Quantitative Proteomics and Phosphoproteomics Reveal TNF-α-Mediated Protein Functions in Hepatocytes
Abstract
:1. Introduction
2. Results
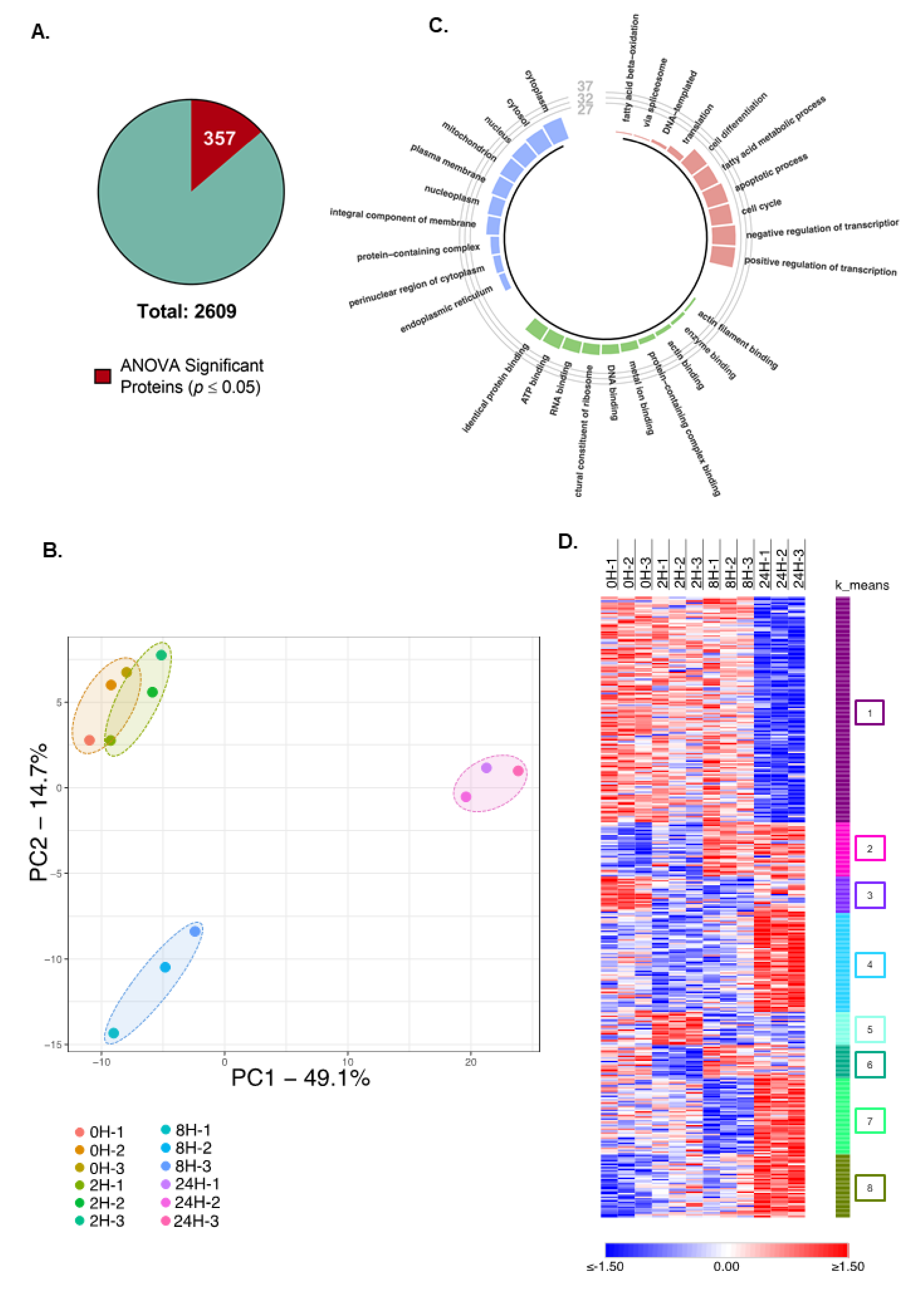
2.1. Time-Resolved Quantitative Proteomic Analysis
2.2. TNFα Treatment Induces an Extensive Modulation of the Proteome Profile of Hepatocytes
2.3. Exposure to TNFα Regulates Protein Synthesis and Cell Cycle Progression
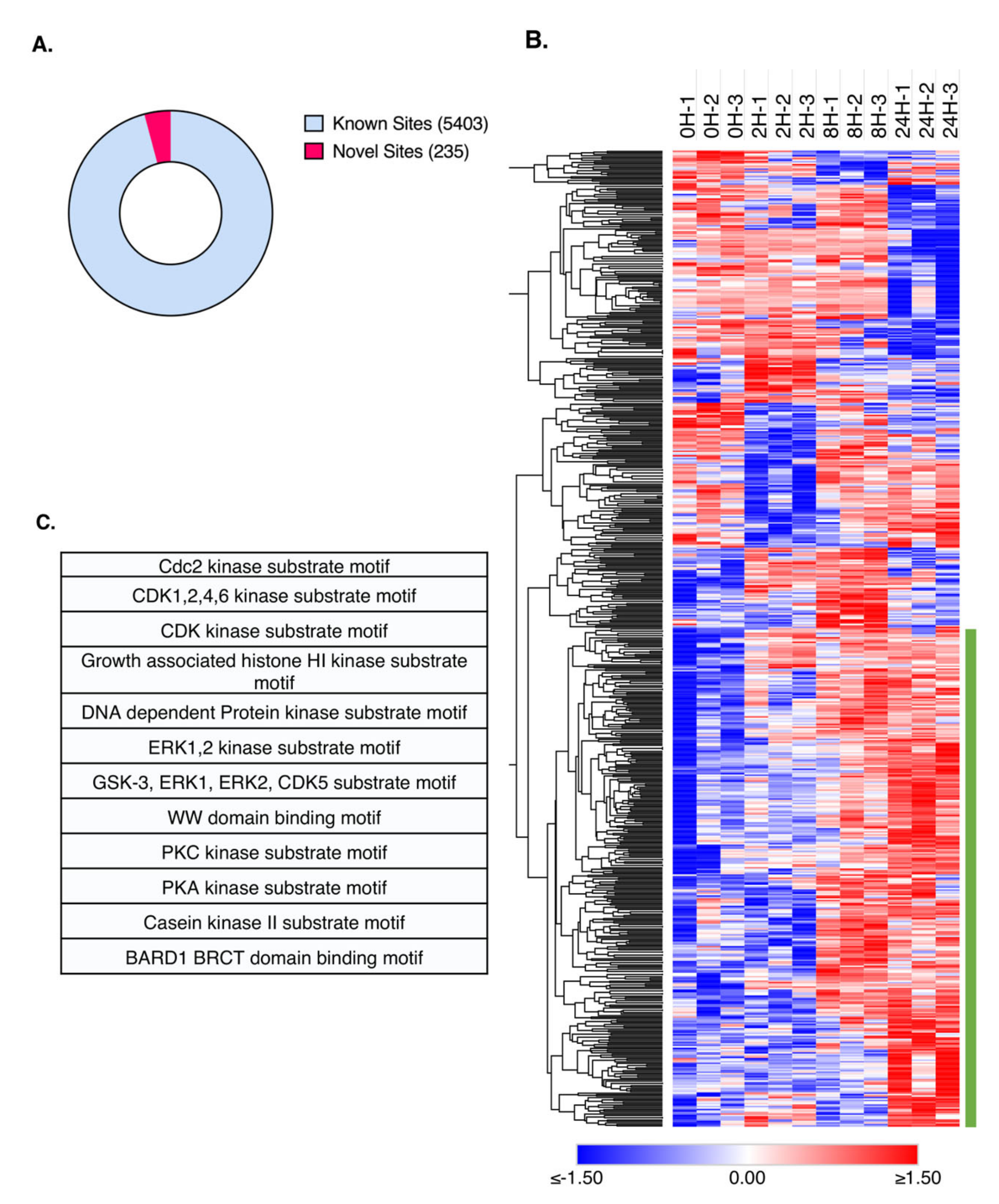
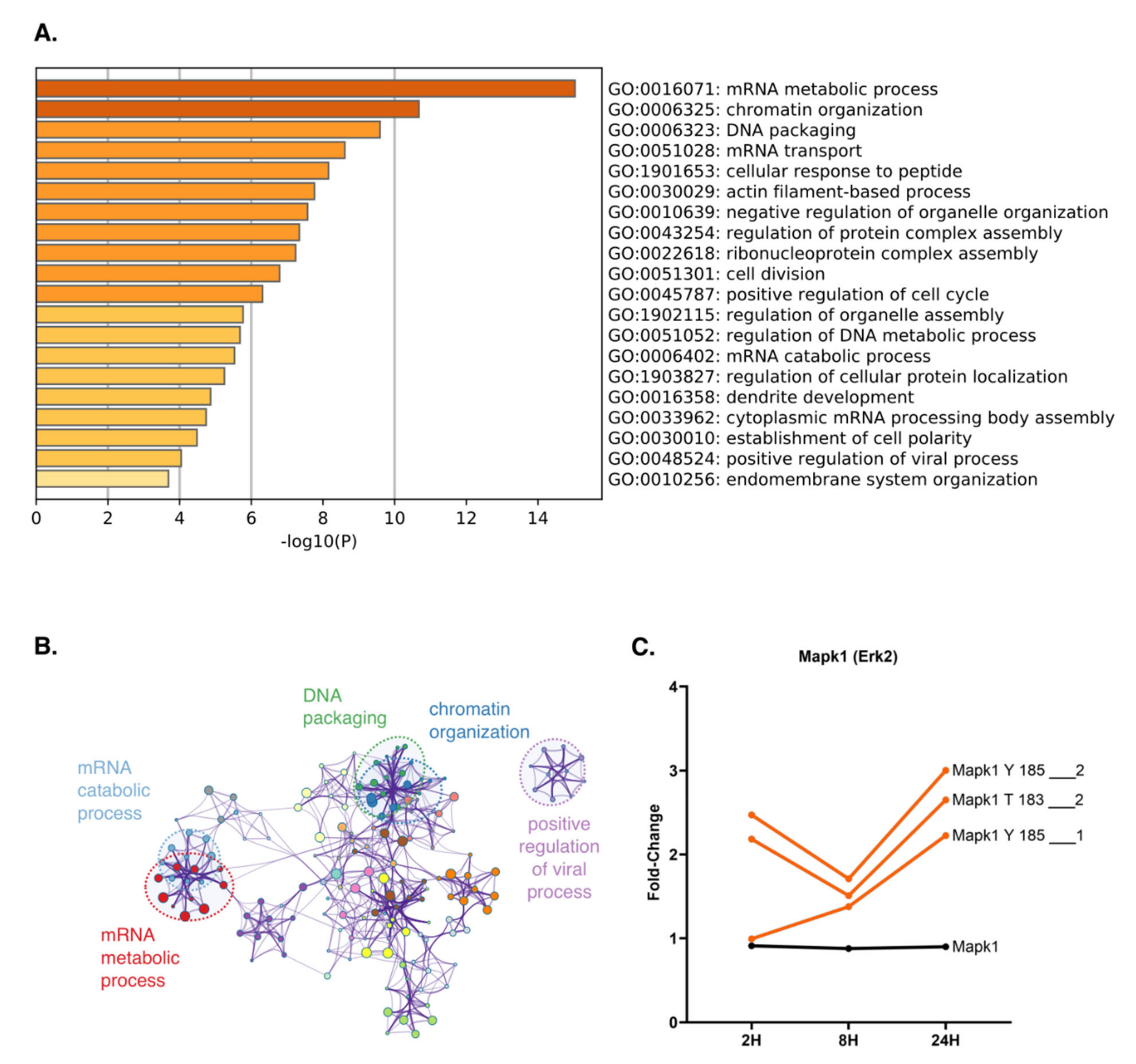
2.4. Time-Resolved Phosphoproteome Analysis Reveals Differential Regulation of Nuclear Proteins in Response to TNFα
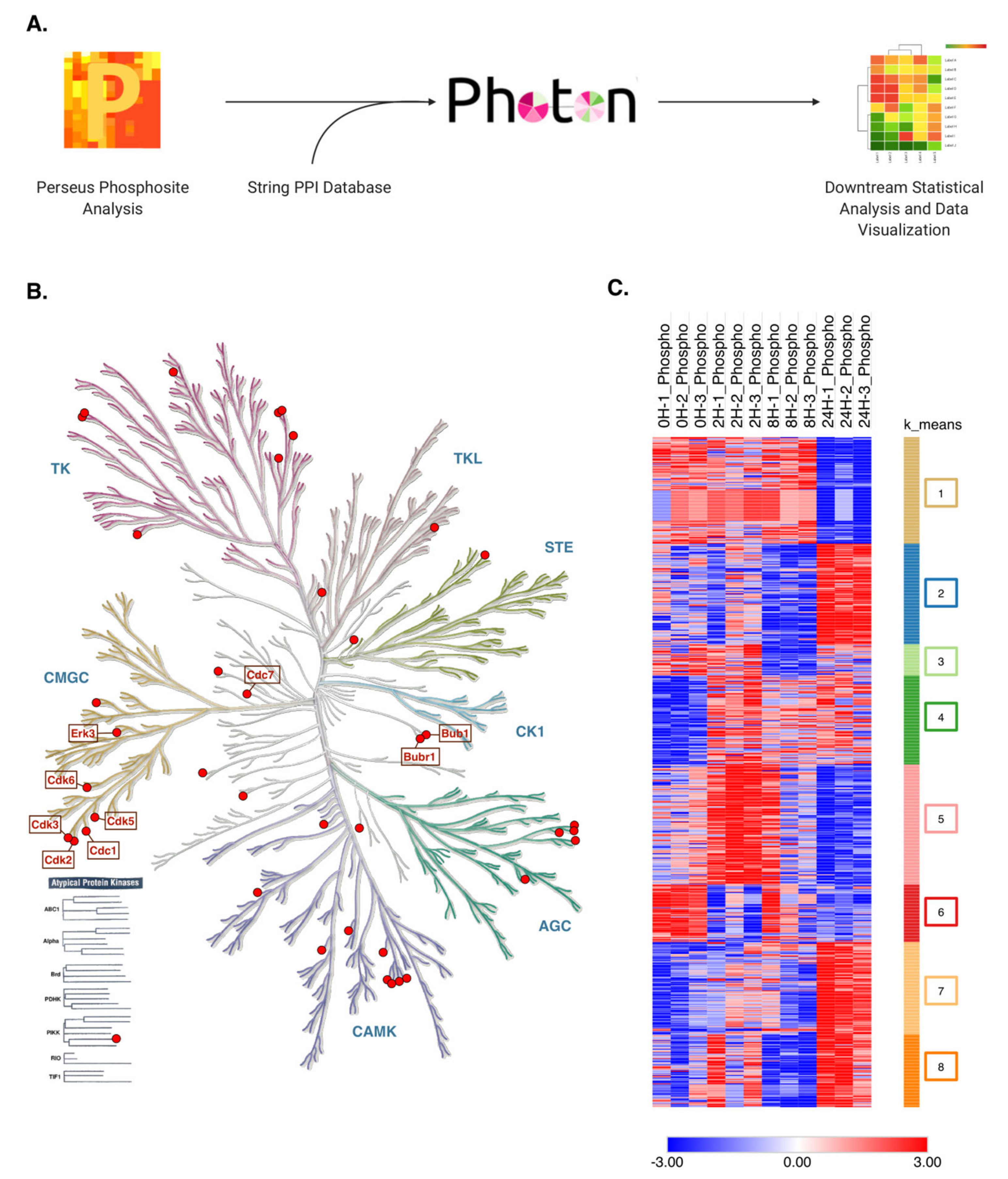
2.5. Key Cell Cycle Regulator Proteins Are Predicted Modulators of Phosphorylation
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Metabolic Assay
4.3. Protein Extraction
4.4. Sample Preparation for MS Analysis
4.5. Mass Spectrometry Analysis
4.6. Protein Identification and Quantification
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Jo, J.; Gavrilova, O.; Pack, S.; Jou, W.; Mullen, S.; Sumner, A.E.; Cushman, S.W.; Periwal, V. Hypertrophy and/or Hyperplasia: Dynamics of Adipose Tissue Growth. PLoS Comput. Biol. 2009, 5, e1000324. [Google Scholar] [CrossRef] [PubMed]
- Hotamisligil, G.S. Molecular mechanisms of insulin resistance and the role of the adipocyte. Int. J. Obes. 2000, 24, S23–S27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hotamisligil, G.S.; Spiegelman, B.M. Tumor Necrosis Factor α: A Key Component of the Obesity-Diabetes Link. Diabetes 1994, 43, 1271–1278. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, R.; Kubota, H.; Yugi, K.; Toyoshima, Y.; Komori, Y.; Soga, T.; Kuroda, S. The selective control of glycolysis, gluconeogenesis and glycogenesis by temporal insulin patterns. Mol. Syst. Biol. 2013, 9, 664. [Google Scholar] [CrossRef] [PubMed]
- Wilcox, G. Insulin and Insulin Resistance. Clin. Biochem. Rev. 2005, 26, 19–39. [Google Scholar] [PubMed]
- Guo, S. Insulin signaling, resistance, and metabolic syndrome: Insights from mouse models into disease mechanisms. J. Endocrinol. 2014, 220, T1–T23. [Google Scholar] [CrossRef] [PubMed]
- Schinner, S.; Scherbaum, W.A.; Bornstein, S.R.; Barthel, A. Molecular mechanisms of insulin resistance. Diabet. Med. 2005, 22, 674–682. [Google Scholar] [CrossRef] [PubMed]
- Boden, G.; She, P.; Mozzoli, M.; Cheung, P.; Gumireddy, K.; Reddy, P.; Xiang, X.; Luo, Z.; Ruderman, N. Free Fatty Acids Produce Insulin Resistance and Activate the Proinflammatory Nuclear Factor- B Pathway in Rat Liver. Diabetes 2005, 54, 3458–3465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hedrick, V.E.; Laland, M.N.; Nakayasu, E.S.; Paul, L.N. Digestion, Purification, and Enrichment of Protein Samples for Mass Spectrometry: Preparation of Protein Samples for MS. Curr. Protoc. Chem. Biol. 2015, 7, 201–222. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef] [PubMed]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Chen, R.; Wang, H.; Liang, F. Mechanisms Linking Inflammation to Insulin Resistance. Int. J. Endocrinol. 2015, 2015, 1–9. [Google Scholar] [CrossRef]
- Hotamisligil, G.S.; Arner, P.; Caro, J.F.; Atkinson, R.L.; Spiegelman, B.M. Increased adipose tissue expression of tumor necrosis factor-alpha in human obesity and insulin resistance. J. Clin. Investig. 1995, 95, 2409–2415. [Google Scholar] [CrossRef] [PubMed]
- Chadt, A.; Scherneck, S.; Joost, H.-G.; Al-Hasani, H. Molecular Links between Obesity and Diabetes: “Diabesity”. In Endotext; Feingold, K.R., Anawalt, B., Boyce, A., Chrousos, G., Dungan, K., Grossman, A., Hershman, J.M., Kaltsas, G., Koch, C., Kopp, P., et al., Eds.; MDText.com, Inc.: South Dartmouth, MA, USA, 2000. [Google Scholar]
- Mohallem, R.; Aryal, U.K. Regulators of TNFα mediated insulin resistance elucidated by quantitative proteomics. Sci. Rep. 2020, 10, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- The Gene Ontology Consortium. The Gene Ontology Resource: 20 years and still Going strong. Nucleic Acids Res. 2019, 47, D330–D338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Black, D.; Bird, M.A.; Hayden, M.; Schrum, L.W.; Lange, P.; Samson, C.; Hatano, E.; Rippe, R.A.; Brenner, D.A.; Behrns, K.E. TNFα-induced hepatocyte apoptosis is associated with alterations of the cell cycle and decreased stem loop binding protein. Surgery 2004, 135, 619–628. [Google Scholar] [CrossRef]
- Galadari, S.; Rahman, A.; Pallichankandy, S.; Galadari, A.; Thayyullathil, F. Role of ceramide in diabetes mellitus: Evidence and mechanisms. Lipids Heal. Dis. 2013, 12, 98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Y.P.; Grill, V. Long term exposure to fatty acids and ketones inhibits B-cell functions in human pancreatic islets of Langerhans. J. Clin. Endocrinol. Metab. 1995, 80, 1584–1590. [Google Scholar] [CrossRef]
- Jornayvaz, F.R.; Shulman, G.I. Diacylglycerol Activation of Protein Kinase Cε and Hepatic Insulin Resistance. Cell Metab. 2012, 15, 574–584. [Google Scholar] [CrossRef] [Green Version]
- Lang, C.H.; Frost, R.A.; Nairn, A.C.; MacLean, D.A.; Vary, T.C. TNF-α impairs heart and skeletal muscle protein synthesis by altering translation initiation. Am. J. Physiol.-Endocrinol. Metab. 2002, 282, E336–E347. [Google Scholar] [CrossRef] [Green Version]
- Amorim, I.S.; Lach, G.; Gkogkas, C.G. The Role of the Eukaryotic Translation Initiation Factor 4E (eIF4E) in Neuropsychiatric Disorders. Front. Genet. 2018, 9, 561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Labib, K.; Tercero, J.A.; Diffley, J. Uninterrupted MCM2-7 Function Required for DNA Replication Fork Progression. Science 2000, 288, 1643–1647. [Google Scholar] [CrossRef] [PubMed]
- Krüger, M.; Kratchmarova, I.; Blagoev, B.; Tseng, Y.-H.; Kahn, C.R.; Mann, M. Dissection of the insulin signaling pathway via quantitative phosphoproteomics. Proc. Natl. Acad. Sci. USA 2008, 105, 2451–2456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hotamisligil, G.S.; Murray, D.L.; Choy, L.N.; Spiegelman, B.M. Tumor necrosis factor alpha inhibits signaling from the insulin receptor. Proc. Natl. Acad. Sci. USA 1994, 91, 4854–4858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chambard, J.C.; Lefloch, R.; Pouysségur, J.; Lenormand, P. ERK implication in cell cycle regulation. Biochim. Biophys. Acta 2007, 1773, 1299–1310. [Google Scholar] [CrossRef]
- Ding, L.; Cao, J.; Lin, W.; Chen, H.; Xiong, X.; Ao, H.; Yu, M.; Lin, J.; Cui, Q. The Roles of Cyclin-Dependent Kinases in Cell-Cycle Progression and Therapeutic Strategies in Human Breast Cancer. Int. J. Mol. Sci. 2020, 21, 1960. [Google Scholar] [CrossRef] [Green Version]
- Giacinti, C.; Giordano, A. RB and cell cycle progression. Oncogene 2006, 25, 5220–5227. [Google Scholar] [CrossRef] [Green Version]
- Hsu, K.-S.; Kao, H.-Y. PML: Regulation and multifaceted function beyond tumor suppression. Cell Biosci. 2018, 8, 1–21. [Google Scholar] [CrossRef] [Green Version]
- Hsu, K.-S.; Guan, B.-J.; Cheng, X.; Guan, D.; Lam, M.; Hatzoglou, M.; Kao, H.-Y. Translational control of PML contributes to TNFα-induced apoptosis of MCF7 breast cancer cells and decreased angiogenesis in HUVECs. Cell Death Differ. 2015, 23, 469–483. [Google Scholar] [CrossRef]
- Rudolph, J.D.; Cox, J. A Network Module for the Perseus Software for Computational Proteomics Facilitates Proteome Interaction Graph Analysis. J. Proteome Res. 2019, 18, 2052–2064. [Google Scholar] [CrossRef] [Green Version]
- Naz, H.; Islam, A.; Ahmad, F.; Hassan, M.I. Calcium/calmodulin-dependent protein kinase IV: A multifunctional enzyme and potential therapeutic target. Prog. Biophys. Mol. Biol. 2016, 121, 54–65. [Google Scholar] [CrossRef] [PubMed]
- Krebs, J. Calmodulin-dependent protein kinase IV: Regulation of function and expression. Biochim. Biophys. Acta Mol. Cell Res. 1998, 1448, 183–189. [Google Scholar] [CrossRef] [Green Version]
- Soderling, T.R. The Ca2+—Calmodulin-dependent protein kinase cascade. Trends Biochem. Sci. 1999, 24, 232–236. [Google Scholar] [CrossRef]
- Kahn, S.E.; Hull, R.L.; Utzschneider, K.M. Mechanisms linking obesity to insulin resistance and type 2 diabetes. Nature 2006, 444, 840–846. [Google Scholar] [CrossRef]
- Lafontan, M. Adipose tissue and adipocyte dysregulation. Diabetes Metab. 2014, 40, 16–28. [Google Scholar] [CrossRef]
- Alipourfard, I.; Datukishvili, N.; Mikeladze, D. TNF-α Downregulation Modifies Insulin Receptor Substrate 1 (IRS-1) in Metabolic Signaling of Diabetic Insulin-Resistant Hepatocytes. Available online: https://www.hindawi.com/journals/mi/2019/3560819/ (accessed on 14 January 2021).
- Zembroski, A.S.; Buhman, K.K.; Aryal, U.K. Proteome and phosphoproteome characterization of liver in the postprandial state from diet-induced obese and lean mice. J. Proteom. 2021, 232, 104072. [Google Scholar] [CrossRef]
- Alberts, B.; Johnson, A.; Lewis, J.; Raff, M.; Roberts, K.; Walter, P. An Overview of the Cell Cycle. In Molecular Biology of the Cell, 4th ed.; Garland Science: New York, NY, USA, 2002. [Google Scholar]
- Schönenberger, F.; Deutzmann, A.; Ferrando-May, E.; Merhof, D. Discrimination of cell cycle phases in PCNA-immunolabeled cells. BMC Bioinform. 2015, 16, 3262. [Google Scholar] [CrossRef] [Green Version]
- Kurki, P.; Vanderlaan, M.; Dolbeare, F.; Gray, J.; Tan, E. Expression of proliferating cell nuclear antigen (PCNA)/cyclin during the cell cycle. Exp. Cell Res. 1986, 166, 209–219. [Google Scholar] [CrossRef]
- Forsburg, S.L. Eukaryotic MCM Proteins: Beyond Replication Initiation. Microbiol. Mol. Biol. Rev. 2004, 68, 109–131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Powell, S.K.; MacAlpine, H.K.; Prinz, J.A.; Li, Y.; Belsky, J.; MacAlpine, D.M. Dynamic loading and redistribution of the Mcm2-7 helicase complex through the cell cycle. EMBO J. 2015, 34, 531–543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, T.-Y.; Huang, H.H.; Wheeler, D.; Xu, Y.; Wells, J.A.; Song, Y.S.; Wiita, A.P. Time-Resolved Proteomics Extends Ribosome Profiling-Based Measurements of Protein Synthesis Dynamics. Cell Syst. 2017, 4, 636–644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kulej, K.; Avgousti, D.C.; Sidoli, S.; Herrmann, C.; Della Fera, A.N.; Kim, E.T.; Garcia, B.A.; Weitzman, M.D. Time-resolved Global and Chromatin Proteomics during Herpes Simplex Virus Type 1 (HSV-1) Infection. Mol. Cell. Proteom. 2017, 16, S92–S107. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Wolf-Yadlin, A.; Ross, P.L.; Pappin, D.; Rush, J.; Lauffenburger, D.A.; White, F. Time-resolved Mass Spectrometry of Tyrosine Phosphorylation Sites in the Epidermal Growth Factor Receptor Signaling Network Reveals Dynamic Modules. Mol. Cell. Proteom. 2005, 4, 1240–1250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1–10. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mohallem, R.; Aryal, U.K. Quantitative Proteomics and Phosphoproteomics Reveal TNF-α-Mediated Protein Functions in Hepatocytes. Molecules 2021, 26, 5472. https://doi.org/10.3390/molecules26185472
Mohallem R, Aryal UK. Quantitative Proteomics and Phosphoproteomics Reveal TNF-α-Mediated Protein Functions in Hepatocytes. Molecules. 2021; 26(18):5472. https://doi.org/10.3390/molecules26185472
Chicago/Turabian StyleMohallem, Rodrigo, and Uma K. Aryal. 2021. "Quantitative Proteomics and Phosphoproteomics Reveal TNF-α-Mediated Protein Functions in Hepatocytes" Molecules 26, no. 18: 5472. https://doi.org/10.3390/molecules26185472





