Structural Impacts of Drug-Resistance Mutations Appearing in HIV-2 Protease
Abstract
:1. Introduction
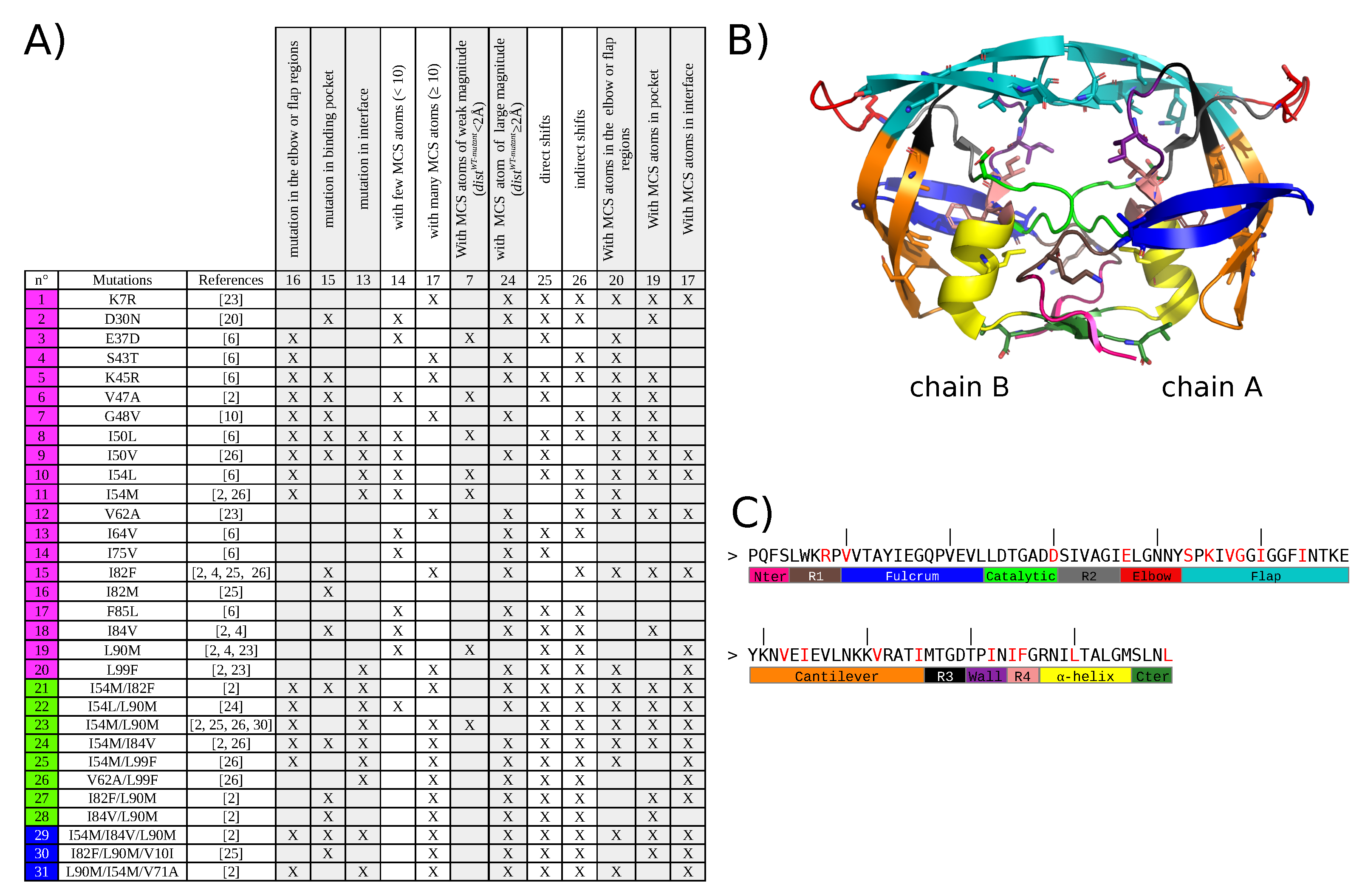
2. Results
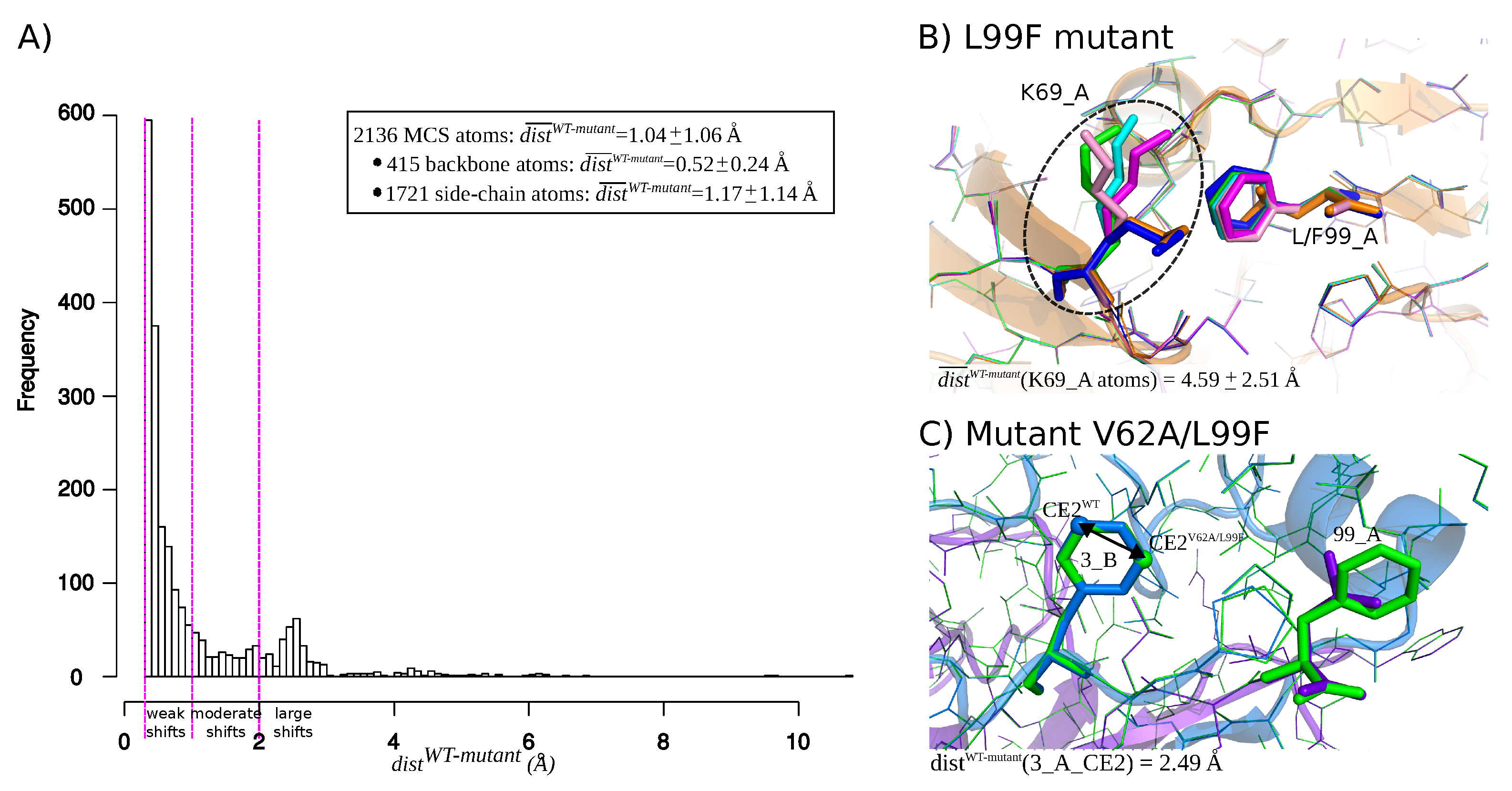
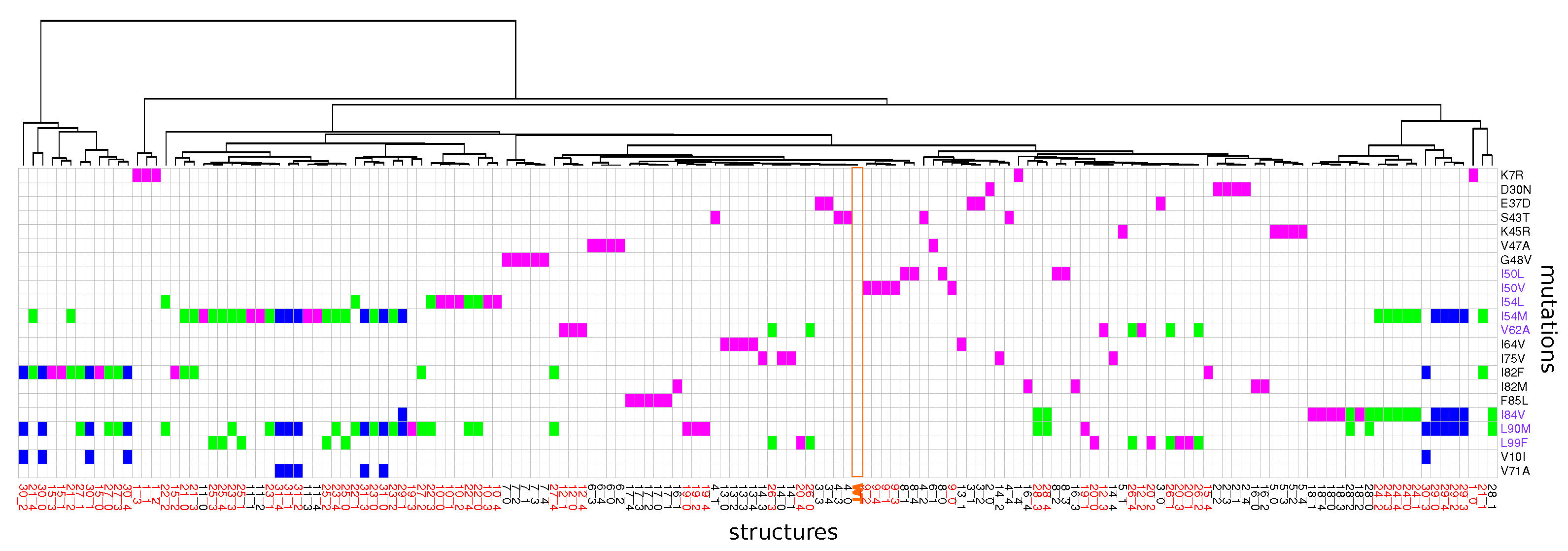
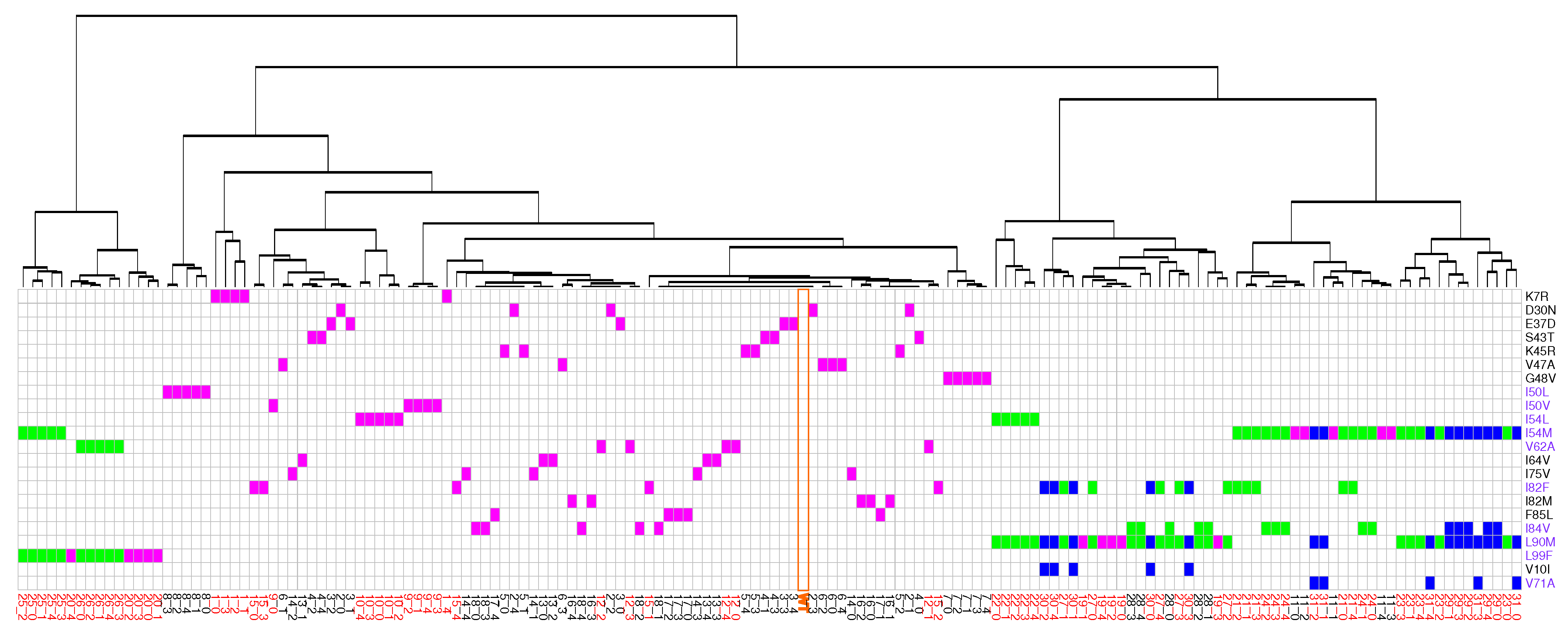
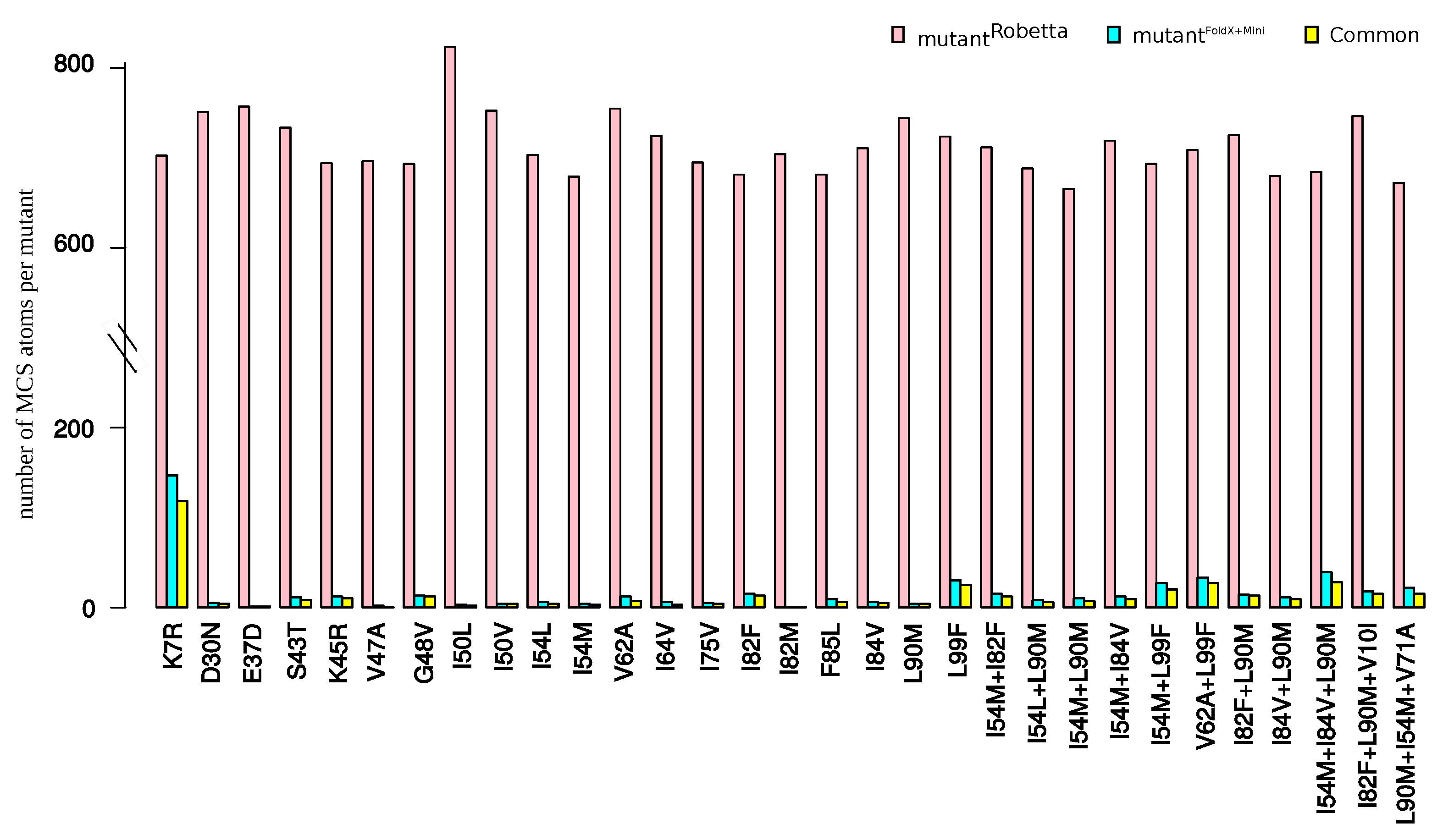
2.1. Identification of Atom Shifts in the Mutant-Structure Set
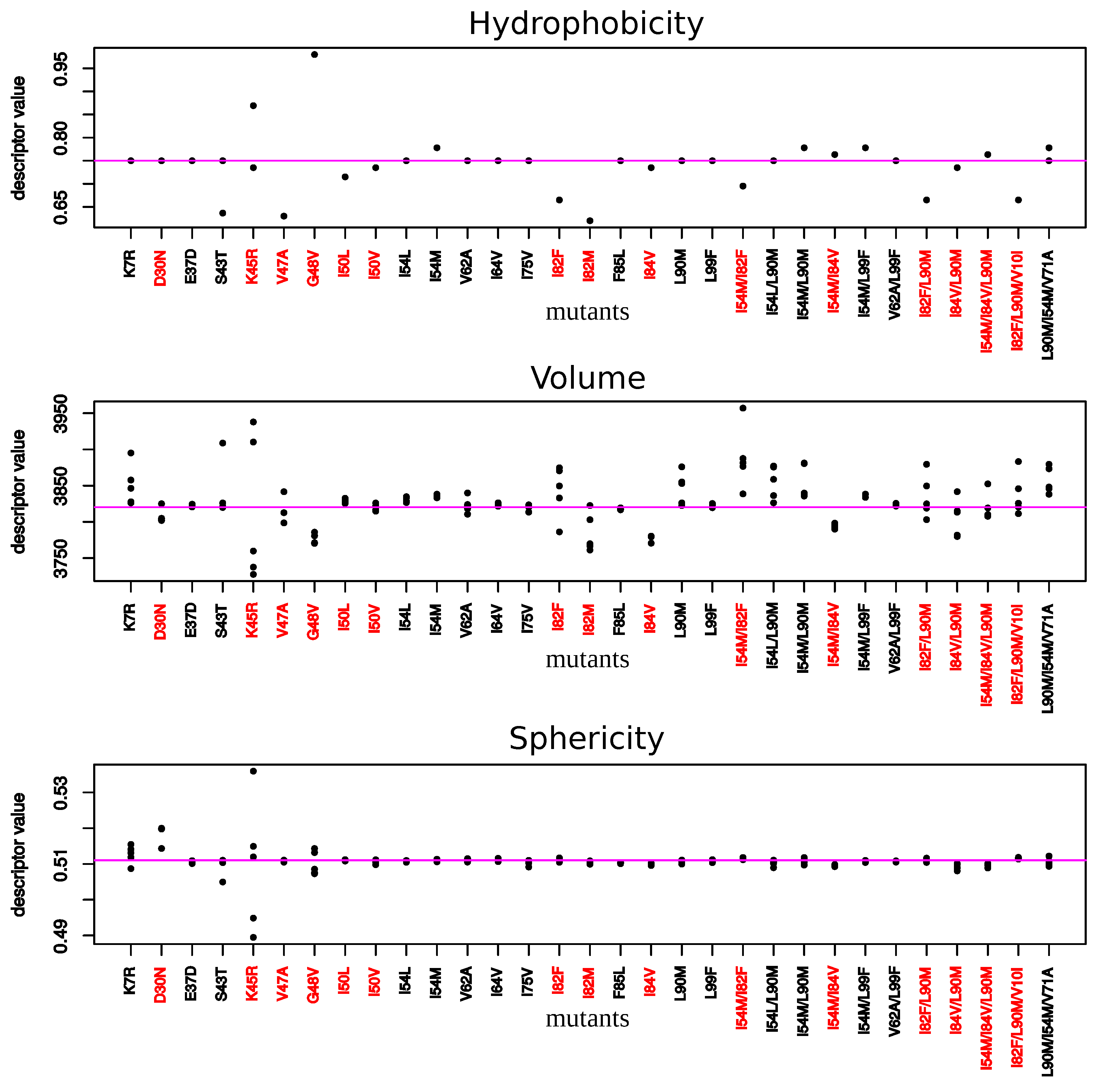
2.2. Impact of Mutations on the Properties of PI-Binding Pocket
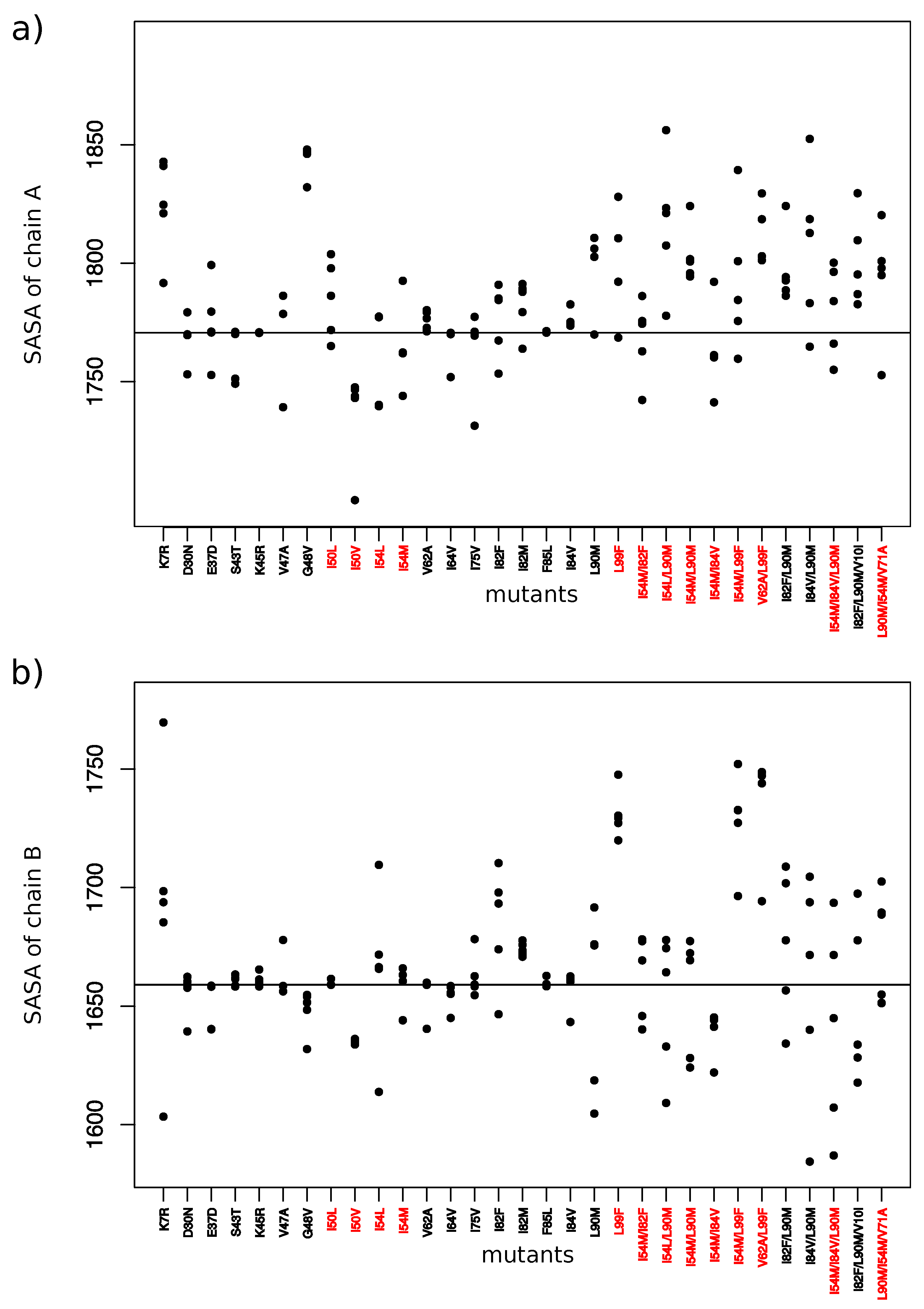
2.3. Impact on Interface
2.4. Impact of Combining Several Mutations Relative to Single Mutant
2.5. Impact of Using Different Structure Modeling Software
3. Discussion
4. Materials and Methods
4.1. Data
4.2. Structure Modeling
4.3. Identification of Shifted Atoms Induced by Mutations
4.4. Comparison of Wild-Type and Mutant Pockets
4.4.1. Pocket Estimation
4.4.2. Comparison of the Conformation of Wild-Type and Mutant Pockets
4.4.3. Comparison of the Properties of Wild-Type and Mutant Pockets
4.5. Interface Comparison
4.5.1. Interface Extraction
4.5.2. Comparison of the Interface Composition
4.5.3. Comparison of the SASA of Interface
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| PR | Protease |
| PR2 | HIV-2 Protease |
| PI | Protease inhibitor |
| MCS atoms | Mutant-Conserved Shifted atoms |
| RMSD | Root Mean Square Deviation |
| DRV | Darunavir |
| LPV | Lopinavir |
| SQV | Saquinavir |
| IDV | Indinavir |
| 3D | Three-dimensional |
| SASA | Solvent Accessible Surface Area |
| PPIC | Protein-Protein Interface Computation |
Appendix A
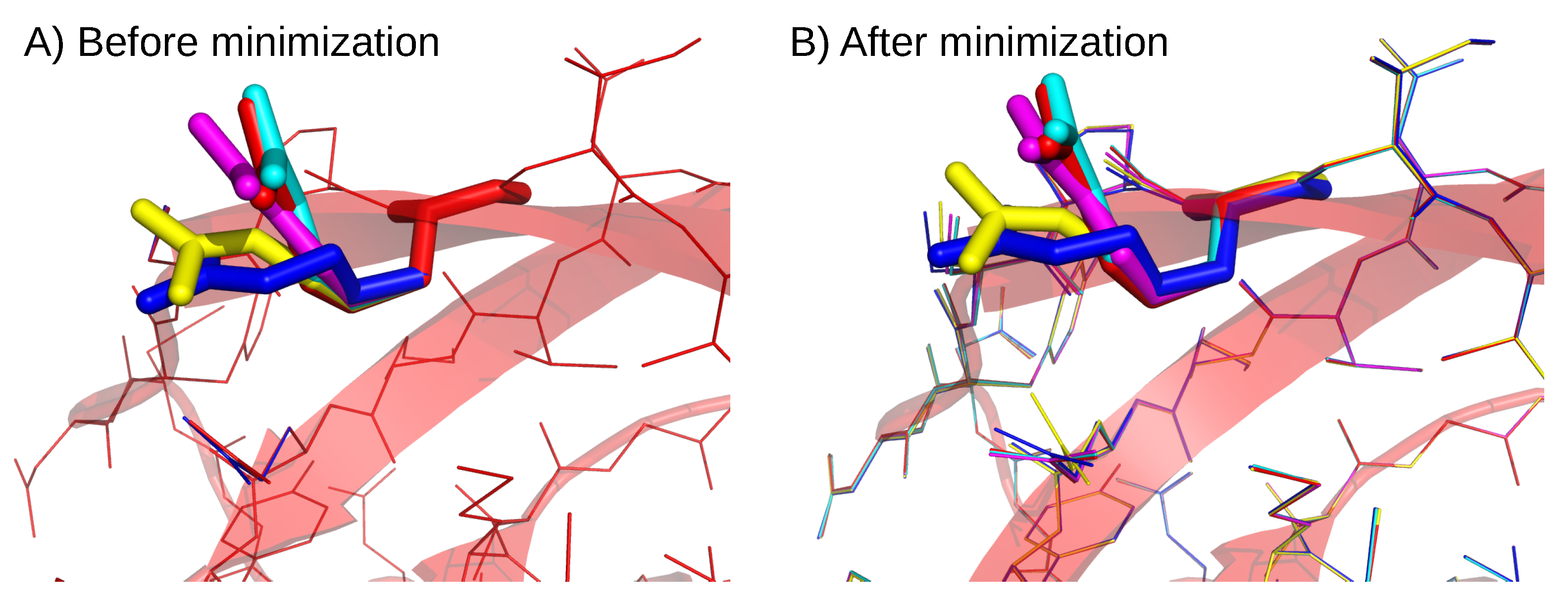
Appendix B
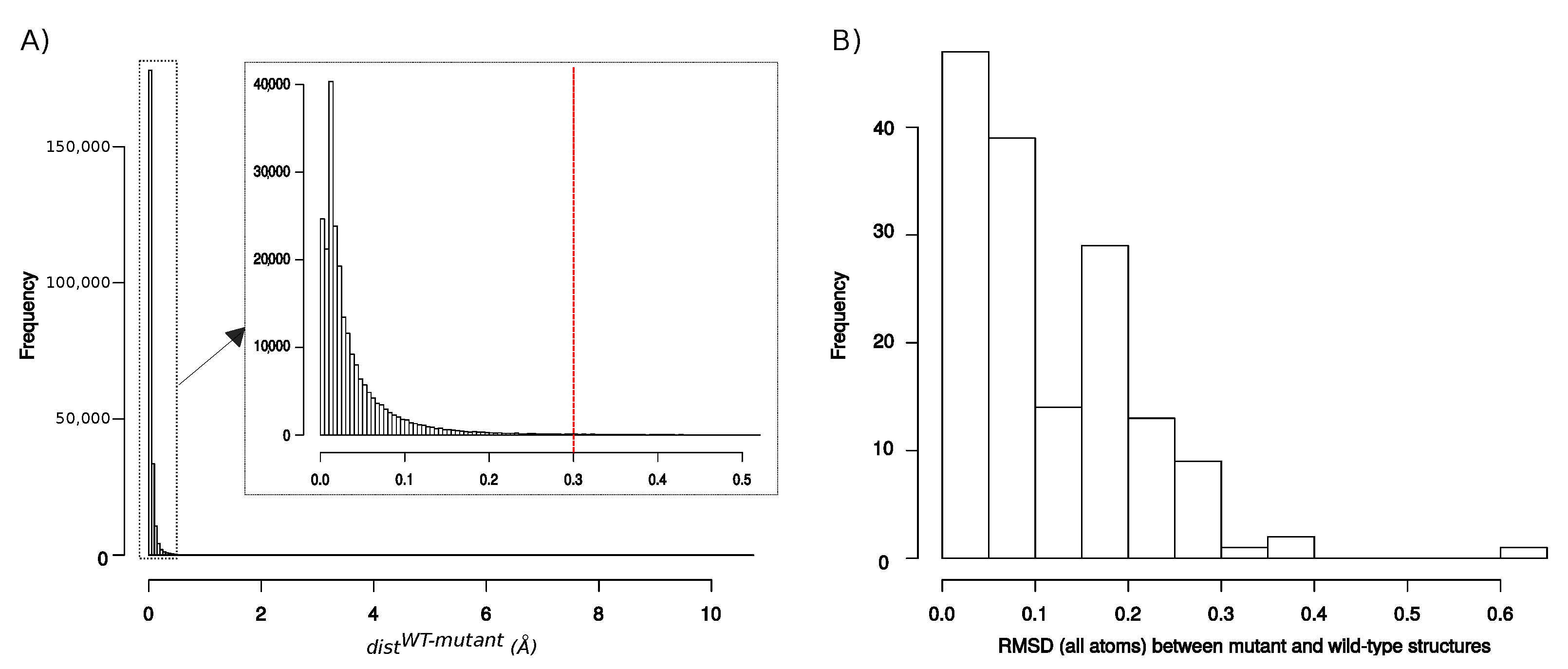
Appendix C
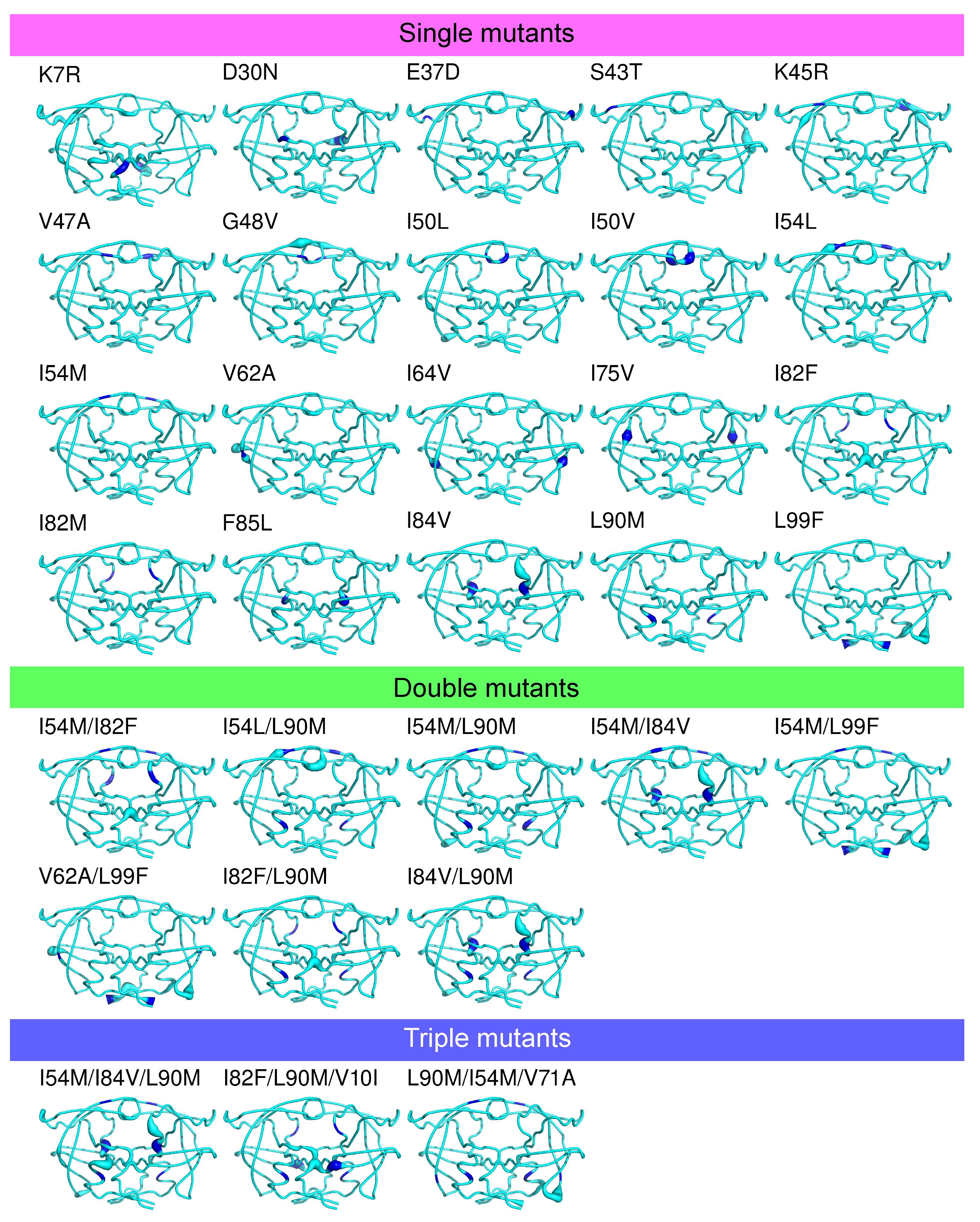
Appendix D
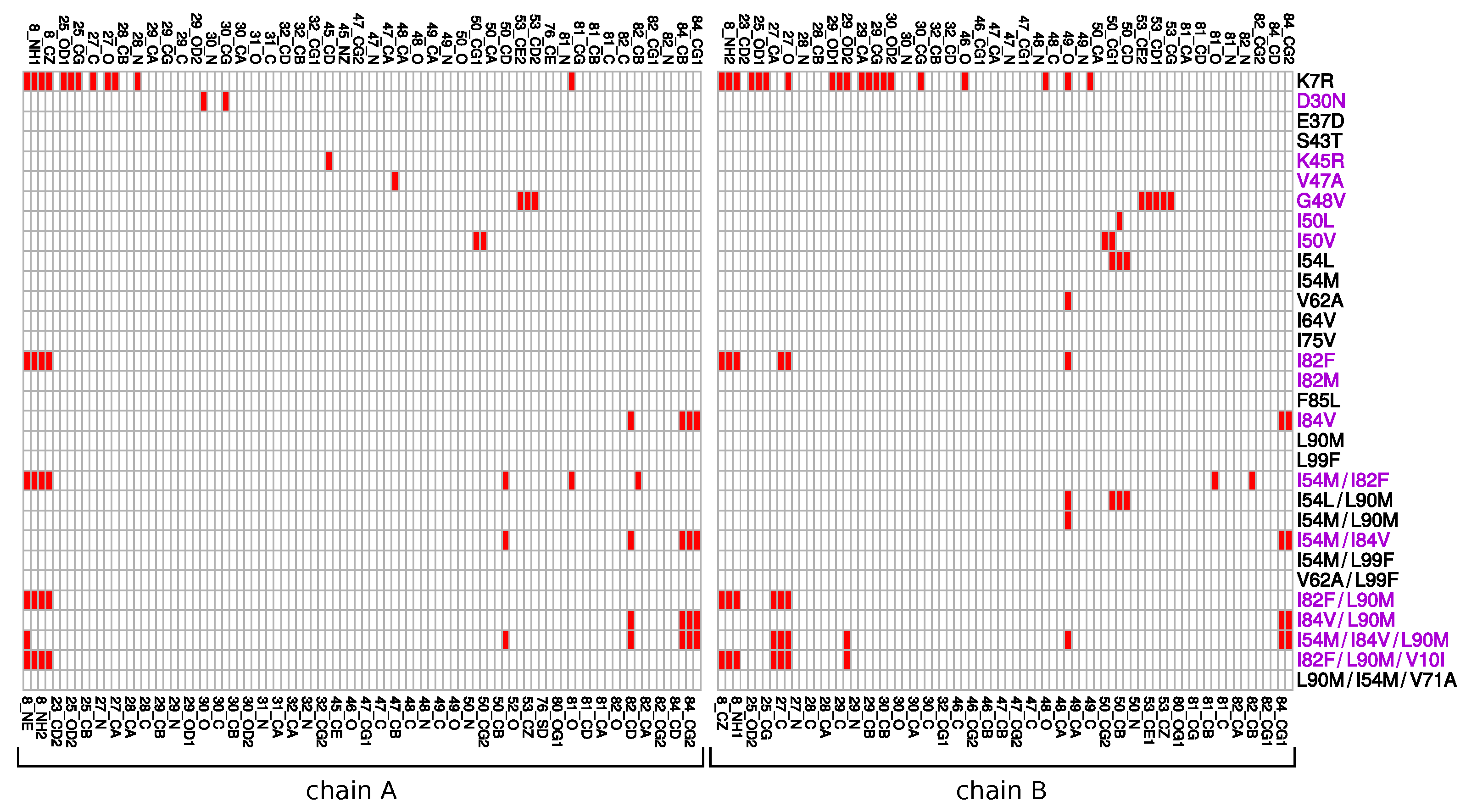
Appendix E
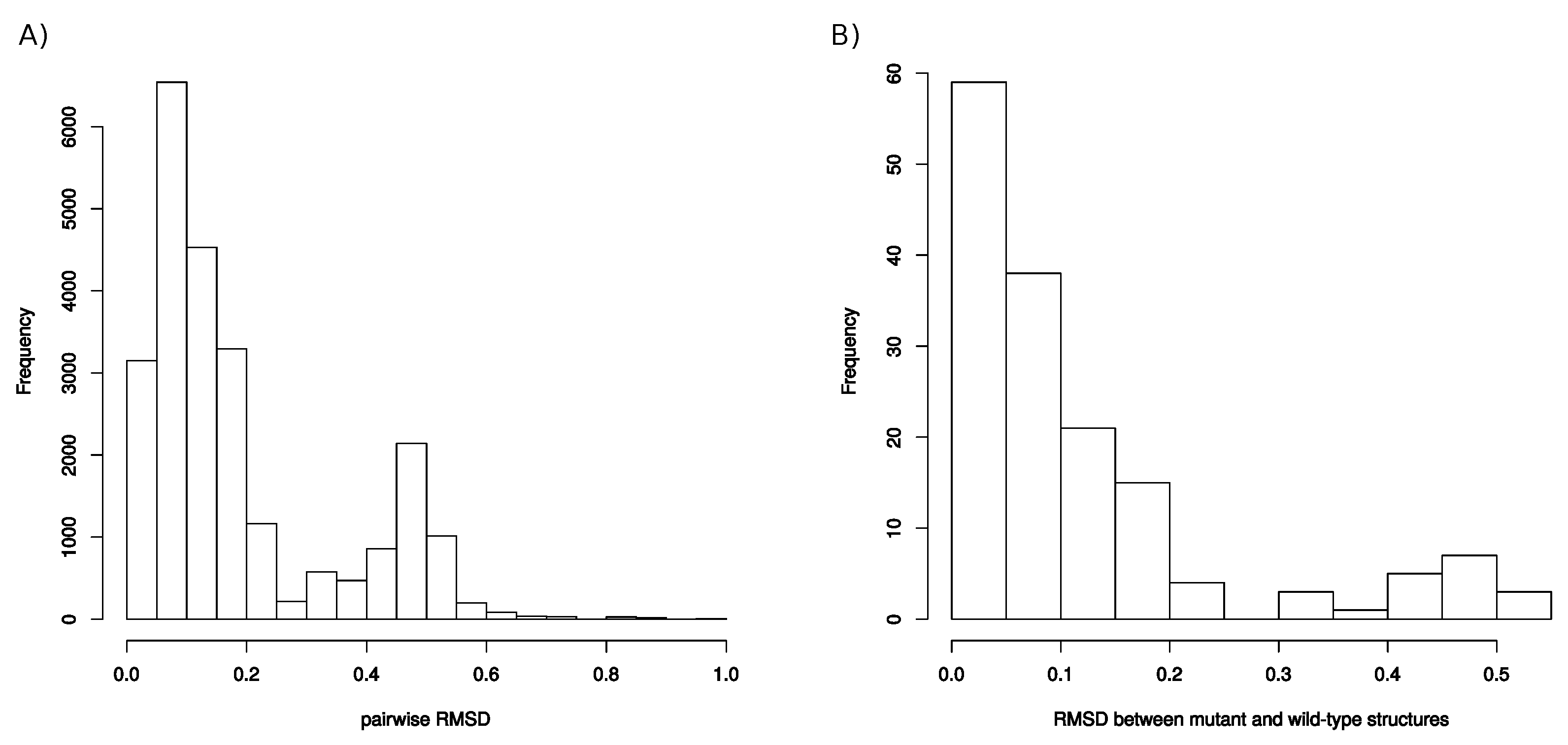
Appendix F
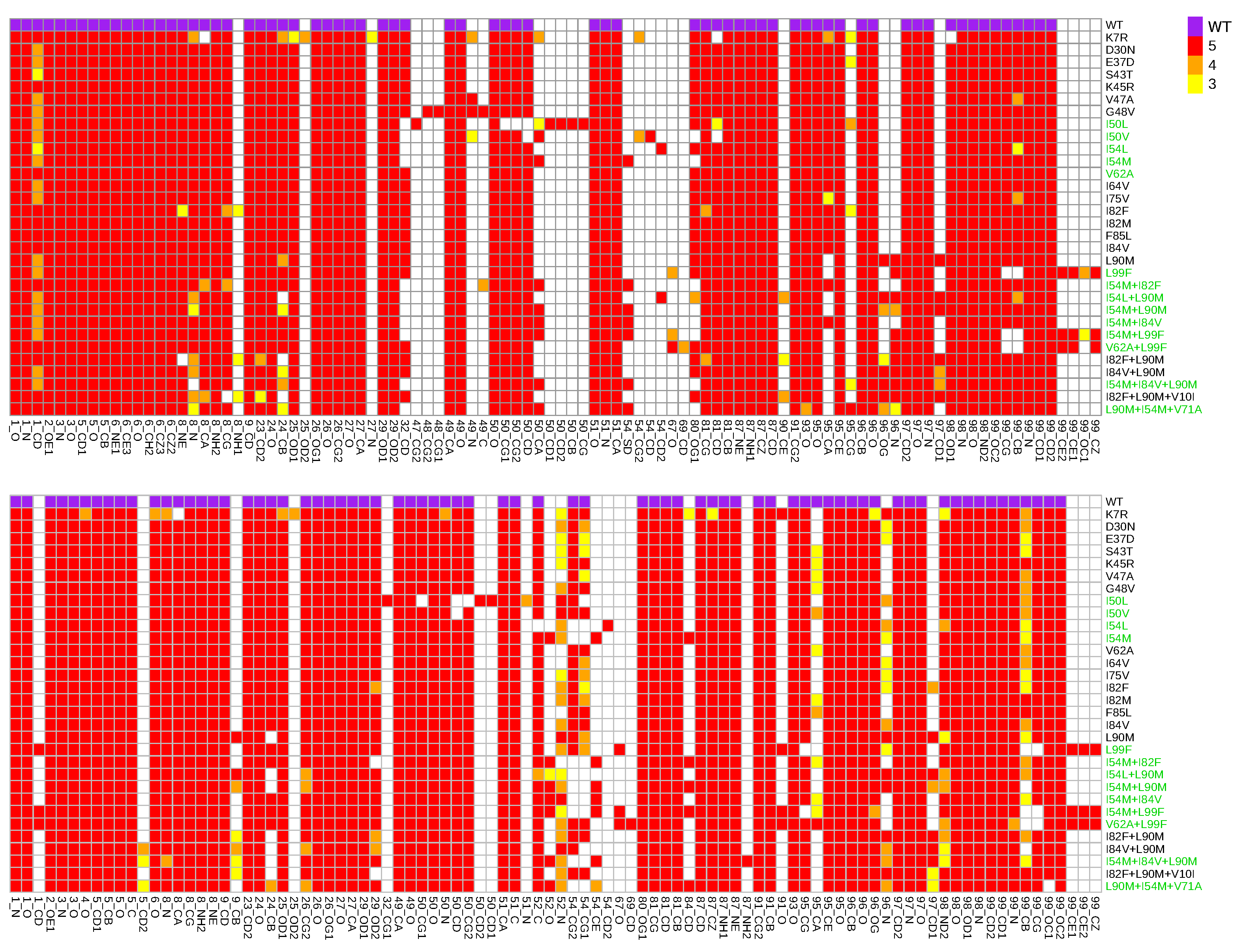
Appendix G
Appendix G.1. Protocol to Model Mutant Structures Using Robetta Software
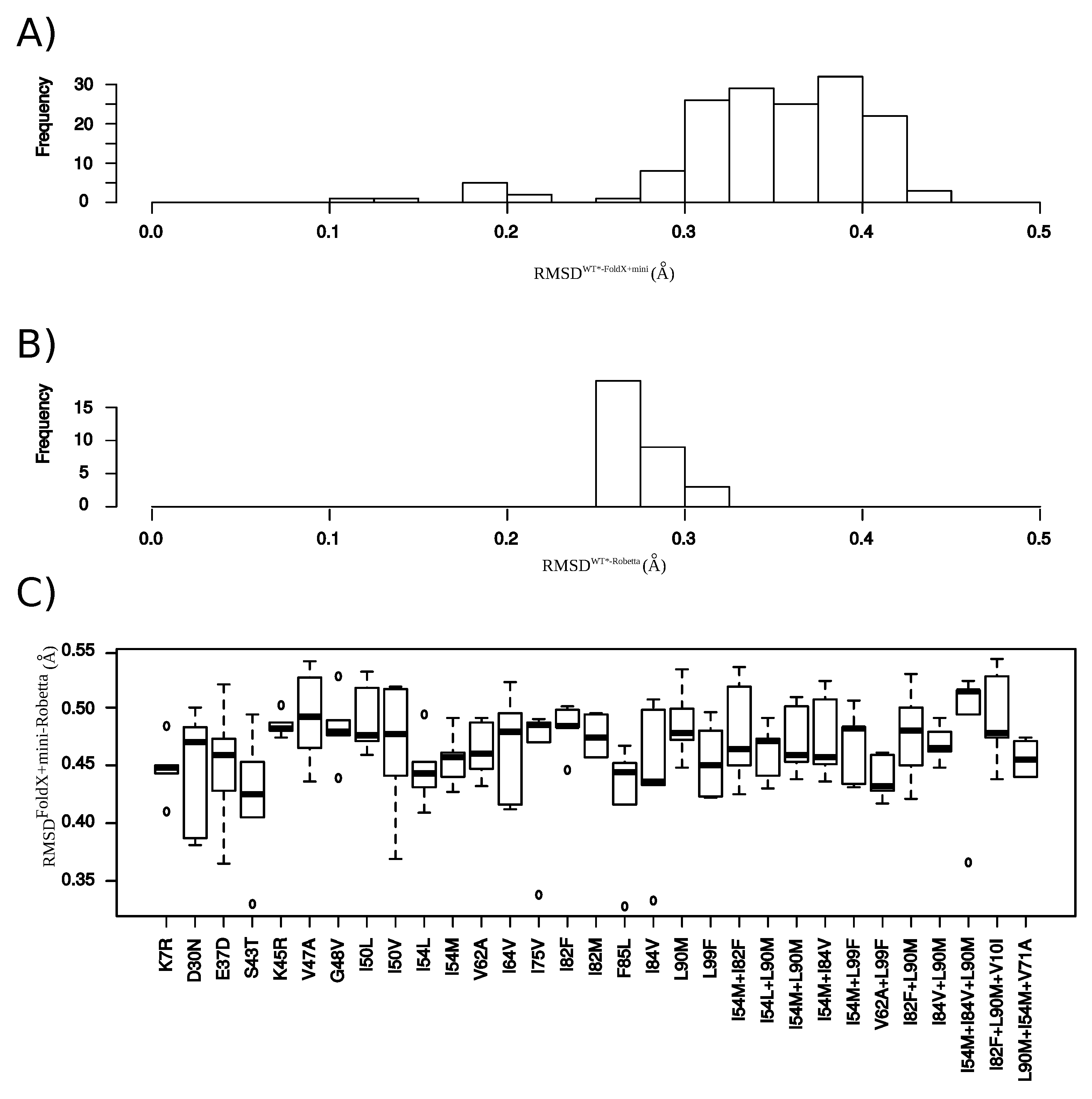
Appendix G.2. Structural Comparison between the and Structures
Appendix G.3. Detection of Structural Rearrangements in the Set of Structures
Appendix H
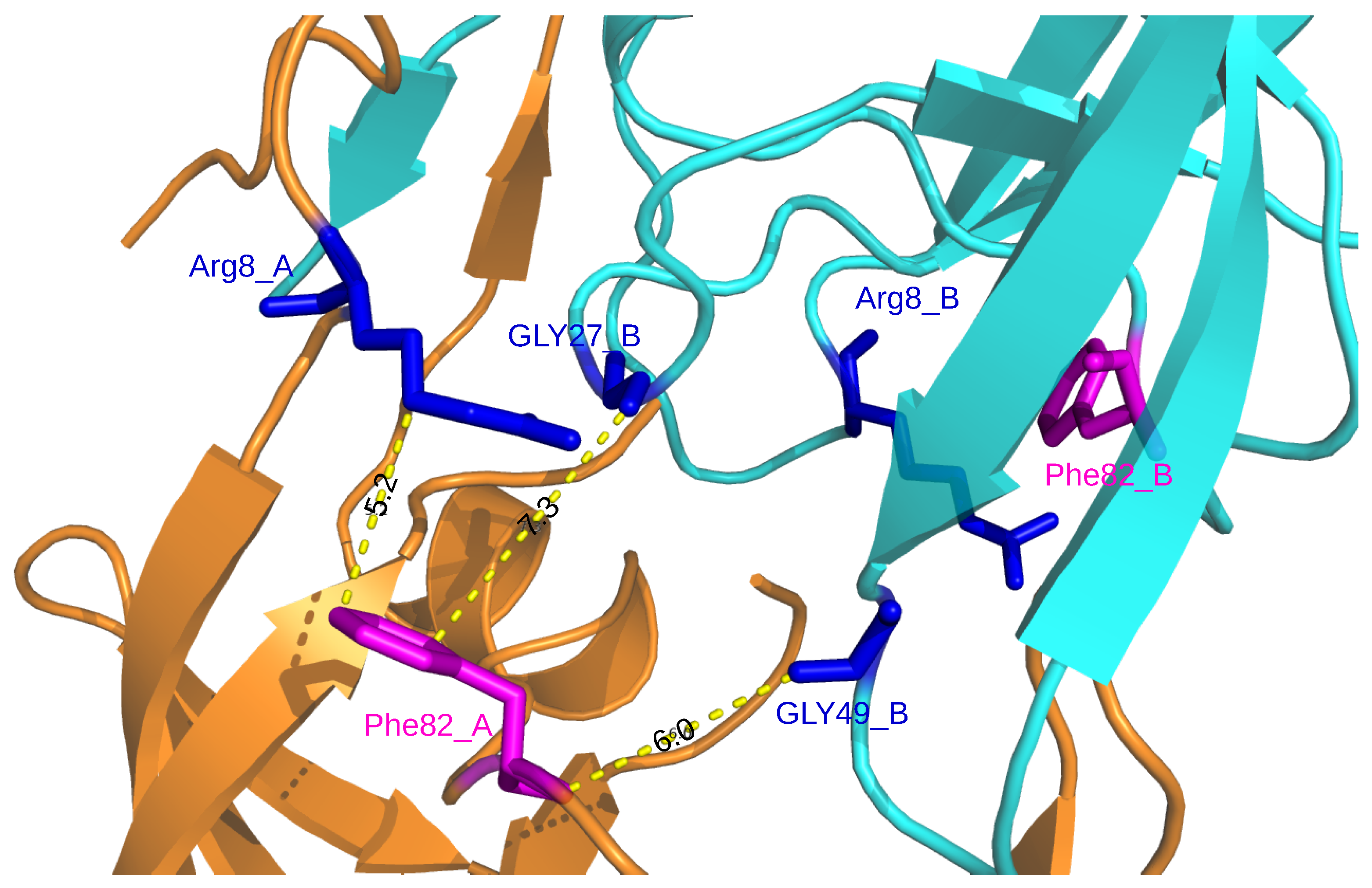
References
- Brower, E.; Bacha, U.M.; Kawasaki, Y.; Freire, E. Inhibition of HIV-2 protease by HIV-1 protease inhibitors in clinical use. Chem. Biol. Drug. Des. 2008, 71, 298–305. [Google Scholar] [CrossRef]
- Raugi, D.; Smith, R.; Ba, S.; Toure, M.; Traore, F.; Sall, F.; Pan, C.; Blankenship, L.; Montano, A.; Olson, J.; et al. Complex patterns of protease inhibitor resistance among antiretroviral treatment-experienced HIV-2 patients from senegal: Implications for second-line therapy. Antimicrob. Agents Chemother. 2013, 57, 2751–2760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raugi, D.N.; Smith, R.A.; Gottlieb, G.S.; the University of Washington-Dakar HIV-2 Study Group. Four Amino Acid Changes in HIV-2 Protease Confer Class-Wide Sensitivity to Protease Inhibitors. J. Virol. 2016, 90, 1062–1069. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Desbois, D.; Roquebert, B.; Peytavin, G.; Damond, F.; Collin, G.; Bénard, A.; Campa, P.; Matheron, S.; Chêne, G.; Brun-Vézinet, F.; et al. In vitro phenotypic susceptibility of human immunodeficiency virus type 2 clinical isolates to protease inhibitors. Antimicrob. Agents Chemother. 2008, 52, 1545–1548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masse, S.; Lu, X.; Dekhtyar, T.; Lu, L.; Koev, G.; Gao, F.; Mo, H.; Kempf, D.; Bernstein, B.; Hanna, G.; et al. In vitro selection and characterization of human immunodeficiency virus type 2 with decreased susceptibility to lopinavir. Antimicrob. Agents Chemother. 2007, 51, 3075–3080. [Google Scholar] [CrossRef] [Green Version]
- Cavaco-Silva, J.; Aleixo, M.; Van Laethem, K.; Faria, D.; Valadas, E.; Gonçalves, M.D.F.; Gomes, P.; Vandamme, A.; Cunha, C.; Camacho, R.J. Mutations selected in HIV-2-infected patients failing a regimen including atazanavir. Antimicrob. Agents Chemother. 2013, 68, 190–192. [Google Scholar] [CrossRef] [Green Version]
- Bénard, A.; Damond, F.; Campa, P.; Peytavin, G.; Descamps, D.; Lascoux-Combes, C.; Taieb, A.; Simon, F.; Autran, B.; Brun-Vézinet, F.; et al. Good response to lopinavir/ritonavir-containing antiretroviral regimens in antiretroviral-naive HIV-2-infected patients. AIDS 2009, 23, 1171–1183. [Google Scholar] [CrossRef] [PubMed]
- Rodés, B.; Sheldon, J.; Toro, C.; Jiménez, V.; Alvarez, M.; Soriano, V. Susceptibility to protease inhibitors in HIV-2 primary isolates from patients failing antiretroviral therapy. Antimicrob. Agents Chemother. 2006, 57, 709–713. [Google Scholar] [CrossRef]
- Ntemgwa, M.; Toni, T.; Brenner, B.; Oliveira, M.; Asahchop, E.; Moisi, D.; Wainberg, M. Nucleoside and nucleotide analogs select in culture for different patterns of drug resistance in human immunodeficiency virus types 1 and 2. Antimicrob. Agents Chemother. 2009, 53, 708–715. [Google Scholar] [CrossRef] [Green Version]
- Menéndez-Arias, L.; Alvarez, M. Antiretroviral therapy and drug resistance in human immunodeficiency virus type 2 infection. Antivir. Res. 2014, 102, 70–86. [Google Scholar] [CrossRef]
- Triki, D.; Cano Contreras, M.; Flatters, D.; Visseaux, B.; Descamps, D.; Camproux, A.; Regad, L. Analysis of the HIV-2 protease’s adaptation to various ligands: Characterization of backbone asymmetry using a structural alphabet. Sci. Rep. 2018, 8, 710. [Google Scholar] [CrossRef] [Green Version]
- Ollitrault, G.; Fartek, S.; Descamps, D.; Camproux, A.; Visseaux, B.; Regad, L. Characterization of HIV-2 protease structure by studying its asymmetry at the different levels of protein description. Symmetry 2019, 10, 644. [Google Scholar] [CrossRef] [Green Version]
- Triki, D.; Kermarrec, M.; Visseaux, B.; Descamps, D.; Flatters, D.; Camproux, A.; Regad, L. Exploration of the effects of sequence variations between HIV-1 and HIV-2 proteases on their three-dimensional structures. J. Biomol. Struct. Dyn. 2020, 38, 5014–5026. [Google Scholar] [CrossRef] [PubMed]
- Gustchina, A.; Weber, I. Comparative analysis of the sequences and structures of HIV-1 and HIV-2 proteases. Proteins Struct. Funct. Bioinform. 1991, 10, 325–339. [Google Scholar] [CrossRef] [PubMed]
- Sardana, V.; Schlabach, A.; Graham, P.; Bush, B.; Condra, J.; Culberson, J.; Gotlib, L.; Graham, D.E.A. Human Immunodeficiency Virus Type 1 Protease Inhibitors: Evaluation of Resistance Engendered by Amino Acid Substitutions in the Enzyme’s Substrate Binding Site. Biochemistry 1994, 33, 2004–2010. [Google Scholar] [CrossRef]
- Hoog, S.; Towler, E.; Zhao, B.; Doyle, M.; Debouck, C.; Abdel-Meguid, S. Human immunodeficiency virus protease ligand specificity conferred by residues outside of the active site cavity. Biochemistry 1996, 35, 10279–10286. [Google Scholar] [CrossRef]
- Tie, Y.; Boross, P.; Wang, Y.; Gaddis, L.; Hussain, A.; Leshchenko, S.; Ghosh, A.; Louis, J.; Harrison, R.; Weber, I. High resolution crystal structures of HIV-1 protease with a potent non-peptide inhibitor (UIC-94017) active against multi-drug-resistant clinical strains. J. Mol. Biol. 2004, 23, 341–352. [Google Scholar] [CrossRef]
- Kovalevsky, A.; Louis, J.; Aniana, A.; Ghosh, A.; Weber, I. Structural evidence for effectiveness of darunavir and two related antiviral inhibitors against HIV-2 protease. J. Mol. Biol. 2008, 384, 178–192. [Google Scholar] [CrossRef] [Green Version]
- Triki, D.; Billot, T.; Flatters, D.; Visseaux, B.; Descamps, D.; Camproux, A.; Regad, L. Exploration of the effect of sequence variations located inside the binding pocket of HIV-1 and HIV-2 proteases. Sci. Rep. 2018, 8, 5789. [Google Scholar] [CrossRef] [Green Version]
- Hightower, M.; Kallas, E.G. Diagnosis, antiretroviral therapy, and emergence of resistance to antiretroviral agents in HIV-2 infection: A review. Braz. J. Infect. Dis. 2003, 7, 7–15. [Google Scholar] [CrossRef] [Green Version]
- Kar, P.; Knecht, V.J. Origin of decrease in potency of darunavir and two related antiviral inhibitors against HIV-2 compared to HIV-1 protease. Phys. Chem. B 2012, 116, 2605–2614. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Liang, Z.; Wang, W.; Yi, C.; Zhang, S.; Zhang, Q. Revealing origin of decrease in potency of darunavir and amprenavir against HIV-2 relative to HIV-1 protease by molecular dynamics simulations. Sci. Rep. 2014, 4, 6872. [Google Scholar] [CrossRef] [Green Version]
- Colson, P.; Henry, M.; Tourres, C.; Lozachmeur, D.; Gallais, H.; Gastaut, J.; Moreau, J.; Tamalet, C. Polymorphism and drug-selected mutations in the protease gene of human immunodeficiency virus type 2 from patients living in southern France. J. Clin. Microbiol. 2004, 42, 570–577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Storto, A.; Visseaux, B.; Bertine, M.; Le Hingrat, Q.; Collin, G.; Damond, F.; Khuong, M.; Blum, L.; Tubiana, R.; Karmochkine, M.; et al. ANRS HIV-2 CO5 cohort. Minority resistant variants are also present in hiv-2-infected antiretroviral-naive patients. J. Antimicrob. Chemother. 2018, 73, 1173–1176. [Google Scholar] [CrossRef] [PubMed]
- Damond, F.; Brun-Vezinet, F.; Matheron, S.; Peytavin, G.; Campa, P.; Pueyo, S.; Mammano, F.; Lastere, S.; Farfara, I.; Simon, F.; et al. Polymorphism of the human immunodeficiency virus type 2 (hiv-2) protease gene and selection of drug resistance mutations in hiv-2-infected patients treated with protease inhibitors. J. Clin. Microbiol. 2005, 43, 484–487. [Google Scholar] [CrossRef] [Green Version]
- Ntemgwa, M.; Brenner, B.; Oliveira, M.; Moisi, D.; Wainberg, M. Variations in reverse transcriptase and RNase H domain mutations in human immunodeficiency virus type 1 clinical isolates are associated with divergent phenotypic resistance to zidovudine. Antimicrob. Agents Chemother. 2007, 51, 604–610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larrouy, L.; Vivot, A.; Charpentier, C.; Bénard, A.; Visseaux, B.; Damond, F.; Matheron, S.; Chene, G.; Brun-Vezinet, F.; Descamps, D. ANRS CO5 HIV-2 Cohort. Impact of gag genetic determinants on virological outcome to boosted lopinavir-containing regimen in hiv-2-infected patients. AIDS 2013, 27, 69–80. [Google Scholar] [CrossRef]
- Jallow, S.; Alabi, A.; Sarge-Njie, R.; Peterson, K.; Whittle, H.; Corrah, T.; Jaye, A.; Cotten, M.; Vanham, G.; McConkey, S.; et al. Virological response to highly active antiretroviral therapy in patients infected with human immunodeficiency virus type 2 (hiv-2) and in patients dually infected with hiv-1 and hiv-2 in the Gambia and emergence of drug-resistant variants. J. Clin. Microbiol. 2009, 47, 2200–2208. [Google Scholar] [CrossRef] [Green Version]
- Mahdi, M.; Szojka, Z.; Mótyán, J.; Tözsér, J. Inhibition Profiling of Retroviral Protease Inhibitors Using an HIV-2 Modular System. Viruses 2015, 71, 6152–6162. [Google Scholar] [CrossRef] [Green Version]
- Charpentier, C.; Visseaux, B.; Bénard, A.; Peytavin, G.; Damond, F.; Roy, C.; Taieb, A.; Chêne, G.; Matheron, S.; Brun-Vézinet, F.; et al. Transmitted drug resistance in French HIV-2-infected patients. AIDS 2013, 27, 1671–1684. [Google Scholar] [CrossRef]
- Laville, P.; Fartek, S.; Cerisier, N.; Flatters, D.; Petitjean, M.; Regad, L. Impacts of drug resistance mutations on the structural asymmetry of the HIV-2 protease. BMC Mol. Cell Biol. 2020, 51, 46. [Google Scholar] [CrossRef] [PubMed]
- Guerois, R.; Nielsen, J.E.; Serrano, L. Predicting changes in the stability of proteins and protein complexes: A study of more than 1000 mutations. J. Mol. Biol. 2002, 320, 369–387. [Google Scholar] [CrossRef]
- Tie, Y.; Wang, Y.; Boross, P.; Chiu, T.; Ghosh, A.; Tozser, J.; Louis, J.; Harrison, R.; Weber, I. Critical differences in HIV-1 and HIV-2 protease specificity for clinical inhibitors. Protein Sci. 2012, 21, 339–350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sadiq, S.K.; de Fabritiis, G. Explicit solvent dynamics and energetics of HIV-1 protease flap opening and closing. Proteins Struct. Funct. Bioinform. 2010, 78, 2873–2885. [Google Scholar] [CrossRef] [PubMed]
- Triki, D.; Fartek, S.; Visseaux, B.; Descamps, D.; Camproux, A.; Regad, L. Characterizing the structural variability of HIV-2 protease upon the binding of diverse ligands using a structural alphabet approach. J. Biomol. Struct. Dyn. 2018, 28, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Kovalevsky, A.; Tie, Y.; Ghosh, A.; Harrison, R.; Weber, I. Effect of flap mutations on structure of HIV-1 protease and inhibition by saquinavir and darunavir. J. Mol. Biol. 2008, 381, 102–115. [Google Scholar] [CrossRef]
- Hubbard, S.; Thornton, J. NACCESS. In Computer Program, Department of Biochemistry and Molecular Biology; University College London: London, UK, 1993. [Google Scholar]
- Koh, Y.; Nakata, H.; Maeda, K.; Ogata, H.; Bilcer, G.; Devasamudram, T.; Kincaid, J.; Boross, P.; Wang, Y.F.; Tie, Y.; et al. Novel bis-tetrahydrofuranylurethane-containing nonpeptidic protease inhibitor (PI) UIC-94017 (TMC114) with potent activity against multi-PI-resistant human immunodeficiency virus in vitro. Antimicrob. Agents Chemother. 2003, 47, 3123–3129. [Google Scholar] [CrossRef] [Green Version]
- Ghosh, A.K.; Dawson, Z.L.; Mitsuya, H. Darunavir, a conceptually new HIV-1 protease inhi-bitor for the treatment of drug-resistant HIV. Bioorg. Med. Chem. 2007, 15, 7576–7580. [Google Scholar] [CrossRef] [Green Version]
- Tong, L.; Pav, S.; Pargellis, C.; Dô, F.; Lamarre, D.; Anderson, P. Crystal structure of human immunodeficiency virus (HIV) type 2 protease in complex with a reduced amide inhibitor and comparison with HIV-1 protease structures. Proc. Natl. Acad. Sci. USA 1993, 90, 8387–8391. [Google Scholar] [CrossRef] [Green Version]
- Mulichak, A.M.; Hui, J.; Tomasselli, A.; Heinrikson, R.; Curry, K.; Tomich, C.; Thaisrivongs, S.; Sawyer, T.; Watenpaugh, K. The crystallographic structure of the protease from human immunodeficiency virus type 2 with two synthetic peptidic transition state analog inhibitors. J. Biol. Chem. 1993, 268, 13103–13109. [Google Scholar] [CrossRef]
- Tong, L.; Pav, S.; Mui, S.; Lamarre, D.; Yoakim, C.; Beaulieu, P.; Anderson, P. Crystal structures of HIV-2 protease in complex with inhibitors containing the hydroxyethylamine dipeptide isostere. Structure 1995, 3, 33–40. [Google Scholar] [CrossRef] [Green Version]
- Priestle, J.; Fassler, A.; Rösel, J.; Tintelnot-Blomley, M.; Strop, P.; Grütter, M. Comparative analysis of the X-ray structures of HIV-1 and HIV-2 proteases in complex with CGP 53820, a novel pseudosymmetric inhibitor. Structure 1995, 3, 381–389. [Google Scholar] [CrossRef] [Green Version]
- Kovalevsky, A.; Liu, F.; Leshchenko, S.; Ghosh, A.K.; Louis, J.; Harrison, R.; Weber, I. Ultra-high resolutioncrystal structure of HIV-1 protease mutant reveals two binding sites forclinical inhibitor TMC114. J. Mol. Biol. 2006, 363, 161–173. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Wang, Y.; Shen, C.H.; Agniswamy, J.; Rao, K.; Xu, C.; Ghosh, A.; Harrison, R.; Weber, I. Novel P2 tris-tetrahydrofuran group in antiviral compound 1 (GRL-0519) fills the S2 binding pocket of selected mutants of HIV-1protease. J. Med. Chem. 2013, 56, 1074–1083. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, C.; Wang, Y.; Kovalevsky, A.; Harrison, R.; Weber, I. Amprenavir complexes with HIV-1 protease andits drug-resistant mutants altering hydrophobic clusters. FEBS 2010, 277, 3699–3714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scott, W.; Schiffer, C. Curling of flap tips in HIV-1 protease as a mechanism for substrate entry and tolerance of drug resistance. Structure 2000, 8, 1259–1265. [Google Scholar] [CrossRef]
- Hong, L.; Zhang, X.; Hartsuck, J.; Tang, J. Crystal structure of an in vivo HIV-1 protease mutant in complex with saquinavir: Insights into the mechanisms of drug resistance. Protein Sci. 2000, 9, 1898–1904. [Google Scholar] [CrossRef] [Green Version]
- Agniswamy, J.; Louis, J.; Roche, J.; Harrison, R.; Weber, I. Structural Studies of a Rationally Selected Multi-Drug Resistant HIV-1 Protease Reveal Synergistic Effect of Distal Mutations on Flap Dynamics. PLoS ONE 2016, 11, e0168616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, Y.; Yilmaz, N.; Myint, W.; Ishima, R.; Schiffer, C. Differential Flap Dynamics in Wild-Type and a Drug Resistant Variant of HIV-1 Protease Revealed by Molecular Dynamics and NMR Relaxation. J. Chem. Theory Comput. 2012, 8, 3452–3462. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Chang, Y.C.E.; Louis, J.; Wang, Y.; Harrison, Y.; Weber, I. Structures of darunavir-resistant HIV-1 protease mutant reveal atypical binding of darunavir to wide open flaps. ACS Chem. Biol. 2014, 9, 1351–1358. [Google Scholar] [CrossRef]
- Logsdon, B.; Vickrey, J.; Martin, P.; Proteasa, G.; Koepke, J.; Terlecky, S.R.; Wawrzak, Z.; Winters, M.A.; Merigan, T.C.; Kovari, L.C. Crystal structures of a multidrug-resistant humanimmunodeficiency virus type 1 protease reveal an expanded active-sitecavity. J. Virol. 2004, 78, 3123–3132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, C.; Chang, Y.; Agniswamy, J.; Harrison, R.; Weber, I. Conformational variation of an extreme drug resistant mutant of HIV protease. J. Mol. Graph. Model. 2015, 62, 87–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muzammil, S.; Armstrong, A.; Kang, L.; Jakalian, A.; Bonneau, P.; Schmelmer, V.; Amzel, L.; Freire, E. Unique thermodynamic response of tipranavir to human immunodeficiency virus type 1 protease drug resistance mutations. J. Virol. 2007, 81, 5144–5154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yeddi, R.; Proteasa, G.; Martinez, J.; Vickrey, J.; Martin, P.; Wawrzak, Z.; Liu, Z.; Kovari, I.; Kovaria, L. Contribution of the 80’s loop of HIV-1 protease to the multidrug-resistant mechanism: Crystallographic study of MDR769 HIV-1 protease variants. Acta Crystallogr. D Biol. Crystallogr. 2011, 67, 524–532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, P.; Vickrey, J.; Proteasa, G.; Jimenez, Y.L.; Wawrzak, Z.; Winters, M.; Merigan, T.; Kovari, L. Wide-open 1.3 Å structure of a multidrug-resistant HIV-1 protease as a drug target. Structure 2005, 13, 1887–1895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kovalevsky, A.; Tie, Y.; Liu, F.; Boross, P.; Wang, Y.; Leshchenko, S.; Ghosh, A.; Harrison, R.; Weber, I.T. Effectiveness of Nonpeptide Clinical Inhibitor TMC-114 on HIV-1 Protease with Highly Drug Resistant Mutations D30N, I50V, and L90M. J. Med. Chem. 2006, 49, 1379–1387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prabu-jeyabalan, M.; Nalivaika, E.; King, N.; Schiffer, C. Viability of a drug-resistant human immunodeficiency virus type 1 protease variant: Structural insights for better antiviral therapy. Society 2003, 77, 1306–1315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weber, I.; Agniswamy, J. HIV-1 Protease: Structural Perspectives on Drug Resistance. Viruses 2009, 1, 1110–1136. [Google Scholar] [CrossRef] [PubMed]
- Meher, B.; Wang, Y. Interaction of I50V mutant and I50L/A71V double mutant HIV-protease with inhibitor TMC114 (darunavir): Molecular dynamics simulation and binding free energy studies. J. Phys. Chem. 2012, 116, 1884–1900. [Google Scholar] [CrossRef] [PubMed]
- Mittal, S.; Bandaranayake, R.; King, N.; Prabu-Jeyabalan, M.; Nalam, M.; Nalivaika, E.; Yilmaz, N.; Schiffer, C. Structural and thermodynamic basis of amprenavir/darunavir and atazanavir resistance in HIV-1 protease with mutations at residue 50. J. Virol. 2013, 87, 4176–4184. [Google Scholar] [CrossRef] [Green Version]
- Lockbaum, G.; Leidner, F.; Rusere, L.; Henes, M.; Kosovrasti, K.; Nachum, G.; Nalivaika, E.; Ali, A.; Yilmaz, N.; Schiffer, C. Structural Adaptation of Darunavir Analogues against Primary Mutations in HIV-1 Protease. ACS Infect. Dis. 2019, 5, 316–325. [Google Scholar] [CrossRef] [PubMed]
- Mahalingam, B.; Louis, J.; Reed, C.; Adomat, J.; Krouse, J.; Wang, Y.; Harrison, R.; Weber, I. Structural and kinetic analysis of drug resistant mutants of HIV-1 protease. Eur. J. Biochem. 1999, 263, 238–245. [Google Scholar] [CrossRef] [PubMed]
- Mahalingam, B.; Wang, Y.; Boross, P.; Tozser, J.; Louis, J.; Harrison, R.; Weber, I. Crystal structures of HIV protease V82A and L90M mutants reveal changes in the indinavir-binding site. Eur. J. Biochem. 2004, 271, 1516–1524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, D.; Gulnik, S.; Gustchina, E.; Yu, B.; Shao, W.; Qoronfleh, W.; Nathan, A.; Erickson, J. Drug resistance mutations can effect dimer stability of HIV-1 protease at neutral pH. Protein Sci. 1999, 8, 1702–1707. [Google Scholar] [CrossRef]
- Abraham, M.; Murtolad, T.; Schulz, R.; Pálla, S.; Smith, J.; Hess, B.; Lindahl, E. Gromacs: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Robertson, A.; Jensen, J.H. Very fast empirical prediction and rationalization of protein pK a values. Proteins 2005, 61, 704–721. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An N log (N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef] [Green Version]
- Perrier, M.; Castain, L.; Regad, L.; Todesco, E.; Landman, R.; Visseaux, B.; Yazdanpanah, Y.; Rodriguez, C.; Joly, V.; Calvez, V.; et al. HIV-1 protease, Gag and gp41 baseline substitutions associated with virological response to a PI-based regimen. J. Antimicrob. Chemother. 2019, 74, 1679–1692. [Google Scholar] [CrossRef]
- The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC. Available online: https://pymol.org/2/ (accessed on 20 January 2021).
- Borrel, A.; Regad, L.; Xhaard, H.; Petitjean, M.; Camproux, A. PockDrug: A Model for Predicting Pocket Druggability That Overcomes Pocket Estimation Uncertainties. J. Chem. Inf. Model. 2015, 55, 882–895. [Google Scholar] [CrossRef]
- Cerisier, N.; Regad, L.; Triki, D.; Camproux, A.; Petitjean, M. Cavity versus ligand shape descriptors: Application to urokinase binding pockets. J. Comput. Biol. 2017, 24, 1134–1137. [Google Scholar] [CrossRef]
- Ozeel, V.; Perrier, A.; Vanet, A.; Petitjean, M. The Symmetric Difference Distance: A New Way to Evaluate the Evolution of Interfaces along Molecular Dynamics Trajectories; Application to Influenza Hemagglutinin. Symmetry 2019, 11, 662. [Google Scholar] [CrossRef] [Green Version]
- Laville, P.; Martin, J.; Launay, G.; Regad, L.; Camproux, A.; de Vries, S.; Petitjean, M. A non-parametric method to compute protein–protein and protein–ligands interfaces. Application to HIV-2 protease–inhibitors complexes. bioRxiv 2018. [Google Scholar] [CrossRef]
- Eppstein, D.; Paterson, M.; Yao, F. On nearest-neighbor graphs. Discret. Comput. Geom. 1997, 17, 263–282. [Google Scholar] [CrossRef] [Green Version]
- Song, Y.; DiMaio, F.; Wang, R.Y.R.; Kim, D.; Miles, C.; Brunette, T.; Thompson, J.; Baker, D. High-Resolution Comparative Modeling with RosettaCM. Structure 2013, 21, 1735–1742. [Google Scholar] [CrossRef] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Laville, P.; Petitjean, M.; Regad, L. Structural Impacts of Drug-Resistance Mutations Appearing in HIV-2 Protease. Molecules 2021, 26, 611. https://doi.org/10.3390/molecules26030611
Laville P, Petitjean M, Regad L. Structural Impacts of Drug-Resistance Mutations Appearing in HIV-2 Protease. Molecules. 2021; 26(3):611. https://doi.org/10.3390/molecules26030611
Chicago/Turabian StyleLaville, Pierre, Michel Petitjean, and Leslie Regad. 2021. "Structural Impacts of Drug-Resistance Mutations Appearing in HIV-2 Protease" Molecules 26, no. 3: 611. https://doi.org/10.3390/molecules26030611





