Targeted Downregulation of MYC through G-quadruplex Stabilization by DNAi
Abstract
:1. Introduction
2. Result
2.1. Biophysical Characterization of DNAi Binding to the MYC G4
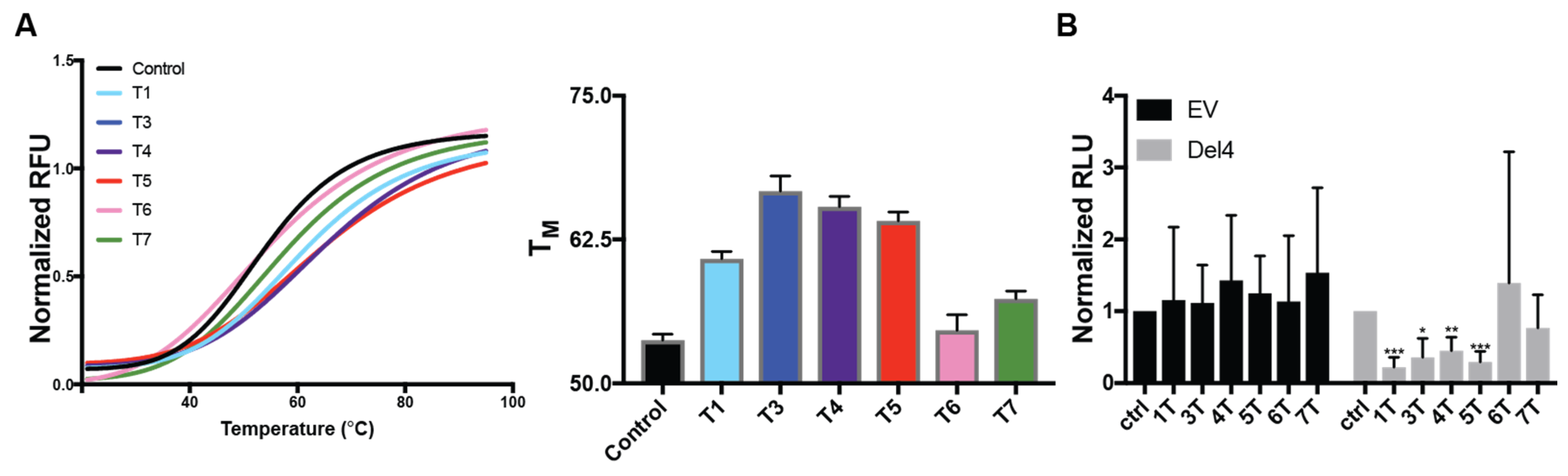
2.2. DNAi Therapeutic Potential
2.3. DNAi Cellular Activity
2.4. DNAi 5T Selectivity and Specificity for the MYC G4
2.5. DNAi 5T Recognition of the MYC G4
3. Discussion
4. Materials and Methods
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Dang, C.V. Enigmatic MYC conducts an Unfolding Systems Biology Symphoma. Genes Cancer 2010, 1, 526–531. [Google Scholar] [CrossRef] [PubMed]
- Baudino, T.A.; McKay, C.; Pendeville-Samain, H.; Nilsson, J.A.; Maclean, K.H.; White, E.L.; Davis, A.C.; Ihle, J.N.; Cleveland, J.L. c-Myc is essential for vasculogenesis and angiogenesis during development and tumor progression. Genes Dev. 2002, 16, 2530–2543. [Google Scholar] [CrossRef] [Green Version]
- Bauters, C.; de Groote, P.; Adamantidis, M.; Delcayre, C.; Hamon, M.; Lablanche, J.M.; Bertrand, M.E.; Dupuis, B.; Swynghedauw, B. Proto-oncogene expression in rabbit aorta after wall injury. First marker of the cellular process leading to restenosis after angioplasty? Eur. Heart J. 1992, 13, 556–559. [Google Scholar] [CrossRef] [PubMed]
- Biro, S.; Fu, Y.M.; Yu, Z.X.; Epstein, S.E. Inhibitory effects of antisense oligodeoxynucleotides targeting c-myc mRNA on smooth muscle cell proliferation and migration. Proc. Natl. Acad. Sci. USA 1993, 90, 654–658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crouch, D.H.; Fincham, V.J.; Frame, M.C. Targeted proteolysis of the focal adhesion kinase pp125 FAK during c-MYC-induced apoptosis is suppressed by integrin signalling. Oncogene 1996, 12, 2689–2696. [Google Scholar] [PubMed]
- Gavioli, R.; Frisan, T.; Vertuani, S.; Bornkamm, G.W.; Masucci, M.G. c-myc overexpression activates alternative pathways for intracellular proteolysis in lymphoma cells. Nat. Cell Biol. 2001, 3, 283–288. [Google Scholar] [CrossRef] [PubMed]
- Mai, S.; Fluri, M.; Siwarski, D.; Huppi, K. Genomic instability in MycER-activated Rat1A-MycER cells. Chromosome Res. 1996, 4, 365–371. [Google Scholar] [CrossRef] [PubMed]
- Martin, S.J.; O’Brien, G.A.; Nishioka, W.K.; McGahon, A.J.; Mahboubi, A.; Saido, T.C.; Green, D.R. Proteolysis of fodrin (non-erythroid spectrin) during apoptosis. J. Biol. Chem. 1995, 270, 6425–6428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Hagan, R.C.; Ohh, M.; David, G.; de Alboran, I.M.; Alt, F.W.; Kaelin, W.G., Jr.; DePinho, R.A. Myc-enhanced expression of Cul1 promotes ubiquitin-dependent proteolysis and cell cycle progression. Genes Dev. 2000, 14, 2185–2191. [Google Scholar] [CrossRef] [Green Version]
- Taylor, C.; Jalava, A.; Mai, S. c-Myc dependent initiation of genomic instability during neoplastic transformation. Curr. Top. Microbiol. Immunol. 1997, 224, 201–207. [Google Scholar]
- Taylor, C.; Mai, S. c-Myc-associated genomic instability of the dihydrofolate reductase locus in vivo. Cancer Detect. Prev. 1998, 22, 350–356. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.Y.; Grove, L.; Datta, N.S.; Long, M.W.; Prochownik, E.V. C-myc overexpression and p53 loss cooperate to promote genomic instability. Oncogene 1999, 18, 1177–1184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cotter, F.E.; Zucca, E. Altered gene expression and oncogenesis of B-cell neoplasia. Ann. Oncol. 1991, 2, 335–342. [Google Scholar] [CrossRef]
- Kato, G.J.; Dang, C.V. Function of the c-Myc oncoprotein. FASEB J. 1992, 6, 3065–3072. [Google Scholar] [CrossRef] [PubMed]
- Lutz, W.; Leon, J.; Eilers, M. Contributions of Myc to tumorigenesis. Biochim. Biophys. Acta 2002, 1602, 61–71. [Google Scholar] [CrossRef]
- Nesbit, C.E.; Tersak, J.M.; Prochownik, E.V. MYC oncogenes and human neoplastic disease. Oncogene 1999, 18, 3004–3016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adams, J.M.; Gerondakis, S.; Webb, E.; Corcoran, L.M.; Cory, S. Cellular myc oncogene is altered by chromosome translocation to an immunoglobulin locus in murine plasmacytomas and is rearranged similarly in human Burkitt lymphomas. Proc. Natl. Acad. Sci. USA 1983, 80, 1982–1986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adams, J.M.; Harris, A.W.; Pinkert, C.A.; Corcoran, L.M.; Alexander, W.S.; Cory, S.; Palmiter, R.D.; Brinster, R.L. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature 1985, 318, 533–538. [Google Scholar] [CrossRef]
- Cory, S.; Gerondakis, S.; Corcoran, L.M.; Bernard, O.; Webb, E.; Adams, J.M. Activation of the c-myc oncogene in B and T lymphoid tumors. Curr. Top. Microbiol. Immunol. 1984, 113, 161–165. [Google Scholar]
- Dalla-Favera, R.; Bregni, M.; Erikson, J.; Patterson, D.; Gallo, R.C.; Croce, C.M. Human c-myc onc gene is located on the region of chromosome 8 that is translocated in Burkitt lymphoma cells. Proc. Natl. Acad. Sci. USA 1982, 79, 7824–7827. [Google Scholar] [CrossRef] [Green Version]
- Frost, M.; Newell, J.; Lones, M.A.; Tripp, S.R.; Cairo, M.S.; Perkins, S.L. Comparative immunohistochemical analysis of pediatric Burkitt lymphoma and diffuse large B-cell lymphoma. Am. J. Clin. Path. 2004, 121, 384–392. [Google Scholar] [CrossRef]
- Rimsza, L.M.; Leblanc, M.L.; Unger, J.M.; Miller, T.P.; Grogan, T.M.; Persky, D.O.; Martel, R.R.; Sabalos, C.M.; Seligmann, B.; Braziel, R.M.; et al. Gene expression predicts overall survival in paraffin-embedded tissues of diffuse large B-cell lymphoma treated with R-CHOP. Blood 2008, 112, 3425–3433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vita, M.; Henriksson, M. The Myc oncoprotein as a therapeutic target for human cancer. Semin. Cancer Biol. 2006, 16, 318–330. [Google Scholar] [CrossRef]
- Hernandez, L.; Hernandez, S.; Bea, S.; Pinyol, M.; Ferrer, A.; Bosch, F.; Nadal, A.; Fernandez, P.L.; Palacin, A.; Montserrat, E.; et al. c-myc mRNA expression and genomic alterations in mantle cell lymphomas and other nodal non-Hodgkin’s lymphomas. Leukemia 1999, 13, 2087–2093. [Google Scholar] [CrossRef] [Green Version]
- Chen, R.; Sanchez, J.; Rosen, S.T. Clinical Management Updates in Mantle Cell Lymphoma. Oncology 2016, 30, 353–360. [Google Scholar]
- Dunleavy, K. Aggressive B cell Lymphoma: Optimal Therapy for MYC-positive, Double-Hit, and Triple-Hit DLBCL. Curr. Treat. Options Oncol. 2015, 16, 58. [Google Scholar] [CrossRef] [PubMed]
- Mossafa, H.; Damotte, D.; Jenabian, A.; Delarue, R.; Vincenneau, A.; Amouroux, I.; Jeandel, R.; Khoury, E.; Martelli, J.M.; Samson, T.; et al. Non-Hodgkin’s lymphomas with Burkitt-like cells are associated with c-Myc amplification and poor prognosis. Leuk. Lymphoma 2006, 47, 1885–1893. [Google Scholar] [CrossRef] [PubMed]
- Nagy, B.; Lundan, T.; Larramendy, M.L.; Aalto, Y.; Zhu, Y.; Niini, T.; Edgren, H.; Ferrer, A.; Vilpo, J.; Elonen, E.; et al. Abnormal expression of apoptosis-related genes in haematological malignancies: Overexpression of MYC is poor prognostic sign in mantle cell lymphoma. Br. J. Haematol. 2003, 120, 434–441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sewastianik, T.; Prochorec-Sobieszek, M.; Chapuy, B.; Juszczynski, P. MYC deregulation in lymphoid tumors: Molecular mechanisms, clinical consequences and therapeutic implications. Biochim. Biophys. Acta 2014, 1846, 457–467. [Google Scholar] [CrossRef]
- Shachaf, C.M.; Gentles, A.J.; Elchuri, S.; Sahoo, D.; Soen, Y.; Sharpe, O.; Perez, O.D.; Chang, M.; Mitchel, D.; Robinson, W.H.; et al. Genomic and proteomic analysis reveals a threshold level of MYC required for tumor maintenance. Cancer Res. 2008, 68, 5132–5142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soucek, L.; Whitfield, J.; Martins, C.P.; Finch, A.J.; Murphy, D.J.; Sodir, N.M.; Karnezis, A.N.; Swigart, L.B.; Nasi, S.; Evan, G.I. Modelling Myc inhibition as a cancer therapy. Nature 2008, 455, 679–683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brooks, T.A.; Hurley, L.H. The role of supercoiling in transcriptional control of MYC and its importance in molecular therapeutics. Nat. Rev. 2009, 9, 849–861. [Google Scholar] [CrossRef] [PubMed]
- Brooks, T.A.; Kendrick, S.; Hurley, L. Making sense of G-quadruplex and i-motif functions in oncogene promoters. FEBS J. 2010, 277, 3459–3469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brooks, T.A.; Hurley, L.H. Targeting MYC Expression through G-Quadruplexes. Genes Cancer 2010, 1, 641–649. [Google Scholar] [CrossRef] [PubMed]
- Hao, T.; Gaerig, V.C.; Brooks, T.A. Nucleic Acid clamps engineered to detect and stabilize the MYC G-quadruplex: A new therapeutic approach. Nucleic Acids Res. 2016, 44, 11013–11023. [Google Scholar] [CrossRef] [Green Version]
- Kaiser, C.E.; Gokhale, V.; Yang, D.; Hurley, L.H. Gaining insights into the small molecule targeting of the G-Quadruplex in the c-MYC promoter using NMR and an allele-specific transcriptional assay. Top. Curr. Chem. 2013, 330, 1–21. [Google Scholar] [CrossRef] [Green Version]
- Morgan, R.K.; Brooks, T.A. Targeting Promoter G-quadruplexes for Transcriptional Control. In Small-Molecule Transcription Factor Inhibitors in Oncology; Rahman, M.T., Thurston, D.E., Eds.; RSC Drug Discovery: London, UK, 2018. [Google Scholar]
- Felsenstein, K.M.; Saunders, L.B.; Simmons, J.K.; Leon, E.; Calabrese, D.R.; Zhang, S.; Michalowski, A.; Gareiss, P.; Mock, B.A.; Schneekloth, J.S., Jr. Small Molecule Microarrays Enable the Identification of a Selective, Quadruplex-Binding Inhibitor of MYC Expression. ACS Chem. Biol. 2016, 11, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, J.E.; Bambury, R.M.; Van Allen, E.M.; Drabkin, H.A.; Lara, P.N., Jr.; Harzstark, A.L.; Wagle, N.; Figlin, R.A.; Smith, G.W.; Garraway, L.A.; et al. A phase II trial of AS1411 (a novel nucleolin-targeted DNA aptamer) in metastatic renal cell carcinoma. Investig. New Drugs 2014, 32, 178–187. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Di Antonio, M.; McKinney, S.; Mathew, V.; Ho, B.; O’Neil, N.J.; Santos, N.D.; Silvester, J.; Wei, V.; Garcia, J.; et al. CX-5461 is a DNA G-quadruplex stabilizer with selective lethality in BRCA1/2 deficient tumours. Nat. Commun. 2017, 8, 14432. [Google Scholar] [CrossRef]
- Le, H.T.; Miller, M.C.; Buscaglia, R.; Dean, W.L.; Holt, P.A.; Chaires, J.B.; Trent, J.O. Not all G-quadruplexes are created equally: An investigation of the structural polymorphism of the c-Myc G-quadruplex-forming sequence and its interaction with the porphyrin TMPyP4. Org. Biomol. Chem. 2012, 10, 9393–9404. [Google Scholar] [CrossRef] [Green Version]
- Brown, R.V.; Danford, F.L.; Gokhale, V.; Hurley, L.H.; Brooks, T.A. Demonstration that drug-targeted down-regulation of MYC in non-Hodgkins lymphoma is directly mediated through the promoter G-quadruplex. J. Biol. Chem. 2011, 286, 41018–41027. [Google Scholar] [CrossRef] [Green Version]
- Sun, D.; Hurley, L.H. The importance of negative superhelicity in inducing the formation of G-quadruplex and i-motif structures in the c-Myc promoter: Implications for drug targeting and control of gene expression. J. Med. Chem. 2009, 52, 2863–2874. [Google Scholar] [CrossRef] [Green Version]
- Ambrus, A.; Chen, D.; Dai, J.; Jones, R.A.; Yang, D. Solution structure of the biologically relevant G-quadruplex element in the human c-MYC promoter. Implications for G-quadruplex stabilization. Biochemistry 2005, 44, 2048–2058. [Google Scholar] [CrossRef]
- Dai, J.; Carver, M.; Mathad, R.; Hurley, L.H.; Yang, D. Structure of 2:1 quindoline–MYC G-quadruplex. Ligand-induced reorientation and insights into drug design. Nat. Chem. 2011, 133, 17673–17680. [Google Scholar]
- Boddupally, P.V.; Hahn, S.; Beman, C.; De, B.; Brooks, T.A.; Gokhale, V.; Hurley, L.H. Anticancer activity and cellular repression of c-MYC by the G-quadruplex-stabilizing 11-piperazinylquindoline is not dependent on direct targeting of the G-quadruplex in the c-MYC promoter. J. Med. Chem. 2012, 55, 6076–6086. [Google Scholar] [CrossRef] [Green Version]
- De Cian, A.; Guittat, L.; Kaiser, M.; Sacca, B.; Amrane, S.; Bourdoncle, A.; Alberti, P.; Teulade-Fichou, M.P.; Lacroix, L.; Mergny, J.L. Fluorescence-based melting assays for studying quadruplex ligands. Methods 2007, 42, 183–195. [Google Scholar] [CrossRef]
- Morgan, R.K.; Psaras, A.M.; Lassiter, Q.; Raymer, K.; Brooks, T.A. G-quadruplex deconvolution with physiological mimicry enhances primary screening: Optimizing the FRET Melt(2) assay. Biochim. Biophys. Acta Gene Regul. Mech. 2020, 1863, 194478. [Google Scholar] [CrossRef] [PubMed]
- Duarte, A.R.; Cadoni, E.; Ressurreicao, A.S.; Moreira, R.; Paulo, A. Design of Modular G-quadruplex Ligands. ChemMedChem 2018, 13, 869–893. [Google Scholar] [CrossRef] [PubMed]
- Shukkur Ebrahim, A.; Kandouz, M.; Liddane, A.; Sabbagh, H.; Hou, Y.; Li, C.; Al-Katib, A. PNT2258, a novel deoxyribonucleic acid inhibitor, induces cell cycle arrest and apoptosis via a distinct mechanism of action: A new class of drug for non-Hodgkin’s lymphoma. Oncotarget 2016, 7, 42374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tolcher, A.W.; Rodrigueza, W.V.; Rasco, D.W.; Patnaik, A.; Papadopoulos, K.P.; Amaya, A.; Moore, T.D.; Gaylor, S.K.; Bisgaier, C.L.; Sooch, M.P.; et al. A phase 1 study of the BCL2-targeted deoxyribonucleic acid inhibitor (DNAi) PNT2258 in patients with advanced solid tumors. Cancer Chemother. Pharmacol. 2014, 73, 363–371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dang, C.V. MYC on the path to cancer. Cell 2012, 149, 22–35. [Google Scholar] [CrossRef] [Green Version]
- Meyer, N.; Penn, L.Z. Reflecting on 25 years with MYC. Nat. Rev. Cancer 2008, 8, 976–990. [Google Scholar] [CrossRef]
- Shachaf, C.M.; Kopelman, A.M.; Arvanitis, C.; Karlsson, A.; Beer, S.; Mandl, S.; Bachmann, M.H.; Borowsky, A.D.; Ruebner, B.; Cardiff, R.D.; et al. MYC inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer. Nature 2004, 431, 1112–1117. [Google Scholar] [CrossRef]
- Delmore, J.E.; Issa, G.C.; Lemieux, M.E.; Rahl, P.B.; Shi, J.; Jacobs, H.M.; Kastritis, E.; Gilpatrick, T.; Paranal, R.M.; Qi, J.; et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 2011, 146, 904–917. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jain, M.; Arvanitis, C.; Chu, K.; Dewey, W.; Leonhardt, E.; Trinh, M.; Sundberg, C.D.; Bishop, J.M.; Felsher, D.W. Sustained loss of a neoplastic phenotype by brief inactivation of MYC. Science 2002, 297, 102–104. [Google Scholar] [CrossRef] [PubMed]
- Carroll, J.S.; Swarbrick, A.; Musgrove, E.A.; Sutherland, R.L. Mechanisms of growth arrest by c-myc antisense oligonucleotides in MCF-7 breast cancer cells: Implications for the antiproliferative effects of antiestrogens. Cancer Res. 2002, 62, 3126–3131. [Google Scholar] [PubMed]
- Christensen, L.A.; Finch, R.A.; Booker, A.J.; Vasquez, K.M. Targeting oncogenes to improve breast cancer chemotherapy. Cancer Res. 2006, 66, 4089–4094. [Google Scholar] [CrossRef] [Green Version]
- Fallah, Y.; Brundage, J.; Allegakoen, P.; Shajahan-Haq, A.N. MYC-Driven Pathways in Breast Cancer Subtypes. Biomolecules 2017, 7, 53. [Google Scholar] [CrossRef]
- Chanvorachote, P.; Sriratanasak, N.; Nonpanya, N. C-myc Contributes to Malignancy of Lung Cancer: A Potential Anticancer Drug Target. Anticancer. Res. 2020, 40, 609–618. [Google Scholar] [CrossRef] [PubMed]





| DNAi | NSC338258 | Mean Hoescht Intensity | Mean FAM Intensity | Hoeschtmean/FAMmean |
|---|---|---|---|---|
| 5T | - | 4.1 | 6.4 | 1.6 |
| 5Tscr | - | 3.9 | 5.6 | 0.9 |
| 5T | + | 2.3 | 7.0 | 3.1 |
| 5Tscr | + | 4.4 | 6.1 | 0.9 |
| Oligonucleotide | 5′-3′ Sequence |
|---|---|
| Pu46/WT | GCGCTTATGGGGAGGGTGGGGAGGGTGGGGAAGGTGGGGAGGAGAC |
| MT5 | GCGCTTATGGGGAGGGTGGGGAGGGTGTGGAAGGTGGGGAGGAGAC |
| MT1,2,3,4 | GCGCTTATGGTGAGTGTGGTGAGTGTGGGGAAGGTGGGGAGGAGAC |
| VEGF | CCGGGGCGGGCCGGGGGCGGGGTCCCGGCGGGGCG |
| KRAS | GCGGGGAGAAGGAGGGGGCCGGGCCGGGCCGGCGGGGGAGGAGCGGGGGCCGGGCC |
| Bcl-2 | AGGGGCGGGCGCGGGAGGAAGGGGGCGGGAGCGGGGC |
| NRAS | GTGGGAGGGGCGGGTCTGGGTGCGGCC |
| Clamp a | CTCCTCCCCACCTTCCCC[C-18]ATAAG |
| 1T | CTCCTCCCCACCTTCCCCTATAAG |
| 3T | CTCCTCCCCACCTTCCCCTTTATAAG |
| 4T | CTCCTCCCCACCTTCCCCTTTTATAAG |
| 5T | CTCCTCCCCACCTTCCCCTTTTTATAAG |
| 5Tscr | GACCTTCCATCTCCCACTCCTTCCTATTA |
| 6T | CTCCTCCCCACCTTCCCCTTTTTTATAAG |
| 7T | CTCCTCCCCACCTTCCCCTTTTTTTATAAG |
| 5T 5′ | CTCCTCCCCACCTTCCCC |
| 5T 3′ | ATAAG |
| 5T 5xT | TTTTT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Psaras, A.M.; Chang, K.T.; Hao, T.; Brooks, T.A. Targeted Downregulation of MYC through G-quadruplex Stabilization by DNAi. Molecules 2021, 26, 5542. https://doi.org/10.3390/molecules26185542
Psaras AM, Chang KT, Hao T, Brooks TA. Targeted Downregulation of MYC through G-quadruplex Stabilization by DNAi. Molecules. 2021; 26(18):5542. https://doi.org/10.3390/molecules26185542
Chicago/Turabian StylePsaras, Alexandra Maria, Katarina T. Chang, Taisen Hao, and Tracy A. Brooks. 2021. "Targeted Downregulation of MYC through G-quadruplex Stabilization by DNAi" Molecules 26, no. 18: 5542. https://doi.org/10.3390/molecules26185542






