Structural and Energetic Characterization of the Denatured State from the Perspectives of Peptides, the Coil Library, and Intrinsically Disordered Proteins
Abstract
:1. Introduction
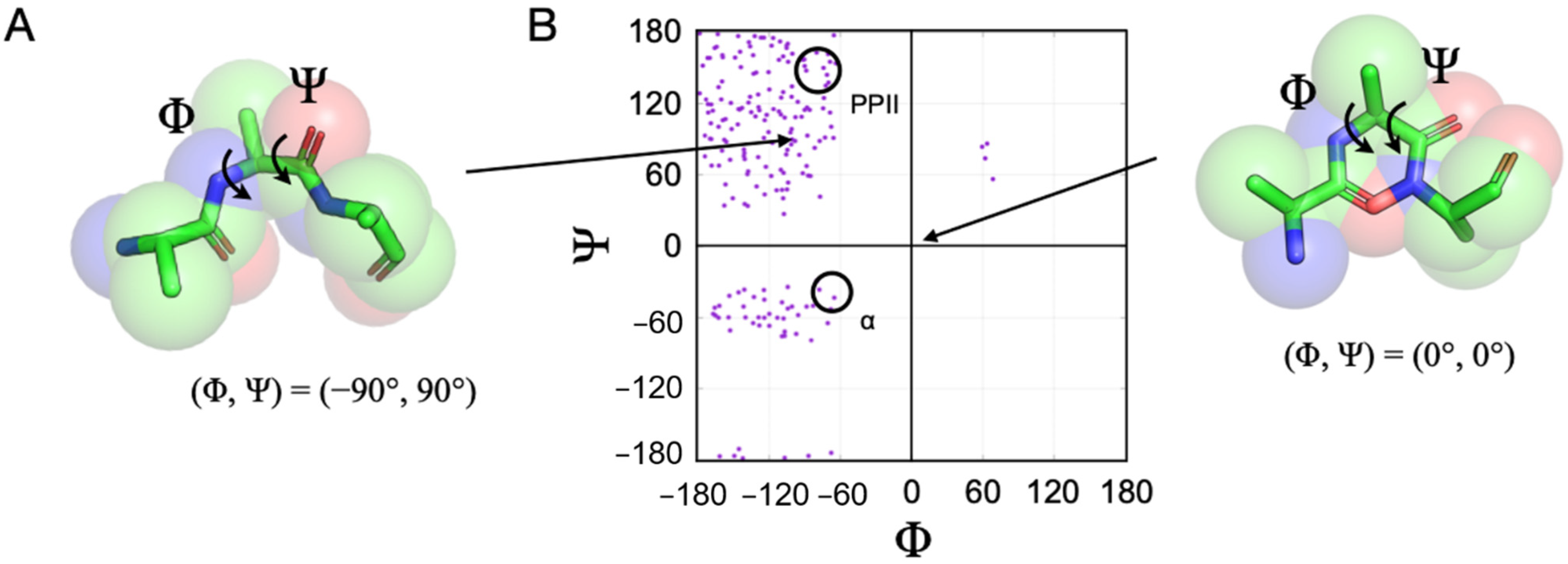
2. Peptide Models of the DSE
3. Protein Coil Library Model of the DSE
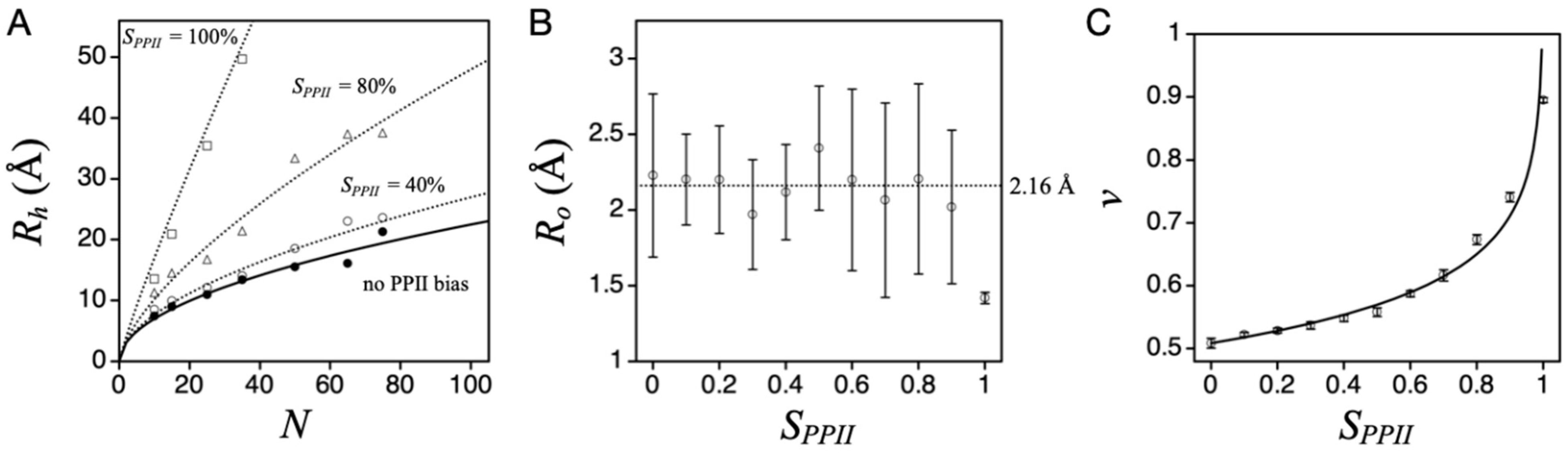
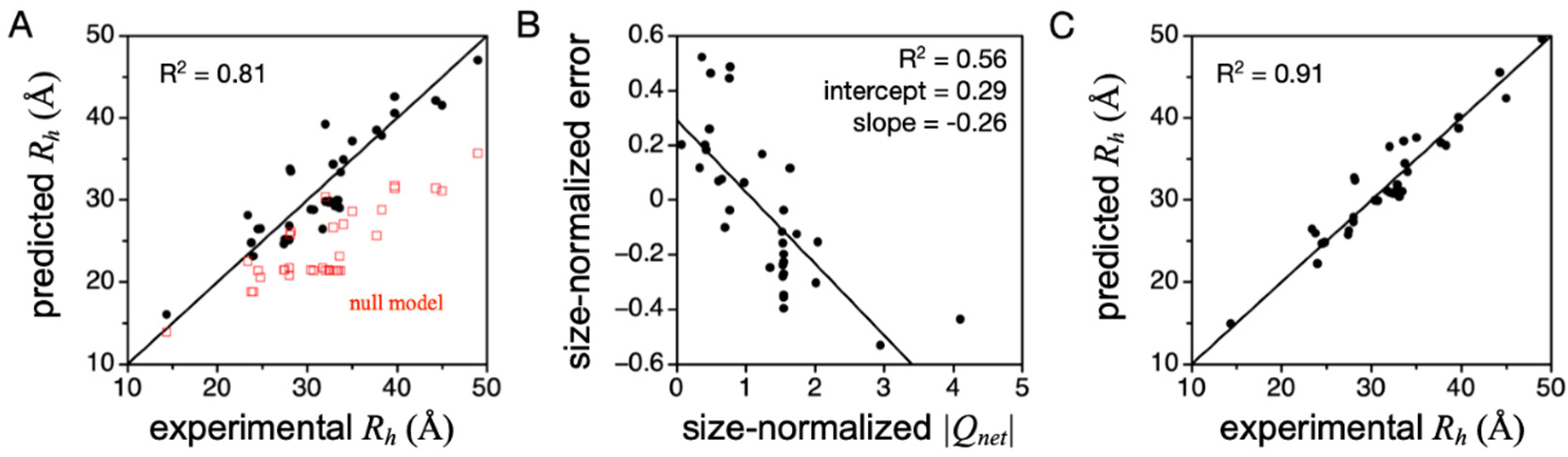
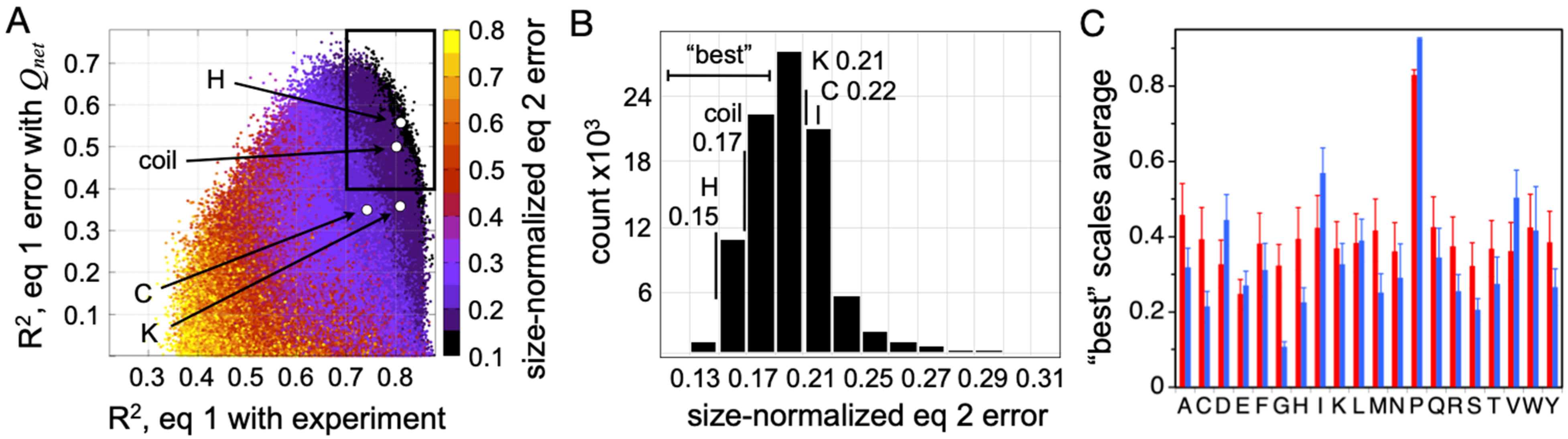
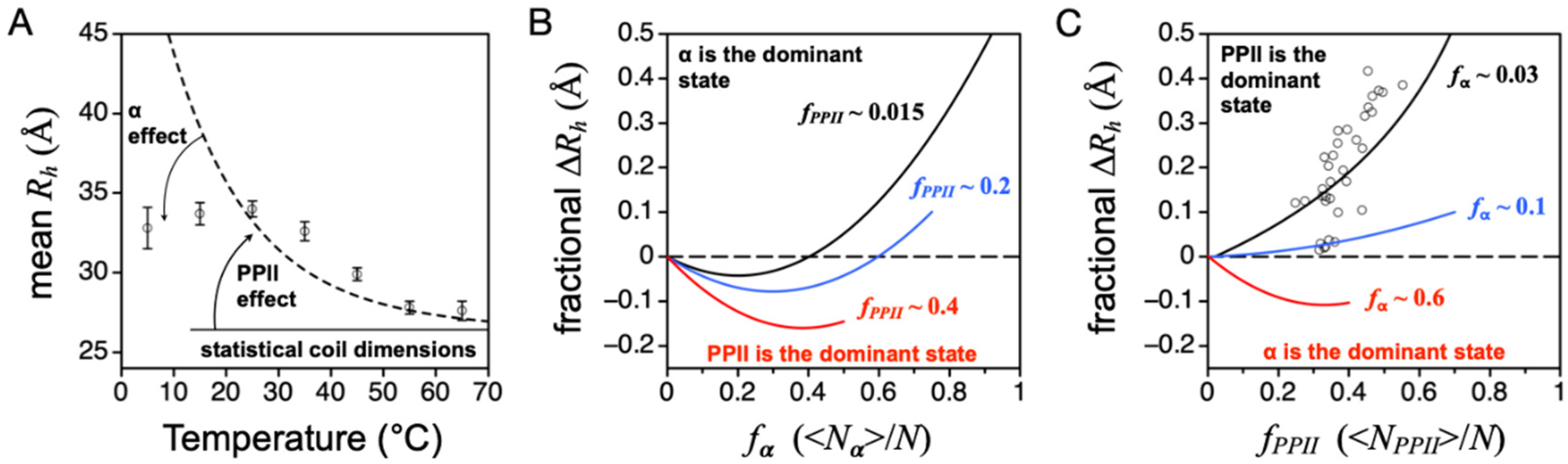
4. IDP Model of the DSE
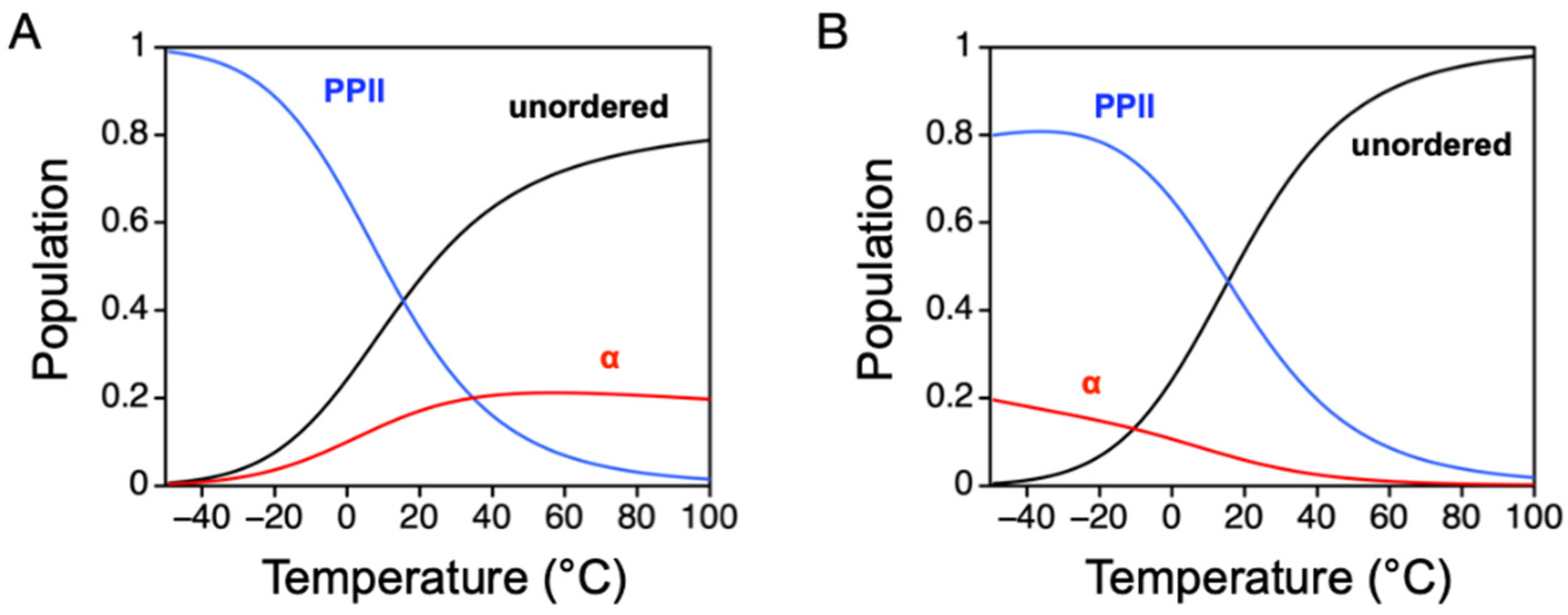
5. Temperature Dependence of Intrinsic α-Helix and PPII Propensities
6. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pace, C.N.; Hermans, J. The Stability of Globular Protein. CRC Crit. Rev. Biochem. 1975, 3, 1–43. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Xu, Y.; Gruebele, M. Temperature Dependence of Protein Folding Kinetics in Living Cells. Proc. Natl. Acad. Sci. USA 2012, 109, 17863–17867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCarney, E.R.; Kohn, J.E.; Plaxco, K.W. Is There or Isn’t There? The Case for (and Against) Residual Structure in Chemically Denatured Proteins. Crit. Rev. Biochem. Mol. Biol. 2005, 40, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.H.; Stites, W.E. Effects of Excluded Volume upon Protein Stability in Covalently Cross-Linked Proteins with Variable Linker Lengths. Biochemistry 2008, 47, 8804–8814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Serrano, L. Comparison between the Phi Distribution of the Amino Acids in the Protein Database and NMR Data Indicates That Amino Acids Have Various Phi Propensities in the Random Coil Conformation. J. Mol. Biol. 1995, 254, 322–333. [Google Scholar] [CrossRef]
- Manson, A.; Whitten, S.T.; Ferreon, J.C.; Fox, R.O.; Hilser, V.J. Characterizing the Role of Ensemble Modulation in Mutation-Induced Changes in Binding Affinity. J. Am. Chem. Soc. 2009, 131, 6785–6793. [Google Scholar] [CrossRef] [Green Version]
- Pan, H.; Lee, J.C.; Hilser, V.J. Binding Sites in Escherichia Coli Dihydrofolate Reductase Communicate by Modulating the Conformational Ensemble. Proc. Natl. Acad. Sci. USA 2000, 97, 12020–12025. [Google Scholar] [CrossRef] [Green Version]
- Wong, K.-B.; Clarke, J.; Bond, C.J.; Neira, J.L.; Freund, S.M.V.; Fersht, A.R.; Daggett, V. Towards a Complete Description of the Structural and Dynamic Properties of the Denatured State of Barnase and the Role of Residual Structure in Folding. J. Mol. Biol. 2000, 296, 1257–1282. [Google Scholar] [CrossRef]
- Kazmirski, S.L.; Wong, K.-B.; Freund, S.M.V.; Tan, Y.-J.; Fersht, A.R.; Daggett, V. Protein Folding from a Highly Disordered Denatured State: The Folding Pathway of Chymotrypsin Inhibitor 2 at Atomic Resolution. Proc. Natl. Acad. Sci. USA 2001, 98, 4349–4354. [Google Scholar] [CrossRef] [Green Version]
- Shortle, D. Staphylococcal Nuclease: A Showcase of m-Value Effects. In Advances in Protein Chemistry; Anfinsen, C.B., Richards, F.M., Edsall, J.T., Eisenberg, D.S., Eds.; Protein Stability; Academic Press: Cambridge, MA, USA, 1995; Volume 46, pp. 217–247. [Google Scholar]
- Shi, Z.; Olson, C.A.; Rose, G.D.; Baldwin, R.L.; Kallenbach, N.R. Polyproline II Structure in a Sequence of Seven Alanine Residues. Proc. Natl. Acad. Sci. USA 2002, 99, 9190–9195. [Google Scholar] [CrossRef] [Green Version]
- Rucker, A.L.; Pager, C.T.; Campbell, M.N.; Qualls, J.E.; Creamer, T.P. Host-Guest Scale of Left-Handed Polyproline II Helix Formation. Proteins 2003, 53, 68–75. [Google Scholar] [CrossRef]
- Ferreon, J.C.; Hilser, V.J. The Effect of the Polyproline II (PPII) Conformation on the Denatured State Entropy. Protein Sci. 2003, 12, 447–457. [Google Scholar] [CrossRef] [PubMed]
- Mezei, M.; Fleming, P.J.; Srinivasan, R.; Rose, G.D. Polyproline II Helix Is the Preferred Conformation for Unfolded Polyalanine in Water. Proteins Struct. Funct. Bioinform. 2004, 55, 502–507. [Google Scholar] [CrossRef] [PubMed]
- Pappu, R.V.; Rose, G.D. A simple model for polyproline II structure in unfolded states of alanine-based peptides. Protein Sci. 2002, 11, 2437–2455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, Z.; Chen, K.; Liu, Z.; Ng, A.; Bracken, W.C.; Kallenbach, N.R. Polyproline II Propensities from GGXGG Peptides Reveal an Anticorrelation with Beta-Sheet Scales. Proc. Natl. Acad. Sci. USA 2005, 102, 17964–17968. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elam, W.A.; Schrank, T.P.; Campagnolo, A.J.; Hilser, V.J. Evolutionary Conservation of the Polyproline II Conformation Surrounding Intrinsically Disordered Phosphorylation Sites. Protein Sci. 2013, 22, 405–417. [Google Scholar] [CrossRef] [Green Version]
- Eker, F.; Cao, X.; Nafie, L.; Schweitzer-Stenner, R. Tripeptides Adopt Stable Structures in Water. A Combined Polarized Visible Raman, FTIR, and VCD Spectroscopy Study. J. Am. Chem. Soc. 2002, 124, 14330–14341. [Google Scholar] [CrossRef]
- Eker, F.; Griebenow, K.; Schweitzer-Stenner, R. Stable Conformations of Tripeptides in Aqueous Solution Studied by UV Circular Dichroism Spectroscopy. J. Am. Chem. Soc. 2003, 125, 8178–8185. [Google Scholar] [CrossRef]
- Weise, C.F.; Weisshaar, J.C. Conformational Analysis of Alanine Dipeptide from Dipolar Couplings in a Water-Based Liquid Crystal. J. Phys. Chem. B 2003, 107, 3265–3277. [Google Scholar] [CrossRef]
- Hinderaker, M.P.; Raines, R.T. An Electronic Effect on Protein Structure. Protein Sci. 2003, 12, 1188–1194. [Google Scholar] [CrossRef]
- Chakrabartty, A.; Kortemme, T.; Baldwin, R.L. Helix Propensities of the Amino Acids Measured in Alanine-Based Peptides without Helix-Stabilizing Side-Chain Interactions. Protein Sci. 1994, 3, 843–852. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reiersen, H.; Clarke, A.R.; Rees, A.R. Short Elastin-like Peptides Exhibit the Same Temperature-Induced Structural Transitions as Elastin Polymers: Implications for Protein Engineering. J. Mol. Biol. 1998, 283, 255–264. [Google Scholar] [CrossRef] [PubMed]
- Minor, D.L.; Kim, P.S. Context Is a Major Determinant of β-Sheet Propensity. Nature 1994, 371, 264–267. [Google Scholar] [CrossRef] [PubMed]
- Dyson, H.J.; Bolinger, L.; Feher, V.A.; Osterhout, J.J., Jr.; Yao, J.; Wright, P.E. Sequence Requirements for Stabilization of a Peptide Reverse Turn in Water Solution. Eur. J. Biochem. 1998, 255, 462–471. [Google Scholar] [CrossRef] [PubMed]
- Hutchinson, E.G.; Thornton, J.M. A Revised Set of Potentials for Beta-Turn Formation in Proteins. Protein Sci. 1994, 3, 2207–2216. [Google Scholar] [CrossRef]
- Swindells, M.B.; MacArthur, M.W.; Thornton, J.M. Intrinsic φ,ψ Propensities of Amino Acids, Derived from the Coil Regions of Known Structures. Nat. Struct. Mol. Biol. 1995, 2. [Google Scholar] [CrossRef]
- Fitzkee, N.C.; Fleming, P.J.; Rose, G.D. The Protein Coil Library: A Structural Database of Nonhelix, Nonstrand Fragments Derived from the PDB. Proteins Struct. Funct. Bioinform. 2005, 58, 852–854. [Google Scholar] [CrossRef]
- Jha, A.K.; Colubri, A.; Zaman, M.H.; Koide, S.; Sosnick, T.R.; Freed, K.F. Helix, Sheet, and Polyproline II Frequencies and Strong Nearest Neighbor Effects in a Restricted Coil Library. Biochemistry 2005, 44, 9691–9702. [Google Scholar] [CrossRef] [Green Version]
- Perskie, L.L.; Street, T.O.; Rose, G.D. Structures, Basins, and Energies: A Deconstruction of the Protein Coil Library. Protein Sci. 2008, 17, 1151–1161. [Google Scholar] [CrossRef]
- Griffiths-Jones, S.R.; Sharman, G.J.; Maynard, A.J.; Searle, M.S. Modulation of Intrinsic Phi, Psi Propensities of Amino Acids by Neighbouring Residues in the Coil Regions of Protein Structures: NMR Analysis and Dissection of a Beta-Hairpin Peptide. J. Mol. Biol. 1998, 284, 1597–1609. [Google Scholar] [CrossRef]
- Smith, L.J.; Bolin, K.A.; Schwalbe, H.; MacArthur, M.W.; Thornton, J.M.; Dobson, C.M. Analysis of Main Chain Torsion Angles in Proteins: Prediction of NMR Coupling Constants for Native and Random Coil Conformations. J. Mol. Biol. 1996, 255, 494–506. [Google Scholar] [CrossRef] [PubMed]
- Mantsyzov, A.B.; Shen, Y.; Lee, J.H.; Hummer, G.; Bax, A. MERA: A Webserver for Evaluating Backbone Torsion Angle Distributions in Dynamic and Disordered Proteins from NMR Data. J. Biomol. NMR 2015, 63, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Roche, J.; Grishaev, A.; Bax, A. Prediction of Nearest Neighbor Effects on Backbone Torsion Angles and NMR Scalar Coupling Constants in Disordered Proteins. Protein Sci. 2018, 27, 146–158. [Google Scholar] [CrossRef] [PubMed]
- Eliezer, D. Biophysical Characterization of Intrinsically Disordered Proteins. Curr. Opin. Struct. Biol. 2009, 19, 23–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kohn, J.E.; Millett, I.S.; Jacob, J.; Zagrovic, B.; Dillon, T.M.; Cingel, N.; Dothager, R.S.; Seifert, S.; Thiyagarajan, P.; Sosnick, T.R.; et al. Random-Coil Behavior and the Dimensions of Chemically Unfolded Proteins. Proc. Natl. Acad. Sci. USA 2004, 101, 12491–12496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- English, L.R.; Voss, S.M.; Tilton, E.C.; Paiz, E.A.; So, S.; Parra, G.L.; Whitten, S.T. Impact of Heat on Coil Hydrodynamic Size Yields the Energetics of Denatured State Conformational Bias. J. Phys. Chem. B 2019, 123, 10014–10024. [Google Scholar] [CrossRef]
- English, L.R.; Tilton, E.C.; Ricard, B.J.; Whitten, S.T. Intrinsic α Helix Propensities Compact Hydrodynamic Radii in Intrinsically Disordered Proteins. Proteins 2017, 85, 296–311. [Google Scholar] [CrossRef] [Green Version]
- Kjaergaard, M.; Nørholm, A.-B.; Hendus-Altenburger, R.; Pedersen, S.F.; Poulsen, F.M.; Kragelund, B.B. Temperature-Dependent Structural Changes in Intrinsically Disordered Proteins: Formation of α-Helices or Loss of Polyproline II? Protein Sci. 2010, 19, 1555–1564. [Google Scholar] [CrossRef]
- Wuttke, R.; Hofmann, H.; Nettels, D.; Borgia, M.B.; Mittal, J.; Best, R.B.; Schuler, B. Temperature-Dependent Solvation Modulates the Dimensions of Disordered Proteins. Proc. Natl. Acad. Sci. USA 2014, 111, 5213–5218. [Google Scholar] [CrossRef] [Green Version]
- Langridge, T.D.; Tarver, M.J.; Whitten, S.T. Temperature Effects on the Hydrodynamic Radius of the Intrinsically Disordered N-Terminal Region of the P53 Protein. Proteins 2014, 82, 668–678. [Google Scholar] [CrossRef]
- English, L.R.; Tischer, A.; Demeler, A.K.; Demeler, B.; Whitten, S.T. Sequence Reversal Prevents Chain Collapse and Yields Heat-Sensitive Intrinsic Disorder. Biophys. J. 2018, 115, 328–340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cowan, P.M.; McGAVIN, S. Structure of Poly-L-Proline. Nature 1955, 176, 501–503. [Google Scholar] [CrossRef]
- Perez, R.B.; Tischer, A.; Auton, M.; Whitten, S.T. Alanine and Proline Content Modulate Global Sensitivity to Discrete Perturbations in Disordered Proteins. Proteins 2014, 82, 3373–3384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tomasso, M.E.; Tarver, M.J.; Devarajan, D.; Whitten, S.T. Hydrodynamic Radii of Intrinsically Disordered Proteins Determined from Experimental Polyproline II Propensities. PLoS Comput. Biol. 2016, 12, e1004686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, K.; Liu, Z.; Kallenbach, N.R. The Polyproline II Conformation in Short Alanine Peptides Is Noncooperative. Proc. Natl. Acad. Sci. USA 2004, 101, 15352–15357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramachandran, G.N.; Ramakrishnan, C.; Sasisekharan, V. Stereochemistry of Polypeptide Chain Configurations. J. Mol. Biol. 1963, 7, 95–99. [Google Scholar] [CrossRef]
- Richards, F.M. Areas, Volumes, Packing and Protein Structure. Annu. Rev. Biophys. Bioeng. 1977, 6, 151–176. [Google Scholar] [CrossRef]
- Whitten, S.T.; Yang, H.-W.; Fox, R.O.; Hilser, V.J. Exploring the Impact of Polyproline II (PII) Conformational Bias on the Binding of Peptides to the SEM-5 SH3 Domain. Protein Sci. 2008, 17, 1200–1211. [Google Scholar] [CrossRef] [Green Version]
- Levinthal, C. How to Fold Graciously. Mössbaun Spectrosc. Biol. Syst. Proc. 1969, 67, 22–24. [Google Scholar]
- Brooks, C.L.; Gruebele, M.; Onuchic, J.N.; Wolynes, P.G. Chemical Physics of Protein Folding. Proc. Natl. Acad. Sci. USA 1998, 95, 11037–11038. [Google Scholar] [CrossRef] [Green Version]
- Craig, P.O.; Lätzer, J.; Weinkam, P.; Hoffman, R.M.B.; Ferreiro, D.U.; Komives, E.A.; Wolynes, P.G. Prediction of Native-State Hydrogen Exchange from Perfectly Funneled Energy Landscapes. J. Am. Chem. Soc. 2011, 133, 17463–17472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bai, Y.; Englander, S.W. Future Directions in Folding: The Multi-State Nature of Protein Structure. Proteins 1996, 24, 145–151. [Google Scholar] [CrossRef]
- Englander, S.W.; Mayne, L. The Case for Defined Protein Folding Pathways. Proc. Natl. Acad. Sci. USA 2017, 114, 8253–8258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tiffany, M.L.; Krimm, S. Circular Dichroism of Poly-L-Proline in an Unordered Conformation. Biopolymers 1968, 6, 1767–1770. [Google Scholar] [CrossRef]
- Tiffany, M.L.; Krimm, S. New Chain Conformations of Poly(Glutamic Acid) and Polylysine. Biopolymers 1968, 6, 1379–1382. [Google Scholar] [CrossRef] [Green Version]
- Tiffany, M.L.; Krimm, S. Effect of Temperature on the Circular Dichroism Spectra of Polypeptides in the Extended State. Biopolymers 1972, 11, 2309–2316. [Google Scholar] [CrossRef] [Green Version]
- Mattice, W.L. The Effect of Temperature and Salt Concentration on the Circular Dichroism Exhibited by Unionized Derivatives of L-Alanine in Aqueous Solution. Biopolymers 1974, 13, 169–183. [Google Scholar] [CrossRef]
- Woody, R. Circular Dichroism and Conformation of Unordered Polypeptides. Adv. Biophys. Chem. 1992, 2, 37–79. [Google Scholar]
- Woody, R. Optical Rotatory Properties of Biopolymers. J. Polym. Sci. Macromol. Rev. 1977, 12, 181–321. [Google Scholar] [CrossRef]
- Karplus, M. Contact Electron-Spin Coupling of Nuclear Magnetic Moments. J. Chem. Phys. 1959, 30, 11–15. [Google Scholar] [CrossRef]
- Asher, S.A.; Mikhonin, A.V.; Bykov, S. UV Raman Demonstrates That Alpha-Helical Polyalanine Peptides Melt to Polyproline II Conformations. J. Am. Chem. Soc. 2004, 126, 8433–8440. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Chen, K.; Santini, P.A.; Shi, Z.; Kallenbach, N.R. The Pentapeptide GGAGG Has PII Conformation. J. Am. Chem. Soc. 2003, 125, 8092–8093. [Google Scholar] [CrossRef] [PubMed]
- McColl, I.H.; Blanch, E.W.; Hecht, L.; Kallenbach, N.R.; Barron, L.D. Vibrational Raman Optical Activity Characterization of Poly(l-Proline) II Helix in Alanine Oligopeptides. J. Am. Chem. Soc. 2004, 126, 5076–5077. [Google Scholar] [CrossRef]
- Schweitzer-Stenner, R.; Eker, F.; Griebenow, K.; Cao, X.; Nafie, L.A. The Conformation of Tetraalanine in Water Determined by Polarized Raman, FT-IR, and VCD Spectroscopy. J. Am. Chem. Soc. 2004, 126, 2768–2776. [Google Scholar] [CrossRef] [PubMed]
- Chakrabartty, A.; Kortemme, T.; Padmanabhan, S.; Baldwin, R.L. Aromatic Side-Chain Contribution to Far-Ultraviolet Circular Dichroism of Helical Peptides and Its Effect on Measurement of Helix Propensities. Biochemistry 1993, 32, 5560–5565. [Google Scholar] [CrossRef] [Green Version]
- Krittanai, C.; Johnson, W.C. Correcting the Circular Dichroism Spectra of Peptides for Contributions of Absorbing Side Chains. Anal. Biochem. 1997, 253, 57–64. [Google Scholar] [CrossRef]
- Chen, K.; Liu, Z.; Zhou, C.; Shi, Z.; Kallenbach, N.R. Neighbor Effect on PPII Conformation in Alanine Peptides. J. Am. Chem. Soc. 2005, 127, 10146–10147. [Google Scholar] [CrossRef]
- Hamburger, J.B.; Ferreon, J.C.; Whitten, S.T.; Hilser, V.J. Thermodynamic Mechanism and Consequences of the Polyproline II (PII) Structural Bias in the Denatured States of Proteins. Biochemistry 2004, 43, 9790–9799. [Google Scholar] [CrossRef]
- Lim, W.A.; Richards, F.M.; Fox, R.O. Structural Determinants of Peptide-Binding Orientation and of Sequence Specificity in SH3 Domains. Nature 1994, 372, 375–379. [Google Scholar] [CrossRef]
- Shi, Z.; Chen, K.; Liu, Z.; Kallenbach, N.R. Conformation of the Backbone in Unfolded Proteins. Chem. Rev. 2006, 106, 1877–1897. [Google Scholar] [CrossRef]
- Creamer, T.P. Left-Handed Polyproline II Helix Formation Is (Very) Locally Driven. Proteins 1998, 33, 218–226. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stapley, B.J.; Creamer, T.P. A Survey of Left-Handed Polyproline II Helices. Protein Sci. 1999, 8, 587–595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Gu, J.; Larson, S.A.; Whitten, S.T.; Hilser, V.J. Denatured-State Energy Landscapes of a Protein Structural Database Reveal the Energetic Determinants of a Framework Model for Folding. J. Mol. Biol. 2008, 381, 1184–1201. [Google Scholar] [CrossRef] [PubMed]
- Marsh, J.A.; Forman-Kay, J.D. Sequence Determinants of Compaction in Intrinsically Disordered Proteins. Biophys. J. 2010, 98, 2383–2390. [Google Scholar] [CrossRef] [Green Version]
- Momany, F.A.; McGuire, R.F.; Burgess, A.W.; Scheraga, H.A. Energy Parameters in Polypeptides. VII. Geometric Parameters, Partial Atomic Charges, Nonbonded Interactions, Hydrogen Bond Interactions, and Intrinsic Torsional Potentials for the Naturally Occurring Amino Acids. J. Phys. Chem. 1975, 79, 2361–2381. [Google Scholar] [CrossRef]
- Mandel, N.; Mandel, G.; Trus, B.L.; Rosenberg, J.; Carlson, G.; Dickerson, R.E. Tuna Cytochrome c at 2.0 A Resolution. III. Coordinate Optimization and Comparison of Structures. J. Biol. Chem. 1977, 252, 4619–4636. [Google Scholar] [CrossRef]
- MacArthur, M.W.; Thornton, J.M. Influence of Proline Residues on Protein Conformation. J. Mol. Biol. 1991, 218, 397–412. [Google Scholar] [CrossRef]
- Lovell, S.C.; Word, J.M.; Richardson, J.S.; Richardson, D.C. The Penultimate Rotamer Library. Proteins 2000, 40, 389–408. [Google Scholar] [CrossRef]
- Iijima, H.; Dunbar, J.B.; Marshall, G.R. Calibration of effective van der Waals atomic contact radii for proteins and peptides. Proteins Struct. Funct. Bioinform. 1987, 2, 330–339. [Google Scholar] [CrossRef]
- Baldwin, R.L. Temperature Dependence of the Hydrophobic Interaction in Protein Folding. Proc. Natl. Acad. Sci. USA 1986, 83, 8069–8072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murphy, K.P.; Freire, E. Thermodynamics of Structural Stability and Cooperative Folding Behavior in Proteins. Adv. Protein Chem. 1992, 43, 313–361. [Google Scholar] [CrossRef] [PubMed]
- Murphy, K.P.; Bhakuni, V.; Xie, D.; Freire, E. Molecular Basis of Co-Operativity in Protein Folding: III. Structural Identification of Cooperative Folding Units and Folding Intermediates. J. Mol. Biol. 1992, 227, 293–306. [Google Scholar] [CrossRef]
- Lee, K.H.; Xie, D.; Freire, E.; Amzel, L.M. Estimation of Changes in Side Chain Configurational Entropy in Binding and Folding: General Methods and Application to Helix Formation. Proteins 1994, 20, 68–84. [Google Scholar] [CrossRef] [PubMed]
- Xie, D.; Freire, E. Structure Based Prediction of Protein Folding Intermediates. J. Mol. Biol. 1994, 242, 62–80. [Google Scholar] [CrossRef] [PubMed]
- Gómez, J.; Hilser, V.J.; Xie, D.; Freire, E. The Heat Capacity of Proteins. Proteins Struct. Funct. Bioinform. 1995, 22, 404–412. [Google Scholar] [CrossRef]
- D’Aquino, J.A.; Gómez, J.; Hilser, V.J.; Lee, K.H.; Amzel, L.M.; Freire, E. The Magnitude of the Backbone Conformational Entropy Change in Protein Folding. Proteins 1996, 25, 143–156. [Google Scholar] [CrossRef]
- Habermann, S.M.; Murphy, K.P. Energetics of Hydrogen Bonding in Proteins: A Model Compound Study. Protein Sci. 1996, 5, 1229–1239. [Google Scholar] [CrossRef] [Green Version]
- Luque, I.; Mayorga, O.L.; Freire, E. Structure-Based Thermodynamic Scale of Alpha-Helix Propensities in Amino Acids. Biochemistry 1996, 35, 13681–13688. [Google Scholar] [CrossRef]
- Das, R.K.; Pappu, R.V. Conformations of Intrinsically Disordered Proteins Are Influenced by Linear Sequence Distributions of Oppositely Charged Residues. Proc. Natl. Acad. Sci. USA 2013, 110, 13392–13397. [Google Scholar] [CrossRef] [Green Version]
- Dunker, A.K.; Obradovic, Z.; Romero, P.; Garner, E.C.; Brown, C.J. Intrinsic Protein Disorder in Complete Genomes. Genome Inform. Ser. Workshop Genome Inform. 2000, 11, 161–171. [Google Scholar] [PubMed]
- Wootton, J.C.; Federhen, S. Statistics of Local Complexity in Amino Acid Sequences and Sequence Databases. Comput. Chem. 1993, 17, 149–163. [Google Scholar] [CrossRef]
- Wootton, J.C. Non-Globular Domains in Protein Sequences: Automated Segmentation Using Complexity Measures. Comput. Chem. 1994, 18, 269–285. [Google Scholar] [CrossRef]
- Romero, P.; Obradovic, Z.; Li, X.; Garner, E.C.; Brown, C.J.; Dunker, A.K. Sequence Complexity of Disordered Protein. Proteins Struct. Funct. Bioinform. 2001, 42, 38–48. [Google Scholar] [CrossRef]
- Uversky, V.N. Natively Unfolded Proteins: A Point Where Biology Waits for Physics. Protein Sci. 2002, 11, 739–756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hynes, T.R.; Fox, R.O. The Crystal Structure of Staphylococcal Nuclease Refined at 1.7 A Resolution. Proteins 1991, 10, 92–105. [Google Scholar] [CrossRef]
- Scholtz, J.M.; Marqusee, S.; Baldwin, R.L.; York, E.J.; Stewart, J.M.; Santoro, M.; Bolen, D.W. Calorimetric Determination of the Enthalpy Change for the Alpha-Helix to Coil Transition of an Alanine Peptide in Water. Proc. Natl. Acad. Sci. USA 1991, 88, 2854–2858. [Google Scholar] [CrossRef] [Green Version]
- Dignon, G.L.; Zheng, W.; Kim, Y.C.; Mittal, J. Temperature-Controlled Liquid—Liquid Phase Separation of Disordered Proteins. ACS Cent. Sci. 2019, 5, 821–830. [Google Scholar] [CrossRef] [Green Version]



| Amino Acid | PPII Propensity a | PPII Propensity b | PPII Propensity c |
|---|---|---|---|
| ALA (A) | 0.61 | 0.818 | 0.37 |
| CYS (C) | 0.55 | 0.557 | 0.25 |
| ASP (D) | 0.63 | 0.552 | 0.30 |
| GLU (E) | 0.61 | 0.684 | 0.42 |
| PHE (F) | 0.58 | 0.639 | 0.17 |
| GLY (G) | 0.58 | - | 0.13 |
| HIS (H) | 0.55 | 0.428 | 0.20 |
| ILE (I) | 0.50 | 0.519 | 0.39 |
| LYS (K) | 0.59 | 0.581 | 0.56 |
| LEU (L) | 0.58 | 0.574 | 0.24 |
| MET (M) | 0.55 | 0.498 | 0.36 |
| ASN (N) | 0.55 | 0.667 | 0.27 |
| PRO (P) | 0.67 | - | 1.00 |
| GLN (Q) | 0.66 | 0.654 | 0.53 |
| ARG (R) | 0.61 | 0.638 | 0.38 |
| SER (S) | 0.58 | 0.774 | 0.24 |
| THR (T) | 0.53 | 0.553 | 0.32 |
| VAL (V) | 0.49 | 0.743 | 0.39 |
| TRP (W) | - | 0.764 | 0.25 |
| TYR (Y) | - | 0.630 | 0.25 |
| average | 0.58 | 0.626 | 0.35 |
| Amino Acid | ∆G (kcal mol−1) a | α-Helix Propensity b |
|---|---|---|
| ALA (A) | −0.258 | 0.62 |
| CYS (C) | 0.570 | 0.26 |
| ASP (D) | 0.635 | 0.24 |
| GLU (E) | 0.433 | 0.31 |
| PHE (F) | 0.672 | 0.22 |
| GLY (G) | 1.62 | 0.05 |
| HIS (H) | 0.525 | 0.28 |
| ILE (I) | 0.445 | 0.31 |
| LYS (K) | 0.108 | 0.45 |
| LEU (L) | 0.022 | 0.49 |
| MET (M) | 0.251 | 0.39 |
| ASN (N) | 0.635 | 0.24 |
| PRO (P) | 4 | 0.001 |
| GLN (Q) | 0.314 | 0.36 |
| ARG (R) | −0.047 | 0.52 |
| SER (S) | 0.525 | 0.28 |
| THR (T) | 1.07 | 0.12 |
| VAL (V) | 0.797 | 0.19 |
| TRP (W) | ~0.6 | 0.25 |
| TYR (Y) | ~0.4 | 0.32 |
| average | 0.29 |
| Amino Acid | PPII Propensity a |
|---|---|
| ALA (A) | 0.48 |
| CYS (C) | 0.38 |
| ASP (D) | 0.34 |
| GLU (E) | 0.38 |
| PHE (F) | 0.36 |
| GLY (G) | 0.21 |
| HIS (H) | 0.28 |
| ILE (I) | 0.39 |
| LYS (K) | 0.35 |
| LEU (L) | 0.47 |
| MET (M) | 0.38 |
| ASN (N) | 0.31 |
| PRO (P) | 0.81 |
| GLN (Q) | 0.35 |
| ARG (R) | 0.32 |
| SER (S) | 0.31 |
| THR (T) | 0.24 |
| VAL (V) | 0.34 |
| TRP (W) | 0.35 |
| TYR (Y) | 0.37 |
| average | 0.37 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paiz, E.A.; Lewis, K.A.; Whitten, S.T. Structural and Energetic Characterization of the Denatured State from the Perspectives of Peptides, the Coil Library, and Intrinsically Disordered Proteins. Molecules 2021, 26, 634. https://doi.org/10.3390/molecules26030634
Paiz EA, Lewis KA, Whitten ST. Structural and Energetic Characterization of the Denatured State from the Perspectives of Peptides, the Coil Library, and Intrinsically Disordered Proteins. Molecules. 2021; 26(3):634. https://doi.org/10.3390/molecules26030634
Chicago/Turabian StylePaiz, Elisia A., Karen A. Lewis, and Steven T. Whitten. 2021. "Structural and Energetic Characterization of the Denatured State from the Perspectives of Peptides, the Coil Library, and Intrinsically Disordered Proteins" Molecules 26, no. 3: 634. https://doi.org/10.3390/molecules26030634








