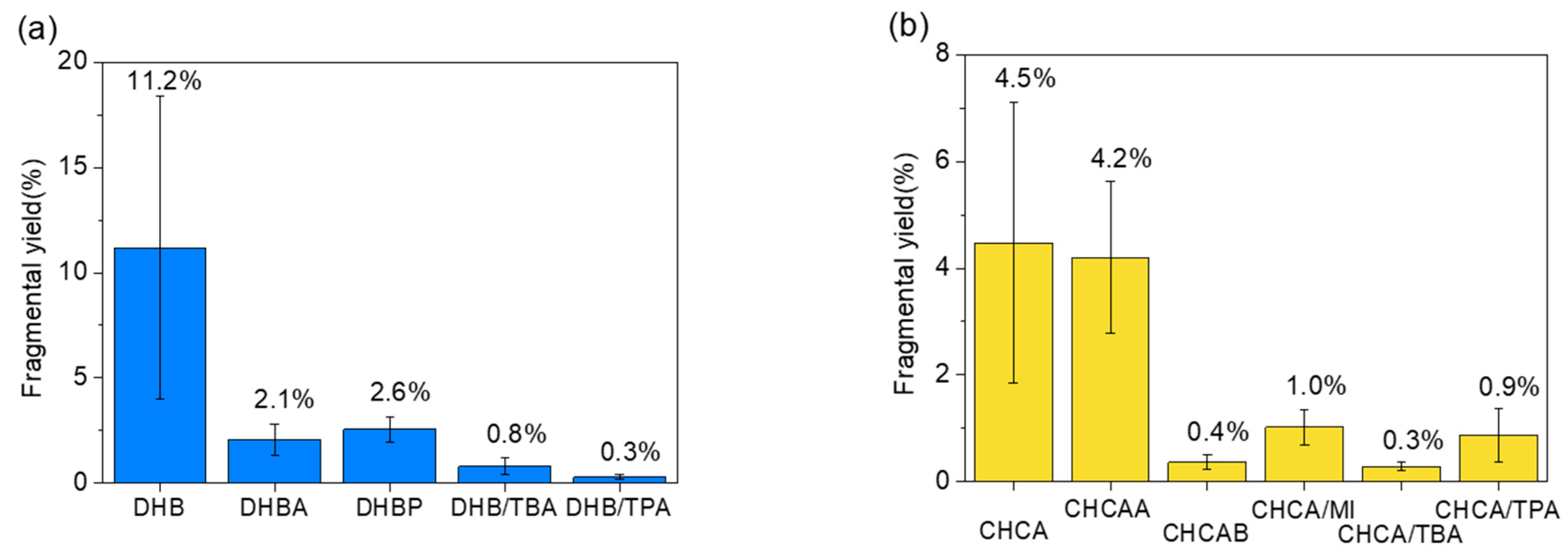
2.1. Fragmentation Yield
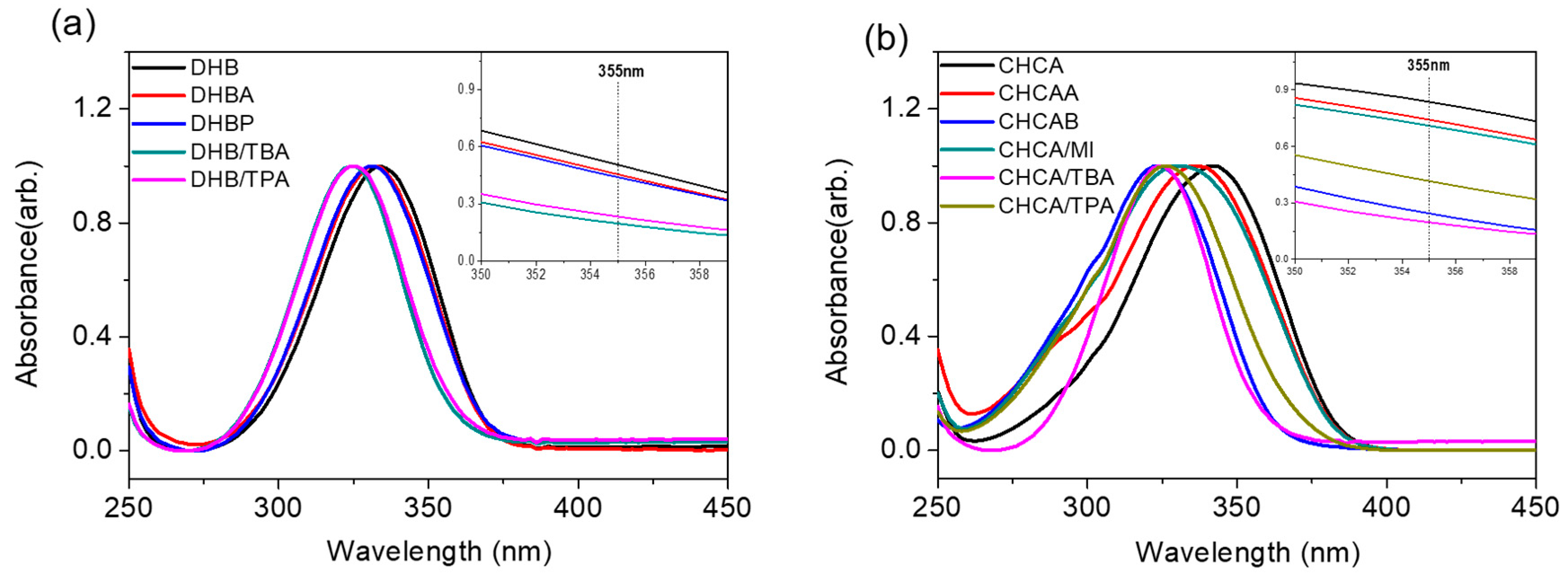
2.2. Possible Ionization Mechanism of ILM
2.3. Homogeneity of Sample Preparations
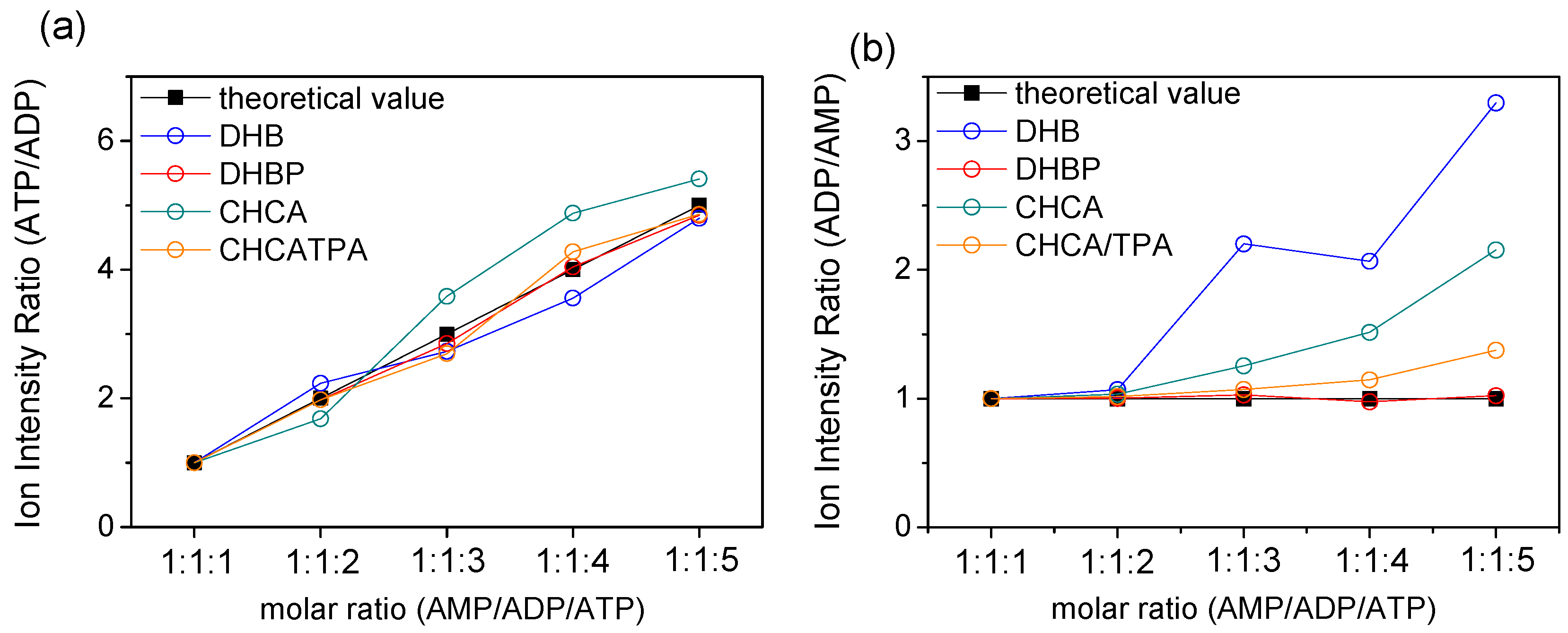
2.4. Quantitative Analysis of ATP
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Synthesis of ILMs and MALDI Sample Preparation
4.3. Instrument
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Atkinson, D.E. Energy charge of adenylate pool as a regulatory parameter interaction with feedback modifiers. Biochemistry 1968, 7, 4030–4034. [Google Scholar] [CrossRef] [PubMed]
- Qian, Y.R.; Wang, X.; Liu, Y.; Li, Y.S.; Colvin, R.A.; Tong, L.Y.; Wu, S.Y.; Chen, X.Z. Extracellular ATP is internalized by macropinocytosis and induces intracellular ATP increase and drug resistance in cancer cells. Cancer Lett. 2014, 351, 242–251. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.F.; Tozzi, F.; Chen, J.Y.; Fan, F.; Xia, L.; Wang, J.R.; Gao, G.; Zhang, A.J.; Xia, X.F.; Brasher, H.; et al. Intracellular ATP levels are a pivotal determinant of chemoresistance in colon cancer cells. Cancer Res. 2012, 72, 304–314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, S.S.; Jacobson, K.N.; McDermott, K.M.; Reddy, S.P.; Cress, A.E.; Tang, H.Y.; Dudek, S.M.; Black, S.M.; Garcia, J.G.N.; Makino, A.; et al. ATP promotes cell survival via regulation of cytosolic Ca2+ and Bcl-2/Bax ratio in lung cancer cells. Am. J. Physiol. Cell Physiol. 2016, 310, C99–C114. [Google Scholar] [CrossRef] [Green Version]
- Circu, M.L.; Maloney, R.E.; Aw, T.Y. Low glucose stress decreases cellular NADH and mitochondrial ATP in colonic epithelial cancer cells: Influence of mitochondrial substrates. Chem. Biol. Interact. 2017, 264, 16–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Towler, M.C.; Hardie, D.G. AMP-activated protein kinase in metabolic control and insulin signaling. Circ. Res. 2007, 100, 328–341. [Google Scholar] [CrossRef]
- Hardie, D.G. AMP-activated protein kinase-an energy sensor that regulates all aspects of cell function. Genes Dev. 2011, 25, 1895–1908. [Google Scholar] [CrossRef] [Green Version]
- Hardie, D.G. Keeping the home fires burning: AMP-activated protein kinase. J. R. Soc. Interface 2018, 15. [Google Scholar] [CrossRef]
- Queiroz, E.; Puukila, S.; Eichler, R.; Sampaio, S.C.; Forsyth, H.L.; Lees, S.J.; Barbosa, A.M.; Dekker, R.F.H.; Fortes, Z.B.; Khaper, N. Metformin induces apoptosis and cell cycle arrest mediated by oxidative stress, AMPK and FOXO3a in MCF-7 breast cancer cells. PLoS ONE 2014, 9, e98207. [Google Scholar] [CrossRef]
- Ke, R.; Xu, Q.C.; Li, C.; Luo, L.Y.; Huang, D.Q. Mechanisms of AMPK in the maintenance of ATP balance during energy metabolism. Cell Biol. Int. 2018, 42, 384–392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fenn, J.B.; Mann, M.; Meng, C.K.; Wong, S.F.; Whitehouse, C.M. Electrospray ionization for mass spectrometry of large biomolecules. Science 1989, 246, 64–71. [Google Scholar] [CrossRef] [PubMed]
- Karas, M.; Bahr, U.; Hillenkamp, F. UV laser matrix desorption ionization mass-spectrometry of proteins in the 100000 dalton range. Int. J. Mass Spectrom. 1989, 92, 231–242. [Google Scholar] [CrossRef]
- Karas, M.; Bachmann, D.; Bahr, U.; Hillenkamp, F. Matrix-assisted ultraviolet laser desorption of non-volatile compounds. Int. J. Mass Spectrom. Ion Process. 1987, 78, 53–68. [Google Scholar] [CrossRef]
- Koichi Tanaka, H.W.; Ido, Y.; Akita, S.; Yoshida, Y.; Yoshida, T. Protein and polymer analyses up to m/z 100 000 by laser Ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 1988, 2, 151–153. [Google Scholar] [CrossRef]
- Barber, M.; Bordoli, R.S.; Sedgwick, R.D.; Tyler, A.N. Tyler Fast atom bombardment of solids (F.A.B.): A new ion source for mass spectrometry. J. Chem. Soc. Chem. Commun. 1981, 325–327. [Google Scholar] [CrossRef]
- Busch, K.L.; Unger, S.E.; Vincze, A.; Cooks, R.G.; Keough, T. Desorption ionization mass spectrometry: Sample preparation for secondary ion mass spectrometry, laser desorption, and field desorption. J. Am. Chem. Soc. 1982, 104, 1507–1511. [Google Scholar] [CrossRef]
- Karas, M.; Bahr, U.; Strupat, K.; Hillenkamp, F.; Tsarbopoulos, A.; Pramanik, B.N. Matrix dependence of metastable fragmentation of glycoproteins in MALDI TOF mass-spectrometry. Anal. Chem. 1995, 67, 675–679. [Google Scholar] [CrossRef]
- Schnolzer, M.; Lehmann, W.D. Identification of modified peptides by metastable fragmentation in MALDI mass spectrometry. Int. J. Mass Spectrom. 1997, 169, 263–271. [Google Scholar] [CrossRef]
- Kailemia, M.J.; Ruhaak, L.R.; Lebrilla, C.B.; Amster, I.J. Oligosaccharide analysis by mass spectrometry: A review of recent developments. Anal. Chem. 2014, 86, 196–212. [Google Scholar] [CrossRef] [Green Version]
- Nelson, C.M.; Tang, L.Z.W.; Smith, L.M.; Crellin, K.; Berry, J.; Beauchamp, J.L. Fragmentation mechanisms of oligonucleotides in MALDI mass spectrometry. SPIE 1996, 2680, 247–255. [Google Scholar]
- Juhasz, P.; Costello, C.E. Matrix-assisted laser desorption ionization time-of-flight mass-spectrometry of underivatized and permethylated gangliosides. J. Am. Soc. Mass Spectrom. 1992, 3, 785–796. [Google Scholar] [CrossRef] [Green Version]
- Abdelhamid, H.N. Ionic liquid-assisted laser desorption/ionization-mass spectrometry: Matrices, microextraction, and separation. Methods Protoc. 2018, 1, 23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mank, M.; Stahl, B.; Boehm, G. 2,5-dihydroxybenzoic acid butylamine and other ionic liquid matrixes for enhanced MALDI-MS analysis of biomolecules. Anal. Chem. 2004, 76, 2938–2950. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.L.; Wang, C.C.; Lai, Y.H.; Lee, H.; Lin, J.D.; Lee, Y.T.; Wang, Y.S. Selective enhancement of carbohydrate ion abundances by diamond nanoparticles for mass spectrometric analysis. Anal. Chem. 2013, 85, 3836–3841. [Google Scholar] [CrossRef]
- Huang, M.F.; Chang, H.T. Detection of carbohydrates using surface-assisted laser desorption/ionization mass spectrometry with HgTe nanostructures. Chem. Sci. 2012, 3, 2147–2152. [Google Scholar] [CrossRef]
- Hunter, J.M.; Lin, H.A.; Becker, C.H. Cryogenic frozen solution matrixes for analysis of DNA by time-of-flight mass spectrometry. Anal. Chem. 1997, 69, 3608–3612. [Google Scholar] [CrossRef]
- Liang, C.W.; Chang, P.J.; Lin, Y.J.; Lee, Y.T.; Ni, C.K. High ion yields of carbohydrates from frozen solution by UV-MALDI. Anal. Chem. 2012, 84, 3493–3499. [Google Scholar] [CrossRef]
- Lee, C.; Ni, C.-K. Soft matrix assisted laser desorption/ionization for labile glycoconjugates. J. Am. Soc. Mass Spectrom. 2019, 30, 1455–1463. [Google Scholar] [CrossRef]
- Armstrong, D.W.; Zhang, L.K.; He, L.F.; Gross, M.L. Ionic liquids as matrixes for matrix-assisted laser desorption/ionization mass spectrometry. Anal. Chem. 2001, 73, 3679–3686. [Google Scholar] [CrossRef]
- Crank, J.A.; Armstrong, D.W. Towards a second generation of ionic liquid matrices (ILMs) for MALDI-MS of peptides, proteins, and carbohydrates. J. Am. Soc. Mass Spectrom. 2009, 20, 1790–1800. [Google Scholar] [CrossRef] [Green Version]
- Laremore, T.N.; Murugesan, S.; Park, T.J.; Avci, F.Y.; Zagorevski, D.V.; Linhardt, R.J. Matrix-assisted laser desorption/ionization mass spectrometric analysis of uncomplexed highly sulfated oligosaccharides using ionic liquid matrices. Anal. Chem. 2006, 78, 1774–1779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, X.Y.; Shen, S.S.; Wu, D.T.; Cai, P.F.; Pan, Y.J. Novel ionic liquid matrices for qualitative and quantitative detection of carbohydrates by matrix assisted laser desorption/ionization mass spectrometry. Anal. Chim. Acta 2017, 985, 114–120. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.R.; Deja, S.; Kucejova, B.; Duarte, J.A.G.; McDonald, J.G.; Burgess, S.C. Targeted determination of tissue energy status by LC-MS/MS. Anal. Chem. 2019, 91, 5881–5887. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amantonico, A.; Oh, J.Y.; Sobek, J.; Heinemann, M.; Zenobi, R. Mass spectrometric method for analyzing metabolites in yeast with single cell sensitivity. Angew. Chem. Int. Ed. Engl. 2008, 47, 5382–5385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edwards, J.L.; Kennedy, R.T. Metabolomic analysis of eukaryotic tissue and prokaryotes using negative mode MALDI time-of-flight mass spectrometry. Anal. Chem. 2005, 77, 2201–2209. [Google Scholar] [CrossRef] [PubMed]
- Jansen, R.S.; Rosing, H.; Jan, H.M.S.; Jos, H.B. Mass spectrometry in the quantitative analysis of therapeutic intracellular nucleotide analogs. Mass Spectrom. Rev. 2011, 30, 321–343. [Google Scholar] [CrossRef]
- Abdelhamid, H.N.; Wu, H.F. Furoic and mefenamic acids as new matrices for matrix assisted laser desorption/ionization-(MALDI)-mass spectrometry. Talanta 2013, 115, 442–450. [Google Scholar] [CrossRef]
- Steinhoff, R.F.; Krismer, J.; Eyer, K.; Fagerer, S.R.; Ibanez, A.; Pabst, M.; Zenobi, R. Rapid estimation of the energy charge from cell lysates using matrix-assisted laser desorption/ionization mass spectrometry: Role of in-source fragmentation. Anal. Biochem. 2014, 447, 107–113. [Google Scholar] [CrossRef]
- Zabet-Moghaddam, M.; Heinzle, E.; Tholey, A. Qualitative and quantitative analysis of low molecular weight compounds by ultraviolet matrix-assisted laser desorption/ionization mass spectrometry using ionic liquid matrices. Rapid Commun. Mass Spectrom. 2004, 18, 141–148. [Google Scholar] [CrossRef]
- Huang, Y.F.; Chang, H.T. Analysis of adenosine triphosphate and glutathione through gold nanoparticles assisted laser desorption/ionization mass spectrometry. Anal. Chem. 2007, 79, 4852–4859. [Google Scholar] [CrossRef]
- Lu, I.C.; Lee, C.; Lee, Y.T.; Ni, C.K. Ionization mechanism of matrix-assisted laser desorption/ionization. Annu. Rev. Anal. Chem. 2015, 8, 21–39. [Google Scholar] [CrossRef] [PubMed]
- Knochenmuss, R. The coupled chemical and physical dynamics model of MALDI. Annu. Rev. Anal. Chem. 2016, 9, 365–385. [Google Scholar] [CrossRef] [PubMed]
- Karas, M.; Kruger, R. Ion formation in MALDI: The cluster ionization mechanism. Chem. Rev. 2003, 103, 427–440. [Google Scholar] [CrossRef] [PubMed]
- Lu, I.C.; Chu, K.; Lin, C.-Y.; Wu, S.-Y.; Dyakov, Y.; Chen, J.-L.; Gray-Weale, A.; Lee, Y.-T.; Ni, C.-K. Ion-to-neutral ratios and thermal proton transfer in matrix-assisted laser desorption/ionization. J. Am. Soc. Mass Spectrom. 2015, 26, 1242–1251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.L.; Gross, M.L.; Hsu, F.F. Ionic-liquid matrices for improved analysis of phospholipids by MALDI-TOF mass spectrometry. J. Am. Soc. Mass Spectrom. 2005, 16, 679–682. [Google Scholar] [CrossRef] [Green Version]
Sample Availability: Samples of the compounds are not available from the authors. |



© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, C.-H.; Lee, C.; Wu, Y.-C.; Lu, I.-C. New Strategy to Preserve Phosphate by Ionic Liquid Matrices in Matrix-Assisted Laser Desorption/Ionization: A Case of Adenosine Nucleotides. Molecules 2020, 25, 1217. https://doi.org/10.3390/molecules25051217
Lin C-H, Lee C, Wu Y-C, Lu I-C. New Strategy to Preserve Phosphate by Ionic Liquid Matrices in Matrix-Assisted Laser Desorption/Ionization: A Case of Adenosine Nucleotides. Molecules. 2020; 25(5):1217. https://doi.org/10.3390/molecules25051217
Chicago/Turabian StyleLin, Chih-Hao, Chuping Lee, Yu-Cheng Wu, and I-Chung Lu. 2020. "New Strategy to Preserve Phosphate by Ionic Liquid Matrices in Matrix-Assisted Laser Desorption/Ionization: A Case of Adenosine Nucleotides" Molecules 25, no. 5: 1217. https://doi.org/10.3390/molecules25051217





