Flexible Folding: Disulfide-Containing Peptides and Proteins Choose the Pathway Depending on the Environments
Abstract
:1. Introduction
2. Historical Background
3. Physicochemical Aspects of SS-Coupled Protein Folding
4. Oxidative Folding of Peptides and Proteins
4.1. Two-Disulfide Peptides

4.2. Three-Disulfide Proteins
4.3. Four-Disulfide Proteins
4.4. Oxidative Folding of Proteins with Odd Cys Residues
4.5. Oxidative Folding Pathways of Two Chain Proteins
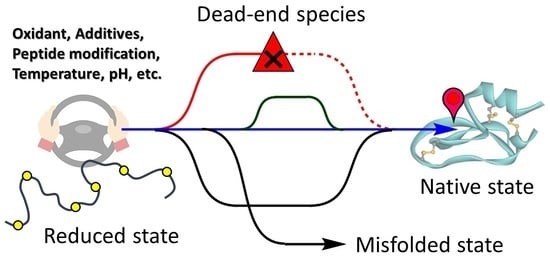
5. Flexibility of the Oxidative Folding Pathways
6. Perspectives and Concluding Remarks
Author Contributions
Funding
Conflicts of Interest
References
- Anfinsen, C.B. The formation and stabilization of protein structure. Biochem. J. 1972, 128, 737–749. [Google Scholar] [CrossRef] [Green Version]
- Anfinsen, C.B. Principles that Govern the Folding of Protein Chains. Science 1973, 181, 223–230. [Google Scholar] [CrossRef] [Green Version]
- Levinthal, C. Are there pathways for protein folding? J. Chim. Phys. Phys.-Chim. Biol. 1968, 65, 44–45. [Google Scholar] [CrossRef]
- Arolas, J.L.; Aviles, F.X.; Chang, J.-Y.; Ventura, S. Folding of small disulfide-rich proteins: Clarifying the puzzle. Trends Biochem. Sci. 2006, 31, 292–301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edsall, J.T. Hsien Wu and the First Theory of Protein Denaturation (1931). In Advances in Protein Chemistry; Anfinsen, C.B., Richards, F.M., Edsall, J.T., Eisenberg, D.S., Eds.; Protein Stability; Academic Press: Cambridge, MA, USA, 1995; Volume 46, pp. 1–5. [Google Scholar] [CrossRef]
- Mossuto, M.F. Disulfide Bonding in Neurodegenerative Misfolding Diseases. Int. J. Cell Biol. 2013, 2013, 318319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hartl, F.U. Protein Misfolding Diseases. Annu. Rev. Biochem. 2017, 86, 21–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khoury, G.A.; Smadbeck, J.; Kieslich, C.A.; Floudas, C.A. Protein folding and de novo protein design for biotechnological applications. Trends Biotechnol. 2014, 32, 99–109. [Google Scholar] [CrossRef] [Green Version]
- Rigoldi, F.; Donini, S.; Redaelli, A.; Parisini, E.; Gautieri, A. Review: Engineering of thermostable enzymes for industrial applications. APL Bioeng. 2018, 2. [Google Scholar] [CrossRef] [Green Version]
- Englander, S.W.; Mayne, L. The nature of protein folding pathways. Proc. Natl. Acad. Sci. USA 2014, 111, 15873–15880. [Google Scholar] [CrossRef] [Green Version]
- Bulaj, G.; Kortemme, T.; Goldenberg, D.P. Ionization−Reactivity Relationships for Cysteine Thiols in Polypeptides. Biochemistry 1998, 37, 8965–8972. [Google Scholar] [CrossRef]
- Tajc, S.G.; Tolbert, B.S.; Basavappa, R.; Miller, B.L. Direct Determination of Thiol pKa by Isothermal Titration Microcalorimetry. J. Am. Chem. Soc. 2004, 126, 10508–10509. [Google Scholar] [CrossRef] [PubMed]
- Aitken, A.; Learmonth, M. Carboxymethylation of Cysteine Using Iodoacetamide/ Iodoacetic Acid. In The Protein Protocols Handbook; Springer Protocols Handbooks; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2002; pp. 455–456. ISBN 978-1-59259-169-5. [Google Scholar]
- Narayan, M.; Welker, E.; Wedemeyer, W.J.; Scheraga, H.A. Oxidative Folding of Proteins. Acc. Chem. Res. 2000, 33, 805–812. [Google Scholar] [CrossRef] [PubMed]
- Wolynes, P.G.; Onuchic, J.N.; Thirumalai, D. Navigating the folding routes. Science 1995, 267, 1619–1620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Narayan, M. Revisiting the Formation of a Native Disulfide Bond: Consequences for Protein Regeneration and Beyond. Molecules 2020, 25, 5337. [Google Scholar] [CrossRef] [PubMed]
- Rothwarf, D.M.; Scheraga, H.A. Regeneration of bovine pancreatic ribonuclease A. 3. Dependence on the nature of the redox reagent. Biochemistry 1993, 32, 2690–2697. [Google Scholar] [CrossRef] [PubMed]
- Beld, J.; Woycechowsky, K.J.; Hilvert, D. Selenoglutathione: Efficient Oxidative Protein Folding by a Diselenide. Biochemistry 2007, 46, 5382–5390. [Google Scholar] [CrossRef]
- Arai, K.; Dedachi, K.; Iwaoka, M. Rapid and Quantitative Disulfide Bond Formation for a Polypeptide Chain Using a Cyclic Selenoxide Reagent in an Aqueous Medium. Chem. Eur. J. 2011, 17, 481–485. [Google Scholar] [CrossRef]
- Arai, K.; Iwaoka, M. Oxidative Protein Folding Using trans-3,4-Dihydroxyselenolane Oxide, Chapter 14. In Functional Disulphide Bonds: Methods and Protocols; Methods in Molecular Biology; Hogg, P., Ed.; Humana Press: Totowa, NJ, USA, 2019; ISBN 978-1-59259-169-5. [Google Scholar]
- Arai, K.; Noguchi, M.; Singh, B.G.; Priyadarsini, K.I.; Fujio, K.; Kubo, Y.; Takayama, K.; Ando, S.; Iwaoka, M. A water-soluble selenoxide reagent as a useful probe for the reactivity and folding of polythiol peptides. FEBS Open Bio 2013, 3, 55–64. [Google Scholar] [CrossRef] [Green Version]
- Iwaoka, M.; Kumakura, F.; Yoneda, M.; Nakahara, T.; Henmi, K.; Aonuma, H.; Nakatani, H.; Tomoda, S. Direct Observation of Conformational Folding Coupled with Disulphide Rearrangement by Using a Water-soluble Selenoxide Reagent—A Case of Oxidative Regeneration of Ribonuclease A under Weakly Basic Conditions. J. Biochem. 2008, 144, 121–130. [Google Scholar] [CrossRef]
- Shi, T.; Rabenstein, D.L. Discovery of a Highly Selective and Efficient Reagent for Formation of Intramolecular Disulfide Bonds in Peptides. J. Am. Chem. Soc. 2000, 122, 6809–6815. [Google Scholar] [CrossRef]
- Narayan, M.; Welker, E.; Scheraga, H.A. Native Conformational Tendencies in Unfolded Polypeptides: Development of a Novel Method To Assess Native Conformational Tendencies in the Reduced Forms of Multiple Disulfide-Bonded Proteins. J. Am. Chem. Soc. 2003, 125, 2036–2037. [Google Scholar] [CrossRef]
- Narayan, M.; Welker, E.; Wanjalla, C.; Xu, G.; Scheraga, H.A. Shifting the Competition between the Intramolecular Reshuffling Reaction and the Direct Oxidation Reaction during the Oxidative Folding of Kinetically Trapped Disulfide-Insecure Intermediates. Biochemistry 2003, 42, 10783–10789. [Google Scholar] [CrossRef]
- Lees, W.J. Small-molecule catalysts of oxidative protein folding. Curr. Opin. Chem. Biol. 2008, 12, 740–745. [Google Scholar] [CrossRef] [PubMed]
- Madar, D.J.; Patel, A.S.; Lees, W.J. Comparison of the oxidative folding of lysozyme at a high protein concentration using aromatic thiols versus glutathione. J. Biotechnol. 2009, 142, 214–219. [Google Scholar] [CrossRef] [PubMed]
- Potempa, M.; Hafner, M.; Frech, C. Mechanism of Gemini Disulfide Detergent Mediated Oxidative Refolding of Lysozyme in a New Artificial Chaperone System. Protein J. 2010, 29, 457–465. [Google Scholar] [CrossRef]
- Metanis, N.; Foletti, C.; Beld, J.; Hilvert, D. Selenoglutathione-Mediated Rescue of Kinetically Trapped Intermediates in Oxidative Protein Folding. Isr. J. Chem. 2011, 51, 953–959. [Google Scholar] [CrossRef]
- Iii, J.C.L.; Andersen, K.A.; Wallin, K.K.; Raines, R.T. Organocatalysts of oxidative protein folding inspired by protein disulfide isomerase. Org. Biomol. Chem. 2014, 12, 8598–8602. [Google Scholar] [CrossRef] [Green Version]
- Reddy, P.S.; Metanis, N. Small molecule diselenide additives for in vitro oxidative protein folding. Chem. Commun. 2016, 52, 3336–3339. [Google Scholar] [CrossRef] [Green Version]
- Arai, K.; Ueno, H.; Asano, Y.; Chakrabarty, G.; Shimodaira, S.; Mugesh, G.; Iwaoka, M. Protein Folding in the Presence of Water-Soluble Cyclic Diselenides with Novel Oxidoreductase and Isomerase Activities. ChemBioChem 2018, 19, 207–211. [Google Scholar] [CrossRef]
- Okada, S.; Matsusaki, M.; Arai, K.; Hidaka, Y.; Inaba, K.; Okumura, M.; Muraoka, T. Coupling effects of thiol and urea-type groups for promotion of oxidative protein folding. Chem. Commun. 2019, 55, 759–762. [Google Scholar] [CrossRef]
- Tsukagoshi, S.; Mikami, R.; Arai, K. Basic Amino Acid Conjugates of 1,2-Diselenan-4-amine with Protein Disulfide Isomerase-like Functions as a Manipulator of Protein Quality Control. Chem. Asian J. 2020, 15, 2646–2652. [Google Scholar] [CrossRef]
- Kersteen, E.A.; Raines, R.T. Catalysis of Protein Folding by Protein Disulfide Isomerase and Small-Molecule Mimics. Antioxid. Redox Signal. 2003, 5, 413–424. [Google Scholar] [CrossRef]
- Pegoraro, S.; Fiori, S.; Rudolph-Böhner, S.; Watanabe, T.X.; Moroder, L. Isomorphous replacement of cystine with selenocystine in endothelin: Oxidative refolding, biological and conformational properties of [Sec3,Sec11,Nle7]-endothelin-11 1Edited by R. Huber. J. Mol. Biol. 1998, 284, 779–792. [Google Scholar] [CrossRef] [PubMed]
- Iwaoka, M.; Arai, K. From Sulfur to Selenium. A New Research Arena in Chemical Biology and Biological Chemistry. Curr. Chem. Biol. 2013, 7, 2–24. [Google Scholar] [CrossRef]
- Pegoraro, S.; Fiori, S.; Cramer, J.; Rudolph-Böhner, S.; Moroder, L. The disulfide-coupled folding pathway of apamin as derived from diselenide-quenched analogs and intermediates. Protein Sci. 1999, 8, 1605–1613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fiori, S.; Pegoraro, S.; Rudolph-Böhner, S.; Cramer, J.; Moroder, L. Synthesis and conformational analysis of apamin analogues with natural and non-natural cystine/selenocystine connectivities. Biopolymers 2000, 53, 550–564. [Google Scholar] [CrossRef]
- Moroder, L.; Musiol, H.-J.; Götz, M.; Renner, C. Synthesis of single- and multiple-stranded cystine-rich peptides. Pept. Sci. 2005, 80, 85–97. [Google Scholar] [CrossRef]
- Walewska, A.; Zhang, M.-M.; Skalicky, J.J.; Yoshikami, D.; Olivera, B.M.; Bulaj, G. Integrated Oxidative Folding of Cysteine/Selenocysteine Containing Peptides: Improving Chemical Synthesis of Conotoxins. Angew. Chem. Int. Ed. 2009, 48, 2221–2224. [Google Scholar] [CrossRef]
- Han, T.S.; Zhang, M.-M.; Gowd, K.H.; Walewska, A.; Yoshikami, D.; Olivera, B.M.; Bulaj, G. Disulfide-Depleted Selenoconopeptides: Simplified Oxidative Folding of Cysteine-Rich Peptides. ACS Med. Chem. Lett. 2010, 1, 140–144. [Google Scholar] [CrossRef]
- de Araujo, A.D.; Callaghan, B.; Nevin, S.T.; Daly, N.L.; Craik, D.J.; Moretta, M.; Hopping, G.; Christie, M.J.; Adams, D.J.; Alewood, P.F. Total Synthesis of the Analgesic Conotoxin MrVIB through Selenocysteine-Assisted Folding. Angew. Chem. Int. Ed. 2011, 50, 6527–6529. [Google Scholar] [CrossRef]
- Walewska, A.; Jaśkiewicz, A.; Bulaj, G.; Rolka, K. Selenopeptide Analogs of EETI-II Retain Potent Trypsin Inhibitory Activities. Chem. Biol. Drug Des. 2011, 77, 93–97. [Google Scholar] [CrossRef] [PubMed]
- Steiner, A.M.; Woycechowsky, K.J.; Olivera, B.M.; Bulaj, G. Reagentless Oxidative Folding of Disulfide-Rich Peptides Catalyzed by an Intramolecular Diselenide. Angew. Chem. Int. Ed. 2012, 51, 5580–5584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Creighton, T.E. Experimental studies of protein folding and unfolding. Prog. Biophys. Mol. Biol. 1979, 33, 231–297. [Google Scholar] [CrossRef]
- Creighton, T.E.; Goldenberg, D.P. Kinetic role of a meta-stable native-like two-disulphide species in the folding transition of bovine pancreatic trypsin inhibitor. J. Mol. Biol. 1984, 179, 497–526. [Google Scholar] [CrossRef]
- Weissman, J.S.; Kim, P.S. Reexamination of the folding of BPTI: Predominance of native intermediates. Science 1991, 253, 1386–1393. [Google Scholar] [CrossRef] [Green Version]
- Weissman, J.S.; Kim, P.S. Kinetic role of nonnative species in the folding of bovine pancreatic trypsin inhibitor. Proc. Natl. Acad. Sci. USA 1992, 89, 9900–9904. [Google Scholar] [CrossRef] [Green Version]
- Weissman, J.S.; Kim, P.S. A kinetic explanation for the rearrangement pathway of BPTI folding. Nat. Struct. Biol. 1995, 2, 1123–1130. [Google Scholar] [CrossRef]
- Creighton, T.E.; Darby, N.J.; Kemmink, J. The roles of partly folded intermediates in protein folding. FASEB J. 1996, 10, 110–118. [Google Scholar] [CrossRef]
- Metanis, N.; Hilvert, D. Strategic Use of Non-Native Diselenide Bridges to Steer Oxidative Protein Folding. Angew. Chem. Int. Ed. 2012, 51, 5585–5588. [Google Scholar] [CrossRef]
- Metanis, N.; Hilvert, D. Harnessing selenocysteine reactivity for oxidative protein folding. Chem. Sci. 2014, 6, 322–325. [Google Scholar] [CrossRef] [Green Version]
- Mousa, R.; Lansky, S.; Shoham, G.; Metanis, N. BPTI folding revisited: Switching a disulfide into methylene thioacetal reveals a previously hidden path. Chem. Sci. 2018, 9, 4814–4820. [Google Scholar] [CrossRef] [Green Version]
- Chatrenet, B.; Chang, J.Y. The folding of hirudin adopts a mechanism of trial and error. J. Biol. Chem. 1992, 267, 3038–3043. [Google Scholar] [PubMed]
- Chatrenet, B.; Chang, J.Y. The disulfide folding pathway of hirudin elucidated by stop/go folding experiments. J. Biol. Chem. 1993, 268, 20988–20996. [Google Scholar]
- Chang, J.-Y. The Properties of Scrambled Hirudins. J. Biol. Chem. 1995, 270, 25661–25666. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thannhauser, T.W.; Rothwarf, D.M.; Scheraga, H.A. Kinetic Studies of the Regeneration of Recombinant Hirudin Variant 1 with Oxidized and Reduced Dithiothreitol. Biochemistry 1997, 36, 2154–2165. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, Y.; Misawa, S.; Sukesada, A.; Ohba, Y.; Hayashi, H. CX-397, a Novel Recombinant Hirudin Analog Having a Hybrid Sequence of Hirudin Variants-1 and -3. Biochem. Biophys. Res. Commun. 1993, 196, 773–779. [Google Scholar] [CrossRef] [PubMed]
- Shimamoto, S.; Fukutsuji, M.; Osumi, T.; Goto, M.; Toyoda, H.; Hidaka, Y. Topological Regulation of the Bioactive Conformation of a Disulfide-Rich Peptide, Heat-Stable Enterotoxin. Molecules 2020, 25, 4798. [Google Scholar] [CrossRef]
- Rothwarf, D.M.; Li, Y.-J.; Scheraga, H.A. Regeneration of Bovine Pancreatic Ribonuclease A: Identification of Two Nativelike Three-Disulfide Intermediates Involved in Separate Pathways. Biochemistry 1998, 37, 3760–3766. [Google Scholar] [CrossRef]
- Welker, E.; Narayan, M.; Volles, M.J.; Scheraga, H.A. Two new structured intermediates in the oxidative folding of RNase A. FEBS Lett. 1999, 460, 477–479. [Google Scholar] [CrossRef] [Green Version]
- Arai, K.; Kumakura, F.; Iwaoka, M. Characterization of Kinetic and Thermodynamic Phases in the Prefolding Process of Bovine Pancreatic Ribonuclease A Coupled with Fast SS Formation and SS Reshuffling. Biochemistry 2010, 49, 10535–10542. [Google Scholar] [CrossRef]
- Arai, K.; Kumakura, F.; Iwaoka, M. Kinetic and thermodynamic analysis of the conformational folding process of SS-reduced bovine pancreatic ribonuclease A using a selenoxide reagent with high oxidizing ability. FEBS Open Bio 2012, 2, 60–70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van den Berg, B.; Chung, E.W.; Robinson, C.V.; Mateo, P.L.; Dobson, C.M. The oxidative refolding of hen lysozyme and its catalysis by protein disulfide isomerase. EMBO J. 1999, 18, 4794–4803. [Google Scholar] [CrossRef] [Green Version]
- van den Berg, B.; Chung, E.W.; Robinson, C.V.; Dobson, C.M. Characterisation of the dominant oxidative folding intermediate of hen lysozyme. J. Mol. Biol. 1999, 290, 781–796. [Google Scholar] [CrossRef]
- Arai, K.; Shibagaki, W.; Shinozaki, R.; Iwaoka, M. Reinvestigation of the Oxidative Folding Pathways of Hen Egg White Lysozyme: Switching of the Major Pathways by Temperature Control. Int. J. Mol. Sci. 2013, 14, 13194–13212. [Google Scholar] [CrossRef] [Green Version]
- Ewbank, J.J.; Creighton, T.E. Structural characterization of the disulfide folding intermediates of bovine alpha-lactalbumin. Biochemistry 1993, 32, 3694–3707. [Google Scholar] [CrossRef]
- Chang, J.-Y. The Folding Pathway of α-Lactalbumin Elucidated by the Technique of Disulfide Scrambling. Isolation of on-pathway and off-pathway intermediates. J. Biol. Chem. 2002, 277, 120–126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, J.-Y.; Li, L. Pathway of Oxidative Folding of α-Lactalbumin: A Model for Illustrating the Diversity of Disulfide Folding Pathways. Biochemistry 2002, 41, 8405–8413. [Google Scholar] [CrossRef] [PubMed]
- Shinozaki, R.; Iwaoka, M. Effects of Metal Ions, Temperature, and a Denaturant on the Oxidative Folding Pathways of Bovine α-Lactalbumin. Int. J. Mol. Sci. 2017, 18, 1996. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iwaoka, M.; Mitsuji, T.; Shinozaki, R. Oxidative folding pathways of bovine milk β-lactoglobulin with odd cysteine residues. FEBS Open Bio 2019, 9, 1379–1391. [Google Scholar] [CrossRef]
- Ikeguchi, M. Transient Non-Native Helix Formation during the Folding of β-Lactoglobulin. Biomolecules 2014, 4, 202–216. [Google Scholar] [CrossRef]
- Arai, K.; Takei, T.; Shinozaki, R.; Noguchi, M.; Fujisawa, S.; Katayama, H.; Moroder, L.; Ando, S.; Okumura, M.; Inaba, K.; et al. Characterization and optimization of two-chain folding pathways of insulin via native chain assembly. Commun. Chem. 2018, 1, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Qiao, Z.-S.; Guo, Z.-Y.; Feng, Y.-M. Putative Disulfide-Forming Pathway of Porcine Insulin Precursor during Its Refolding in Vitro. Biochemistry 2001, 40, 2662–2668. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiao, Z.-S.; Min, C.-Y.; Hua, Q.-X.; Weiss, M.A.; Feng, Y.-M. In Vitro Refolding of Human Proinsulin. Kinetic intermediates, putative disulfide-forming pathway folding initiation site, and potential role of C-peptide in folding process. J. Biol. Chem. 2003, 278, 17800–17809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meienhofer, J.; Schnabel, E.; Bremer, H.; Brinkhoff, O.; Zabel, R.; Sroka, W.; Klostermayer, H.; Brandenburg, D.; Okuda, T.; Zahn, H. Synthesis of insulin and their combination to insulin Synthesis of insulin and their combination to insulin-active preparation. Z. Naturforsch. B 1963, 18, 1120–1121. [Google Scholar] [CrossRef]
- Katsoyannis, P.G.; Fukuda, K.; Tometsko, A.; Suzuki, K.; Tilak, M. The Synthesis of the B-Chain of Insulin and Its Combination with Natural or Synthetis A-Chin to Generate Insulin Activity. J. Am. Chem. Soc. 1964, 86, 930–932. [Google Scholar] [CrossRef]
- Kung, Y.; Du, Y.; Huang, W.; Chen, C.; Ke, L.; Hu, S.; Jiang, R.; Chu, S.; Niu, C.; Hsu, J.; et al. Total synthesis of crystalline insulin. Scientia Sinica 1966, 15, 544–561. [Google Scholar] [CrossRef]
- Arai, K.; Takei, T.; Okumura, M.; Watanabe, S.; Amagai, Y.; Asahina, Y.; Moroder, L.; Hojo, H.; Inaba, K.; Iwaoka, M. Preparation of Selenoinsulin as a Long-Lasting Insulin Analogue. Angew. Chem. Int. Ed. 2017, 56, 5522–5526. [Google Scholar] [CrossRef]
- Weil-Ktorza, O.; Rege, N.; Lansky, S.; Shalev, D.E.; Shoham, G.; Weiss, M.A.; Metanis, N. Substitution of an Internal Disulfide Bridge with a Diselenide Enhances both Foldability and Stability of Human Insulin. Chem. Eur. J. 2019, 25, 8513–8521. [Google Scholar] [CrossRef]
- Polverino de Laureto, P.; Frare, E.; Gottardo, R.; van Dael, H.; Fontana, A. Partly folded states of members of the lysozyme/lactalbumin superfamily: A comparative study by circular dichroism spectroscopy and limited proteolysis. Protein Sci. 2002, 11, 2932–2946. [Google Scholar] [CrossRef] [Green Version]
- Okumura, M.; Watanabe, S.; Inaba, K. CHAPTER 3.3: Structural Insights into Disulfide Bond Formation and Protein Quality Control in the Mammalian Endoplasmic Reticulum. In Oxidative Folding of Proteins; RSC Publishing: Cambridge, UK, 2018; pp. 224–248. [Google Scholar]













| Category | Proteins | Number of SS bonds | Number of Cys Residues | SS-Bond Topology | Section |
|---|---|---|---|---|---|
| 2SS | insulin A-chain | 2 | 4 |  | 4.1 |
| relaxin A-chain | 2 | 4 |  | ||
| endothelin-1 | 2 | 4 |  | ||
| 3SS | BPTI | 3 | 6 |  | 4.2 |
| hirudin | 3 | 6 |  | ||
| enterotoxin | 3 | 6 |  | ||
| 4SS | ribonuclease A | 4 | 8 |  | 4.3 |
| lysozyme | 4 | 8 |  | ||
| α-lactalbumin | 4 | 8 |  | ||
| Odd Cys | β-lactoglobulin | 2 | 5 |  | 4.4 |
| Tow-chain | insulin selenoinsulin | 3 2 + 1SeSe | 4/2 a |  | 4.5 |
| relaxin 2 | 3 | 4/2 a |  |
| pH and Additive | k1 for R → 1SS (mM−1 s−1) | k2 for 1SS → 2SS (mM−1 s−1) | k3 for 2SS → 3SS (mM−1 s−1) |
|---|---|---|---|
| 4.0 | 4.3 ± 0.2 | 2.6 ± 0.1 | 1.2 ± 0.1 |
| 7.0 | 9.8 ± 0.5 | 6.6 ± 0.3 | 2.8 ± 0.3 |
| 8.0 | 18.6 ± 1.0 | 10.0 ± 0.6 | 3.6 ± 0.6 |
| 8.0, +2 M Gdn-HCl | 9.8 ± 0.6 | 5.0 ± 0.3 | 2.7 ± 0.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arai, K.; Iwaoka, M. Flexible Folding: Disulfide-Containing Peptides and Proteins Choose the Pathway Depending on the Environments. Molecules 2021, 26, 195. https://doi.org/10.3390/molecules26010195
Arai K, Iwaoka M. Flexible Folding: Disulfide-Containing Peptides and Proteins Choose the Pathway Depending on the Environments. Molecules. 2021; 26(1):195. https://doi.org/10.3390/molecules26010195
Chicago/Turabian StyleArai, Kenta, and Michio Iwaoka. 2021. "Flexible Folding: Disulfide-Containing Peptides and Proteins Choose the Pathway Depending on the Environments" Molecules 26, no. 1: 195. https://doi.org/10.3390/molecules26010195