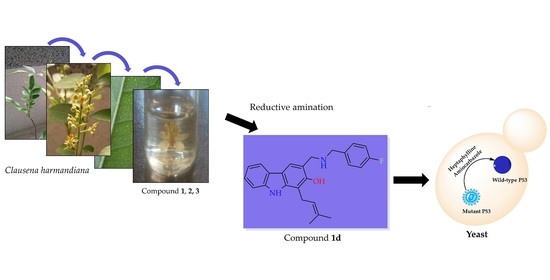
Semi-Synthesis of Small Molecules of Aminocarbazoles: Tumor Growth Inhibition and Potential Impact on p53
Abstract
:1. Introduction
2. Results and Discussion
2.1. Semi-Synthesis of Aminocarbazole Alkaloids by Direct Reductive Amination
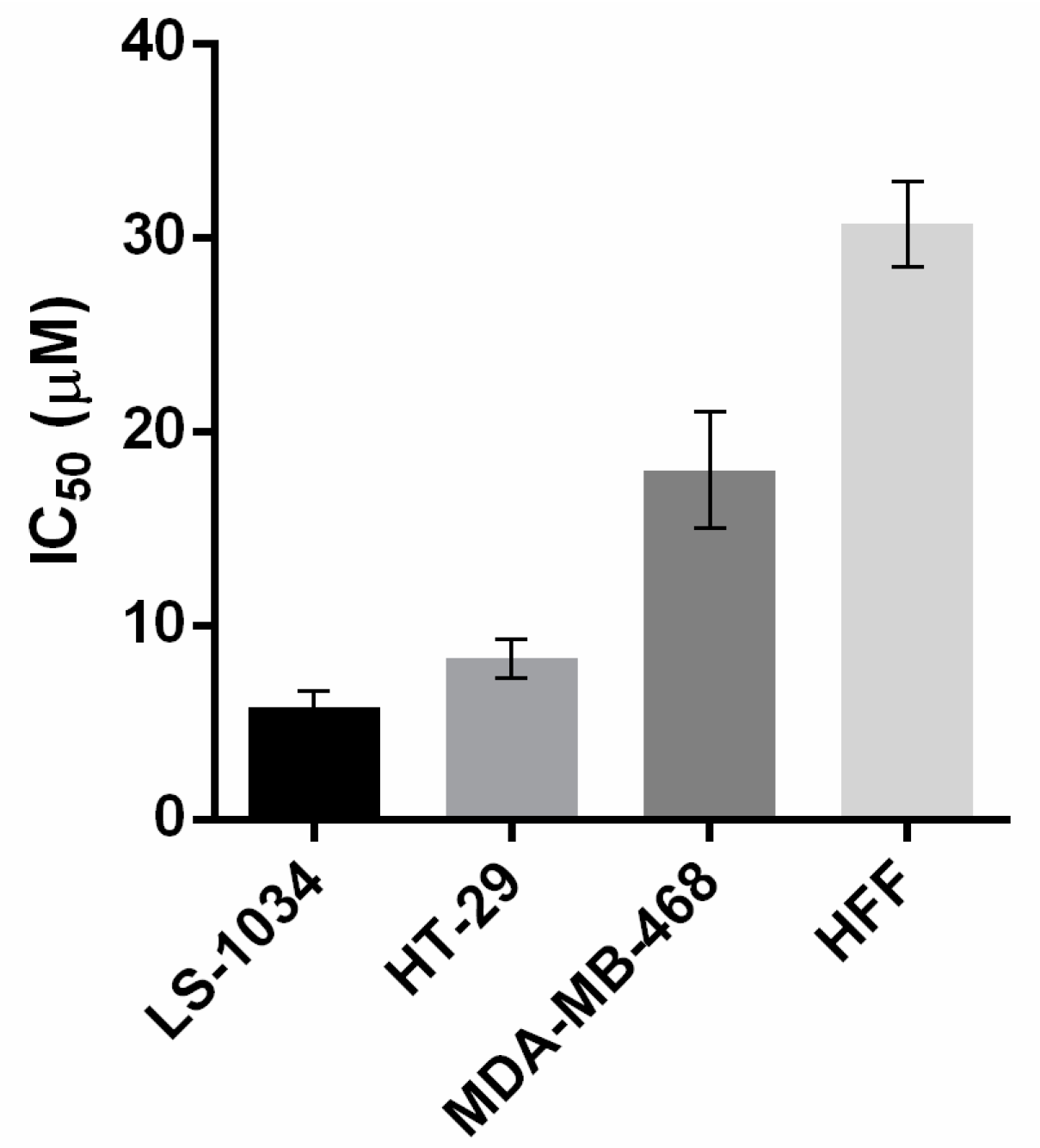
2.2. Aminocarbaozoles Exhibit Tumor Growth-Inhibitory Effect
2.3. Evaluation of Aminocarbazoles Potential Activation of p53 Using a Yeast-Based Screening Assay
3. Materials and Methods
3.1. Isolation
3.2. Purity Determination by HPLC-DAD
3.3. General Semi-Synthesis of the Aminocarbazole Derivatives of Heptaphylline (1), 7-Methoxyheptaphylline (2), and 7-Methoxymukonal (3)
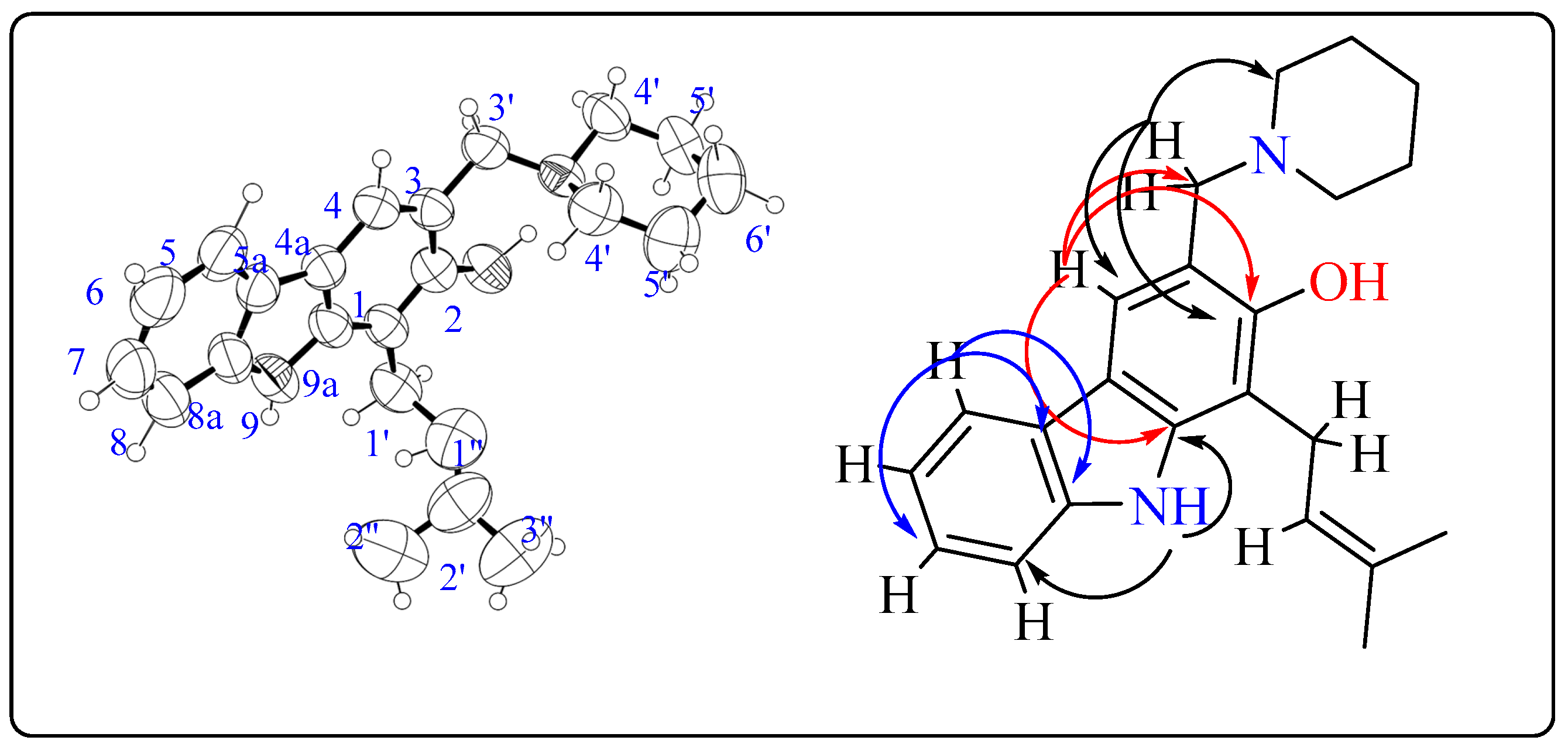
3.4. Crystallography
3.5. Yeast Screening Assay
3.6. Human Tumor Cell Lines and Growths Conditions
3.7. Sulforhodamine B (SRB) Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Xin, Z.-Q.; Lu, J.-J.; Ke, C.-Q.; Hu, C.-X.; Lin, L.-P.; Ye, Y. Constituents from Clausena excavata. Chem. Pharm. Bull. 2008, 56, 827–830. [Google Scholar] [CrossRef] [Green Version]
- Knölker, H.-J.; Reddy, K.R. Isolation and Synthesis of Biologically Active Carbazole Alkaloids. Chem. Rev. 2002, 102, 4303–4428. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.-S.; Huang, S.-C.; Wu, P.-L.; Kuoh, C.-S. Alkaloidal and other constituents from the root bark of Clausena excavata. Phytochemistry 1999, 52, 523–527. [Google Scholar] [CrossRef]
- Das, K.C.; Chakraborty, D.P.; Bose, P.K. Antifungal activity of some constituents ofMurraya koenigii spreng. Cell. Mol. Life Sci. 1965, 21, 340. [Google Scholar] [CrossRef] [PubMed]
- Chakthong, S.; Bindulem, N.; Raknai, S.; Yodwaree, S.; Kaewsanee, S.; Kanjana-Opas, A. Carbazole-pyranocoumarin conjugate and two carbazole alkaloids from the stems of Clausena excavata. Nat. Prod. Res. 2016, 30, 1690–1697. [Google Scholar] [CrossRef] [PubMed]
- Thongthoom, T.; Promsuwan, P.; Yenjai, C. Synthesis and cytotoxic activity of the heptaphylline and 7-methoxyheptaphylline series. Eur. J. Med. Chem. 2011, 46, 3755–3761. [Google Scholar] [CrossRef]
- Thongthoom, T.; Songsiang, U.; Phaosiri, C.; Yenjai, C. Biological activity of chemical constituents from Clausena harmandiana. Arch. Pharmacal Res. 2010, 33, 675–680. [Google Scholar] [CrossRef]
- Yenjai, C.; Sripontan, S.; Sriprajun, P.; Kittakoop, P.; Jintasirikul, A.; Tanticharoen, M.; Thebtaranonth, Y. Coumarins and Carbazoles with Antiplasmodial Activity from Clausena harmandiana. Planta Medica 2000, 66, 277–279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, T.-S.; Huang, S.-C.; Wu, P.-L.; Teng, C.-M. Carbazole alkaloids from Clausena excavata and their biological activity. Phytochemistry 1996, 43, 133–140. [Google Scholar] [CrossRef]
- Maneerat, W.; Phakhodee, W.; Ritthiwigrom, T.; Cheenpracha, S.; Promgool, T.; Yossathera, K.; Deachathai, S.; Laphookhieo, S. Antibacterial carbazole alkaloids from Clausena harmandiana twigs. Fitoterapia 2012, 83, 1110–1114. [Google Scholar] [CrossRef]
- Chakraborty, A.; Saha, C.; Podder, G.; Chowdhury, B.; Bhattacharyya, P. Carbazole alkaloid with antimicrobial activity from clausena heptaphylla. Phytochemistry 1995, 38, 787–789. [Google Scholar] [CrossRef]
- Patel, O.P.S.; Mishra, A.; Maurya, R.; Saini, D.; Pandey, J.; Taneja, I.; Raju, K.S.R.; Kanojiya, S.; Shukla, S.K.; Srivastava, M.N.; et al. Naturally Occurring Carbazole Alkaloids fromMurraya koenigiias Potential Antidiabetic Agents. J. Nat. Prod. 2016, 79, 1276–1284. [Google Scholar] [CrossRef]
- Boonyarat, C.; Yenjai, C.; Vajragupta, O.; Waiwut, P. Heptaphylline induces apoptosis in human colon adenocarcinoma cells through bid and Akt/NF-?B (p65) pathways. Asian Pac. J. Cancer Prev. 2015, 15, 10483–10487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saturnino, C.; Iacopetta, D.; Sinicropi, M.S.; Rosano, C.; Caruso, A.; Caporale, A.; Marra, N.; Marengo, B.; Pronzato, M.A.; Parisi, O.I.; et al. N-Alkyl Carbazole Derivatives as New Tools for Alzheimer’s Disease: Preliminary Studies. Molecules 2014, 19, 9307–9317. [Google Scholar] [CrossRef] [PubMed]
- Krahl, M.P.; Jäger, A.; Krause, T.; Knölker, H.-J. First total synthesis of the 7-oxygenated carbazole alkaloids clauszoline-K, 3-formyl-7-hydroxycarbazole, clausine M, clausine N and the anti-HIV active siamenol using a highly efficient palladium-catalyzed approach. Org. Biomol. Chem. 2006, 4, 3215–3219. [Google Scholar] [CrossRef]
- Wangboonskul, J.D.; Pummangura, S.; Chaichantipyuth, C. Five Coumarins and a Carbazole Alkaloids From the Root Bark of Clausena harmandiana. J. Nat. Prod. 1984, 47, 1058–1059. [Google Scholar] [CrossRef]
- Issa, S.; Walchshofer, N.; Kassab, I.; Termoss, H.; Chamat, S.; Geahchan, A.; Bouaziz, Z. Synthesis and antiproliferative activity of oxazinocarbazole and N,N-bis(carbazolylmethyl)amine derivatives. Eur. J. Med. Chem. 2010, 45, 2567–2577. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-L.; Hung, H.-M.; Lu, C.-M.; Li, K.-C.; Tzeng, C.-C. Synthesis and anticancer evaluation of certain indolo[2,3-b]quinoline derivatives. Bioorg. Med. Chem. 2004, 12, 6539–6546. [Google Scholar] [CrossRef]
- Compain-Batissou, M.; Latreche, D.; Gentili, J.; Walchshofer, N.; Bouaziz, Z. Synthesis and Diels–Alder Reactivity of ortho-Carbazolequinones. Chem. Pharm. Bull. 2004, 52, 1114–1116. [Google Scholar] [CrossRef] [Green Version]
- Joerger, A.C.; Fersht, A.R. Structural Biology of the Tumor Suppressor p53 and Cancer-Associated Mutants. In Advances in Cancer Research; Elsevier BV: Amsterdam, The Netherlands, 2007; Volume 97, pp. 1–23. [Google Scholar]
- Vousden, K.H.; Lane, D.P. p53 in health and disease. Nat. Rev. Mol. Cell Biol. 2007, 8, 275–283. [Google Scholar] [CrossRef] [PubMed]
- Fridman, J.S.; Lowe, S.W. Control of apoptosis by p53. Oncogene 2003, 22, 9030–9040. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muller, P.A.; Vousden, K.H. Mutant p53 in Cancer: New Functions and Therapeutic Opportunities. Cancer Cell 2014, 25, 304–317. [Google Scholar] [CrossRef] [Green Version]
- Yu, X.; Blanden, A.R.; Narayanan, S.; Jayakumar, L.; Lubin, D.; Augeri, D.; Kimball, S.D.; Loh, S.N.; Carpizo, D.R. Small molecule restoration of wildtype structure and function of mutant p53 using a novel zinc-metallochaperone based mechanism. Oncotarget 2014, 5, 8879–8892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Wilcken, R.; Joerger, A.C.; Chuckowree, I.S.; Amin, J.; Spencer, J.; Fersht, A.R. Small molecule induced reactivation of mutant p53 in cancer cells. Nucleic Acids Res. 2013, 41, 6034–6044. [Google Scholar] [CrossRef]
- Boeckler, F.M.; Joerger, A.C.; Jaggi, G.; Rutherford, T.J.; Veprintsev, D.B.; Fersht, A.R. Targeted rescue of a destabilized mutant of p53 by an in silico screened drug. Proc. Natl. Acad. Sci. USA 2008, 105, 10360–10365. [Google Scholar] [CrossRef] [Green Version]
- Bauer, M.R.; Jones, R.N.; Baud, M.G.J.; Wilcken, R.; Boeckler, F.M.; Fersht, A.R.; Joerger, A.C.; Spencer, J. Harnessing Fluorine–Sulfur Contacts and Multipolar Interactions for the Design of p53 Mutant Y220C Rescue Drugs. ACS Chem. Biol. 2016, 11, 2265–2274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rauf, S.M.A.; Endou, A.; Takaba, H.; Miyamoto, A. Effect of Y220C Mutation on p53 and Its Rescue Mechanism: A Computer Chemistry Approach. Protein J. 2013, 32, 68–74. [Google Scholar] [CrossRef]
- Roughley, S.D.; Jordan, A.M. The Medicinal Chemist’s Toolbox: An Analysis of Reactions Used in the Pursuit of Drug Candidates. J. Med. Chem. 2011, 54, 3451–3479. [Google Scholar] [CrossRef]
- Abdel-Magid, A.F.; Carson, K.G.; Harris, B.D.; Maryanoff, C.A.; Shah, R.D. Reductive Amination of Aldehydes and Ketones with Sodium Triacetoxyborohydride. Studies on Direct and Indirect Reductive Amination Procedures1. J. Org. Chem. 1996, 61, 3849–3862. [Google Scholar] [CrossRef] [PubMed]
- Joshi, B.; Kamat, V.; Gawad, D.; Govindachari, T. Structure and synthesis of heptaphylline. Phytochemistry 1972, 11, 2065–2071. [Google Scholar] [CrossRef]
- Joshi, B.; Kamat, V.; Saksena, A.; Govindachari, T. Structure of heptaphylline, a carbazole alkaloid from clausena heptaphylla wt. & arn. Tetrahedron Lett. 1967, 8, 4019–4022. [Google Scholar] [CrossRef]
- Leão, M.; Moreira, S.; Soares, J.; Bessa, C.; Maciel, C.; Ciribilli, Y.; Pereira, C.; Inga, A.; Saraiva, L. Novel simplified yeast-based assays of regulators of p53-MDMX interaction and p53 transcriptional activity. FEBS J. 2013, 280, 6498–6507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jantamat, P.; Weerapreeyakul, N.; Puthongking, P. Cytotoxicity and Apoptosis Induction of Coumarins and Carbazole Alkaloids from Clausena harmandiana. Molecules 2019, 24, 3385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheldrick, G. A short history ofSHELX. Acta Crystallogr. Sect. A Found. Crystallogr. 2008, 64, 112–122. [Google Scholar] [CrossRef] [Green Version]
- Soares, J.; Raimundo, L.; Pereira, N.A.; Monteiro, Â.; Gomes, S.; Bessa, C.; Pereira, C.; Queiroz, G.; Bisio, A.; Fernandes, J.; et al. Reactivation of wild-type and mutant p53 by tryptophanolderived oxazoloisoindolinone SLMP53-1, a novel anticancer small-molecule. Oncotarget 2016, 7, 4326–4343. [Google Scholar] [CrossRef] [PubMed]
- Soares, J.; Raimundo, L.; Pereira, N.A.; dos Santos, D.J.; Pérez, M.; Queiroz, G.; Leão, M.; Santos, M.M.; Saraiva, L. A tryptophanol-derived oxazolopiperidone lactam is cytotoxic against tumors via inhibition of p53 interaction with murine double minute proteins. Pharmacol. Res. 2015, 95–96, 42–52. [Google Scholar] [CrossRef] [PubMed] [Green Version]


 | ||||||
|---|---|---|---|---|---|---|
| Entry | Sub. | Amine Precursors | Products | Solvent | Time (Days) | Yield (%) |
| 1 | 1 | 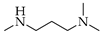 | 1a | THF DCE | 5 3 | 39 49 |
| 2 | 1 | 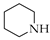 | 1b | THF DCE | 4 3 | 44 90 |
| 3 | 1 | 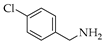 | 1c | THF DCE | 3 - | 31 - |
| 4 | 1 | 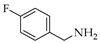 | 1d | THF DCE | 3 3 | 15 42 |
| 5 | 1 | 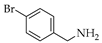 | 1e | THF DCE | 5 3 | 47 64 |
| 6 | 2 | 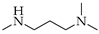 | 2a | THF DCE | 5 4 | 16 51 |
| 7 | 2 |  | 2b | THF DCE | 8 5 | 21 39 |
| 8 | 2 |  | 2c | THF DCE | 4 4 | 15 34 |
| 9 | 2 | 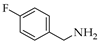 | 2d | THF DCE | 4 4 | 13 30 |
| 10 | 2 | 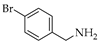 | 2e | THF DCE | 5 3 | 35 86 |
| 11 | 2 | 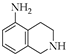 | 2f | THF | 10 | 25 |
| 12 | 3 | 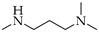 | 3a | THF | 7 | 51 |
| Cell Line | HCT116 (wt) | HT-29 (R273H) | HuH-7 (Y220C) | SW837 (R248W) | MDA-MB-468 (R273H) | A375 | SF-268 (R273H) | LS-1034 (G245S) |
|---|---|---|---|---|---|---|---|---|
| 1 | 4.4 ± 1.1 | 14.0 ± 0.1 | 6.1 ± 0.3 | 7.2 ± 0.4 | <3.13 | - | - | - |
| 2 | 15.0 ± 1.0 | 23.0 ± 6.0 | 21.5 ± 0.5 | 30.5 ± 2.5 | - | 5.0 ± 0.3 | - | - |
| 3 | 4.7 ± 0.5 | 9.4 ± 0.7 | 6.3± 2.2 | 6.1 ± 1.2 | - | - | - | - |
| 1a | > 50 | >50 | - | - | >50 | - | - | - |
| 1b | 18.1 ± 0.9 | 18.0 ± 1.9 | > 50 | 23.0 ± 3.0 | 16.0 ± 2.0 | - | - | - |
| 1c | 22.6 ± 4.5 | - | 30.5 ± 0.5 | 29.0 ± 2.0 | - | - | - | - |
| 1d | 15.5 ± 1.5 | 8.3 ± 1.0 | 28.5 ± 0.5 | 26.0 ± 2.0 | 18.0 ± 3.0 | 24.5 ± 1.5 | 20.0 ± 4.0 | 5.8 ± 0.8 |
| 1e | 30.6 ± 2.8 | - | >50 | 23.0 ± 1.0 | - | 38.2 ± 1.9 | - | - |
| 1f | - | - | - | - | - | - | - | - |
| 2a | 22.3 ± 1.0 | - | 15.5 ± 3.5 | 18.0 ± 0.0 | - | 29.7 ± 1.8 | - | - |
| 2b | 24.0 ± 4.0 | 28.5 ± 1.5 | >50 | >50 | - | >50 | - | - |
| 2c | 25.0 ± 1.8 | - | 45.0 ± 3.0 | >50 | - | 30.7 ± 3.7 | - | - |
| 2d | 16.1 ± 3.5 | - | 23.0 ± 3.0 | 10.8 ± 1.3 | - | 27.8 ± 3.0 | - | - |
| 2e | 27.0 ± 4.4 | - | 33.5 ± 5.5 | 37.5 ± 0.5 | - | 38.3 ± 3.2 | - | - |
| 2f | - | - | - | - | - | - | - | - |
| 2g | - | - | - | - | - | - | - | - |
| 2h | - | - | - | - | - | - | - | - |
| 3a | 22.2± 0.9 | - | 15.5 ± 3.5 | 18.0 ± 0.0 | - | 29.7 ± 1.8 | - | - |
| Etoposide | 0.54 ± 0.1 | 1.52 ± 0.3 | 3.66 ± 0.7 | 0.9 ± 0.07 | 2.07 ± 0.15 | 0.85 ± 0.09 | - | - |
| Mutant p53 | 1 | 1b | 1c | 1d |
|---|---|---|---|---|
| R280K | - | - | - | - |
| Y220C | 71.17 ± 11.53 | - | - | 47.83 ± 5.80 |
| G245D | - | 57.06 ± 13.03 | 34.73 ± 12.50 | 63.43 ± 11.50 |
| R273H | - | - | - | - |
| R175H | - | - | 41.50 ± 16.40 | - |
| R248W | - | - | - | 34.30 ± 2.97 |
| R248Q | - | - | 52.90 ± 9.13 | 46.20 ± 5.27 |
| R273C | - | 47.30 ± 6.07 | - | - |
| R282W | - | 42.33 ± 1.63 | 83.63 ± 7.16 | - |
| G245S | 68.40 ± 12.20 | 47.87 ± 8.83 | 52.20 ± 6.90 | 55.33 ± 4.53 |
| wt p53 | - | 61.70 ± 11.1 | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Long, S.; Loureiro, J.B.; Carvalho, C.; Gales, L.; Saraiva, L.; Pinto, M.M.M.; Puthongking, P.; Sousa, E. Semi-Synthesis of Small Molecules of Aminocarbazoles: Tumor Growth Inhibition and Potential Impact on p53. Molecules 2021, 26, 1637. https://doi.org/10.3390/molecules26061637
Long S, Loureiro JB, Carvalho C, Gales L, Saraiva L, Pinto MMM, Puthongking P, Sousa E. Semi-Synthesis of Small Molecules of Aminocarbazoles: Tumor Growth Inhibition and Potential Impact on p53. Molecules. 2021; 26(6):1637. https://doi.org/10.3390/molecules26061637
Chicago/Turabian StyleLong, Solida, Joana B. Loureiro, Carla Carvalho, Luís Gales, Lucília Saraiva, Madalena M. M. Pinto, Ploenthip Puthongking, and Emília Sousa. 2021. "Semi-Synthesis of Small Molecules of Aminocarbazoles: Tumor Growth Inhibition and Potential Impact on p53" Molecules 26, no. 6: 1637. https://doi.org/10.3390/molecules26061637