Promising Antidiabetic and Antimicrobial Agents Based on Fused Pyrimidine Derivatives: Molecular Modeling and Biological Evaluation with Histopathological Effect
Abstract
:1. Introduction
2. Results and Discussion
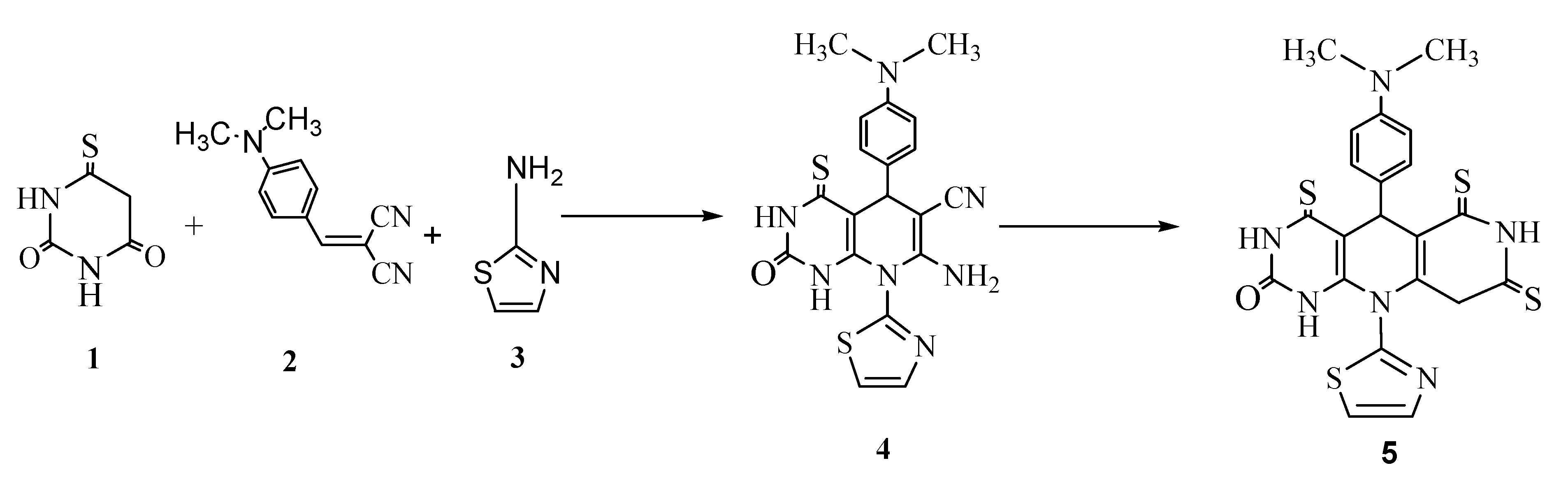
2.1. Chemistry
2.2. Pharmacology
Antidiabetic Study
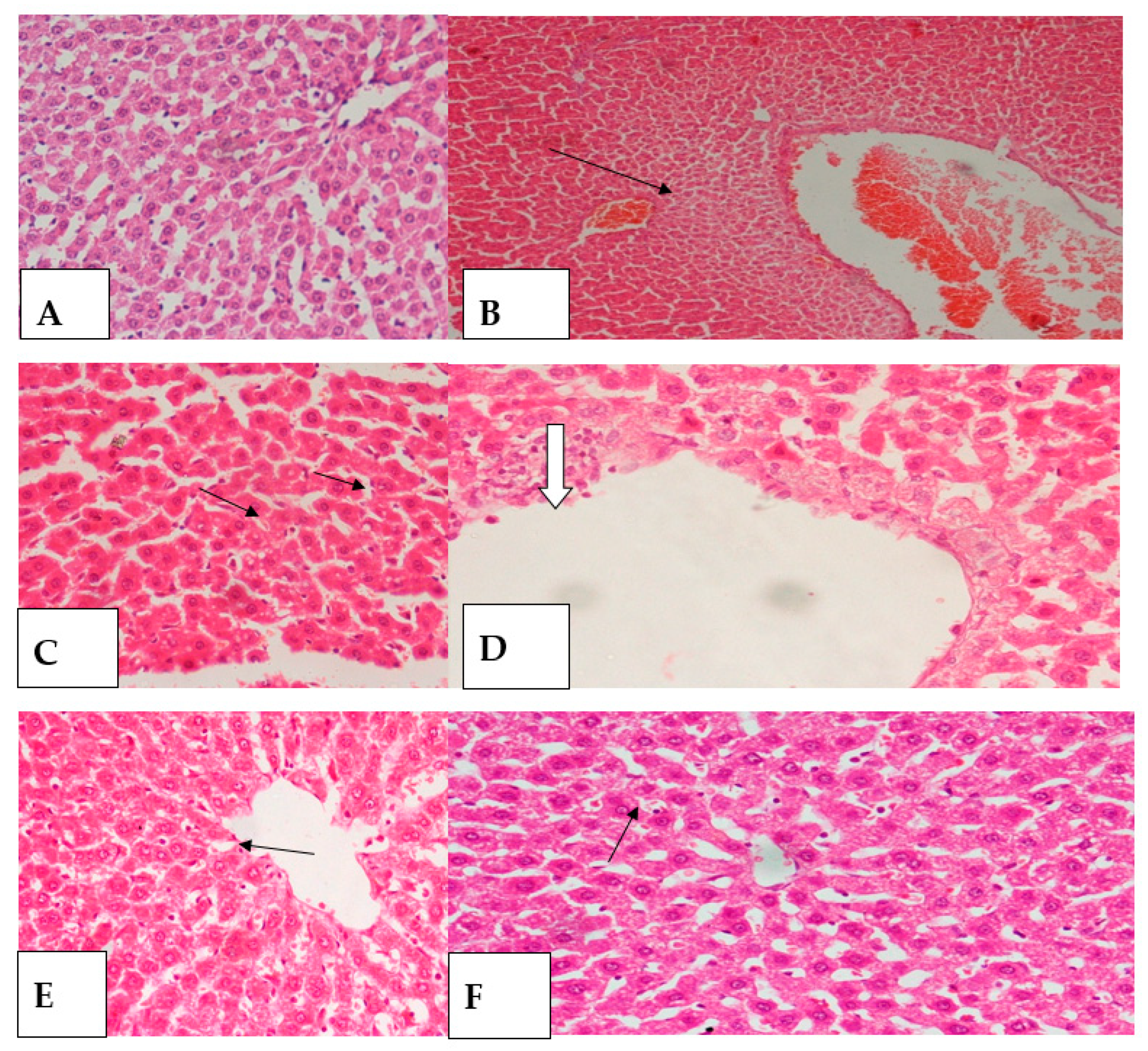
2.3. Histopathology Study
2.4. Antimicrobial Activity
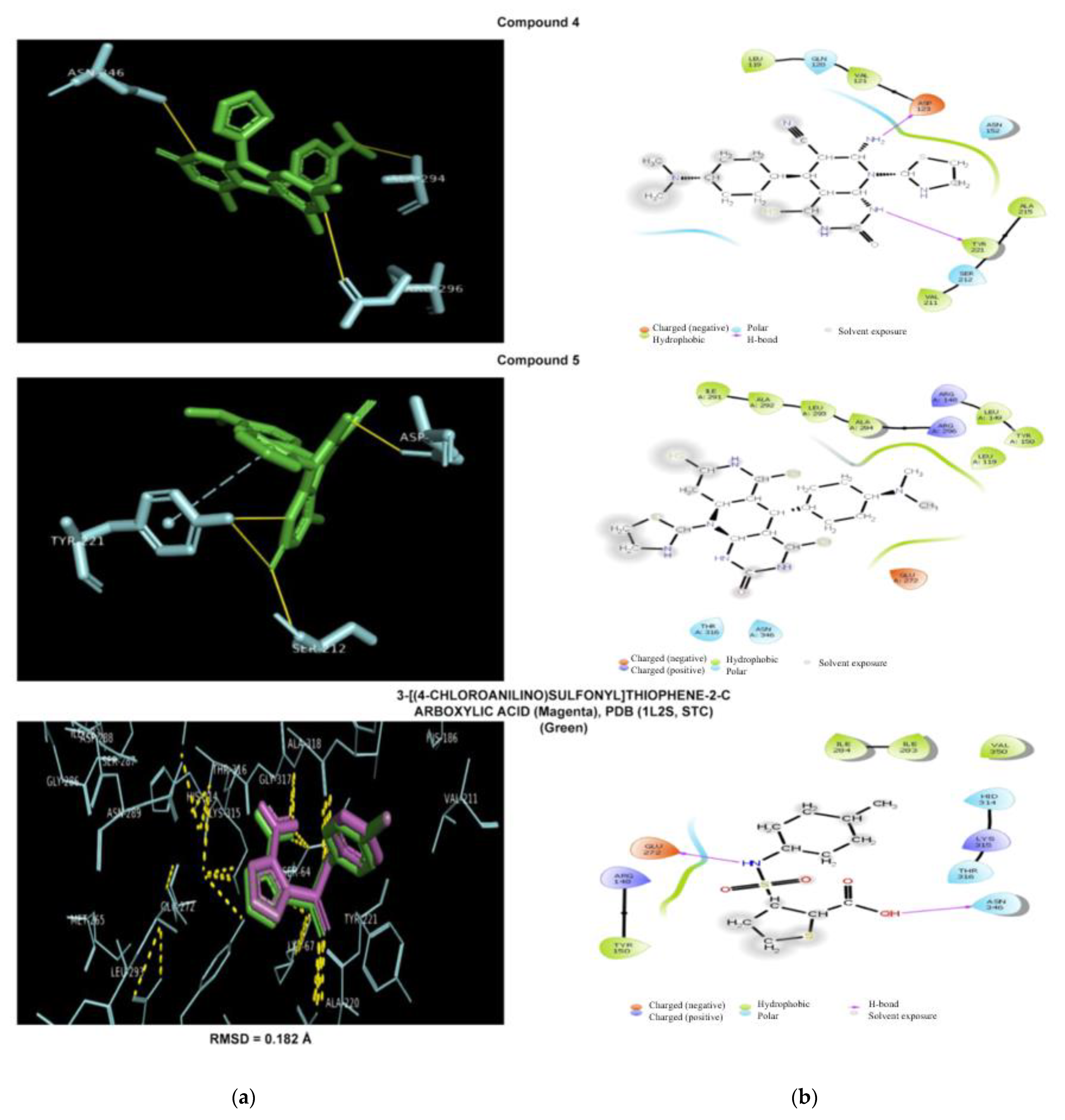
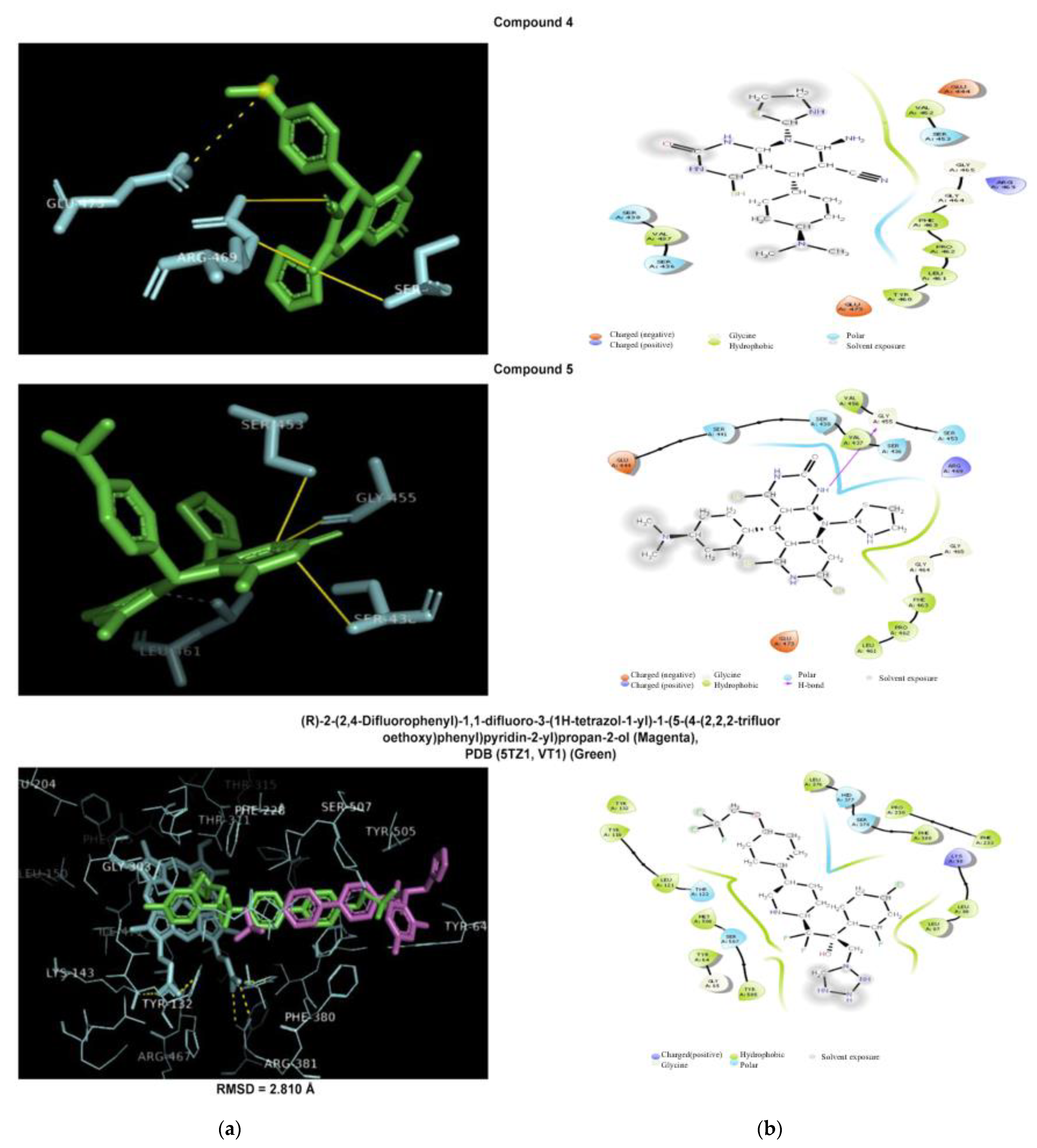
2.5. Molecular Modeling Study
2.5.1. Antibacterial Docking Simulation
2.5.2. Antifungal Docking Simulation
2.5.3. Antidiabetic Docking Simulation
3. Experimental Section
3.1. General Information
3.2. Synthesis of 7-Amino-5-(4-(dimethylamino)-phenyl)-1,2,4,6-hexahydro-2-oxo-8-(thiazolyl)-4-thioxopyrido [2,3-d] pyrimidine-6-carbonitrile (4)
3.3. Synthesis of 5-(4-(Dimethylamino)phenyl)-3,4,6,7,8,9-hexahydro-10-(thiazolyl)-4,6,8-trithioxopyrimido [4,5-b] naphthyridine-2 (1H, 5H, 10H)-one (5)
3.4. Antidiabetic Activity Study
3.4.1. Animals
3.4.2. Reagents and Tested Compounds
3.4.3. Diagnostic Kits
3.4.4. Experimental Design
3.4.5. Biochemical Assays
3.5. Histopathological Tests
3.6. Statistical Analysis
3.7. Antimicrobial Activity Assay
3.8. Molecular Modeling Study
3.8.1. Target Fishing Simulation
3.8.2. Antidiabetic Target Fishing Simulation
3.8.3. Ligands and Targets Docking Simulation
3.8.4. Simulations Using AutoDockvina
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Kenny, H.C.; Abel, E.D. Heart Failure in Type 2 Diabetes Mellitus Impact of Glucose-Lowering Agents, Heart Failure Therapies, and Novel Therapeutic Strategies. Circ. Res. 2019, 124, 121–141. [Google Scholar] [CrossRef] [PubMed]
- American Diabetes Association. 8. Pharmacologic approaches to glycemic treatment: Standards of medical care in diabetes. Diabetes Care 2018, 41, S73–S85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gothai, S.; Ganesan, P.; Park, S.Y.; Fakurazi, S.; Choi, D.K.; Arulselvan, P. Natural phyto-bioactive compounds for the treatment of type 2 diabetes: Inflammation as a target. Nutrients 2016, 8, 461. [Google Scholar] [CrossRef]
- Becan, L.; Wójcicka, A. Synthesis, anti-hepatitis b and c virus activity and antitumor screening of novel thiazolo[4,5-d]-pyrimidine derivatives. Drug Res. 2016, 73, 107–114. [Google Scholar]
- Baxter, A.; Cooper, A.; Kinchin, E.; Moakes, K.; Unitt, J.; Wallace, A. Hit-to-Lead studies: The discovery of potent, orally bioavailable thiazolopyrimidine CXCR2 receptor antagonists. Bioorg. Med. Chem. Lett. 2006, 16, 960–963. [Google Scholar] [CrossRef] [PubMed]
- AbdElhameed, A.A.; El-Gohary, N.S.; El-Bendary, E.R.; Shaaban, M.I.; Bayomi, S.M. New Thiazole and Thiazolopyrimidine Derivatives: Synthesis, Antimicrobial, Antiquorum-Sensingand Antitumor Evaluation. Pharm. Lett. 2018, 10, 55–72. [Google Scholar]
- Sambhaji, P.V.; Digambar, B.K.; Nilesh, K.H.; Mahesh, M.P. An Efficient method for Synthesis of Novel Iminothiazolo Pyrimidines and Plausible Antioxidant Potential. Int. J. Drug Dev. Res. 2013, 5, 128–134. [Google Scholar]
- Babasaheb, K.; Sudhakar, R.B.; Pawan, S.H. Antimicrobial and Anti-Inflammatory Activity Studies of Novel Thiazolopyrimidines. Am. J. Pharm. Tech. Res. 2020, 10. [Google Scholar] [CrossRef]
- Fatima, S.; Sharma, A.; Saxena, R.; Tripathi, R.; Shukla, S.K.; Pandey, S.K.; Tripathi, R.; Tripathi, R.P. One pot efficient diversity oriented synthesis of polyfunctional styryl thiazolopyrimidines and their bio-evaluation as antimalarial and anti-HIV agents. Eur. J. Med. Chem. 2012, 55, 195–204. [Google Scholar] [CrossRef]
- Dong, C.; Zhi-Hua, Z.; Yu Chen, X.Y.; Shi-Ti, Z.; Liang-Jing, Z.; Li-Hong, M.; Fang, L.; Bing-Jie, F. Synthesis of some new thiazolo[3,2-a]pyrimidine derivatives and screening of their in vitro antibacterial and antitubercular activities. Med. Chem. Res. 2016, 25, 292–302. [Google Scholar]
- Kumar, S.; Deep, A.; Narasimhan, B. A Review on Synthesis, Anticancer and Antiviral Potentials of Pyrimidine Derivatives. Current Bioact. Compounds 2019, 15, 289–303. [Google Scholar] [CrossRef]
- Azam, F.; Alkskas, A.I.; Ahmed, A.M. Synthesis of some urea and thiourea derivatives of 3-phenyl/ethyl-2-thioxo-2,3-dihydrothiazolo[4,5-d]pyrimidine and their antagonistic effects on haloperidol-induced catalepsy and oxidative stress in mice. Eur. J. Med. Chem. 2009, 44, 3889–3897. [Google Scholar] [CrossRef] [PubMed]
- Flefel, E.M. Synthesis and anticancer activity of new 1-thia-4-azaspiro[4.5]decane, their derived thiazolopyrimidine and 1,3,4-thiadiazole thioglycosides. Molecules 2017, 22, 170. [Google Scholar] [CrossRef]
- Fatma, A.B.; Mahmound El, H.; Ahmed El, R.; Mohamed Abdel, R. Molecular Modeling and Biological Activities of New Potent Antimicrobial, Anti-Inflammatory and Anti-Nociceptive of 5-Nitro Indoline-2-One Derivatives. Drug Des. 2017, 6, 1–6. [Google Scholar]
- Bassyouni, F.A.; Saleh, T.S.; ElHefnawi, M.M.; Abd El-Moez, S.I.; El-Senousy, W.M.; Abdel-Rehim, M.E. Synthesis, Pharmacological Activity Evaluation and Molecular Modeling of New Polynuclear Heterocyclic Compounds Containing Benzimidazole derivatives. Arch. Pharm. Res. 2012, 12, 2063–2075. [Google Scholar] [CrossRef] [PubMed]
- Tawfik, H.A.; Bassyouni, F.; Gamal-Eldeen, A.M.; Abo-Zeid, M.A.; El-Hamouly, W.S. Tumor Anti–initiating Activity of Some Novel 3,4-Dihydropyrimidinones. Pharmacol. Rep. 2010, 61, 1153–1162. [Google Scholar] [CrossRef] [Green Version]
- Bassyouni, F.A.; Abu-Baker, S.M.; K, M.; Moharam, M.E.; El-Nakkady, S.S.; Rehim, M.A. Synthesis and biological evaluation of some new triazolo[ 1,5 a] quinoline derivatives as anticancer and antimicrobial. RSC Adv. 2014, 4, 24131–24141. [Google Scholar] [CrossRef]
- Bhat, A.R.; Dongre, R.S.; Naikoo, G.A.; Hassan, I.U.; Ara, T. Proficient synthesis of bioactive annulated pyrimidine derivatives. J. Taibah Univ. Sci. 2017, 11, 1047–1069. [Google Scholar] [CrossRef] [Green Version]
- Ziarani, G.M.; Aleali, F.; Lashgari, N. Recent applications of barbituric acid in multicomponent reactions. RSC Adv. 2016, 6, 50895–50922. [Google Scholar] [CrossRef]
- Salama, A.A.; Ibrahim, B.M.; Mahmoud, S.S.; Yassin, N.A.; El-din, A.A.; Hegazy, M.E.; Bassyouni, F. effects of chiliadenus montanus extract on streptozotocin induced diabetes and its liver complication in rats. Plant Arch. 2020, 20, 7301–7308. [Google Scholar]
- Salama, A.A.; Ibrahim, B.M.; Yassin, N.A.; Mahmoud, S.S.; El-Din, A.G.; Shaffie, N.A. Regulatory Effects of Morus alba Aqueous Leaf Extract in Streptozotocin-Induced Diabetic Nephropathy. Pharma Chem. 2017, 9, 46–52. [Google Scholar]
- Soliman, S.M.; MSheta, N.; MMIbrahim, B.; MEl-Shawwa, M.; MAbd El-Halim, S. Novel intranasal drug delivery: Geraniol charged polymeric mixed micelles for targeting cerebral insult as a result of Ischaemia/Reperfusion. Pharmaceutics 2020, 12, 76. [Google Scholar] [CrossRef] [Green Version]
- Ajayi, F.A.; Olorunnisola, O.S.; Adetutu, A.; Olorunfemi, F.G.; Owoade, A.O.; Adegbola, P.; Afolabi, O.K. Antihyperglycaemic and Mode of Action of Thaumatococcus danielli (BENN.) BENTH Ethanol Leave Extract in Streptozotocin-induced Diabetic Rats. AJRIMPS 2019, 6, 1–10. [Google Scholar]
- Ahmed, O.M.; Hussein, A.M.; Ahmed, R.R. Antidiabetic and antioxidant effects of newly synthesized pyrimido[1,6-a]pyrimidinederivatives in neonatal streptozotocin-induced diabetic Rats. Med Chem. 2012, 2, 20–28. [Google Scholar] [CrossRef] [Green Version]
- Mansour, D.F.; Abdallah, H.M.; Ibrahim, B.M.; Hegazy, R.R.; Esmail, R.S.; Abdel-Salam, L.O. The carcinogenic agent diethylnitrosamine induces early oxidative stress, inflammation and proliferation in rat liver, stomach and colon: Protective effect of ginger extract. Asian Pac. J. Cancer Prev. 2019, 20, 2551–2561. [Google Scholar] [CrossRef] [PubMed]
- Mostafa, R.E.; Ibrahim, B.M.; Abdel Jaleel, G.A. Neuro-protective effects of Ginkgo biloba leaves extract on cerebral ischemia-reperfusion injury induced experimentally in ovariectomized rats. Int. J. Pharm. Pharm. Sci. 2016, 8, 237–242. [Google Scholar]
- Abeer, A.A.S.; Noha, N.Y. A Cytoprotectant Effect of Morus alba against Streptozotocin-Induced Diabetic Damage in Rat Brains. Der Pharma Chem. 2017, 9, 24–30. [Google Scholar]
- Kassem, A.A.; Abd El-Alim, S.H.; Basha, M.; Salama, A. Phospholipid complex enriched micelles: A novel drug delivery approach for promoting the antidiabetic effect of repaglinide. Eur. J. Pharm. Sci. 2017, 99, 75–84. [Google Scholar] [CrossRef]
- Salama, A.A.A.; Allam, R.M. Promising targets of chrysin and daidzein in colorectal cancer: Amphiregulin, CXCL1, and MMP-9. Eur. J. Pharmacol. 2021, 892, 173763. [Google Scholar] [CrossRef]
- Salama, A.A.A.; Mostafa, R.; Omara, E. Effects of phosphodiestrase type 5 inhibitors in epinephrine-induced arrhythmia in rats: Involvement of lactate dehydrogenase and creatine kinase downregulation and adiponectin expression. Hum. Exp. Toxicol. 2018, 37, 256–264. [Google Scholar] [CrossRef]
- Salama, A.A.A.; Arbid, M.S. Protective effect of Al-hagi graecorum in alloxan- induced diabetic rats. Der Pharma Chem. 2016, 8, 8–15. [Google Scholar]
- EL-Kassaby, M.; Salama, A.A.A.; Mourad, H.H.; Abdel-Wahha, K.G. Effect of lemon balm (Melissa officinalis) aqueous extract on streptozotocin-induced diabetic rats. Egypt. Pharm. J. 2019, 18, 296–303. [Google Scholar] [CrossRef]
- Afifi, N.A.; Ramadan, A.; El-Eraky, W.; Salama, A.A.A.; Amany, A. El-Fadaly and Azza Hassan. Quercetin protects against thioacetamide induced hepatotoxicity in rats through decreased oxidative stress biomarkers, the inflammatory cytokines; (TNF-α), (NF-κ B) and DNA fragmentation. Der Pharma Chem. 2016, 8, 48–55. [Google Scholar]
- Elhenawy, A.A.; Salama, A.A.A.; Abdel All, M.M.; Alomri, A.A.; Nassar, H.S. Synthesis, Characterization and Discovery Novel Anti-diabetic and Anti-hyperlipidemic Thiazolidinedione Derivatives. Int. J. Pharm. Sci. Rev. Res. 2015, 31, 23. [Google Scholar]
- Ibrahim, B.M.M.; Salama, A.A.A.; Yassin, N.A.; Mahmoud, S.S.; El-Din, A.A.G.; Shaffie, N.A.; Baker, D.H.A. Potential effects of glimepiride and a herbal mixture on hyperglycaemia, hypercholesterolaemia and oxidative stress. Plant Arch. 2020, 20, 2242–2248. [Google Scholar]
- El-Baz, F.K.; Salama, A.; Salama, R.A.A. Dunaliella salina Attenuates Diabetic Neuropathy Induced by STZ in Rats: Involvement of Thioredoxin. BioMed Res. Int. Vol. 2020, 1295492. [Google Scholar] [CrossRef] [Green Version]
- Salama, A.; Hegazy, R.; Hassan, A. Intranasal Chromium Induces Acute Brain and Lung Injuries in Rats: Assessment of Different Potential Hazardous Effects of Environmental and Occupational Exposure to Chromium and Introduction of a Novel Pharmacological and Toxicological Animal Model. PLoS ONE 2016, 20, e0168688. [Google Scholar] [CrossRef] [PubMed]
- Drury, R.A.B.; Wallington, E.A. Cancerson Carleton’s Histological Technique, 4th ed.; Oxford University Press: London, UK, 1980. [Google Scholar]
- Feldman, A.T.; Wolfe, D. Tissue processing and hexatomoxylin and eosin staining. Histopathol. Methods Protoc. 1976, 1180. [Google Scholar] [CrossRef]
- Kumar, S.; Lim, S.M.; Ramasamy, K.; Vasudevan, M.; Shah, S.A.; Selvaraj, M.; Narasimhan, B. Synthesis, molecular docking and biological evaluation of bis-pyrimidine Schiff base derivatives. Chem. Cent. J. 2017, 11, 1–16. [Google Scholar] [CrossRef] [Green Version]
- El-Samahy, M.A.; Mohamed, S.A.; Rehim, M.H.; Mohram, M.E. Synthesis of hybrid paper sheets with enhanced air barrier and antimicrobial properties for food packaging. Carbohydr. Polym. 2017, 168, 212–219. [Google Scholar] [CrossRef]
- Althagafi, I.; El-Metwaly, N.; Farghaly, T.A. New Series of Thiazole Derivatives: Synthesis, Structural Elucidation, Antimicrobial Activity, Molecular Modeling and MOE Docking. Molecules 2019, 24, 1741. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Sze, K.H.; Lu, G.; Ballester, P.J. Machine-learning scoring functions for structure-based drug lead optimization. WIREs Comput. Mol. Sci. 2020, 10, e1465. [Google Scholar] [CrossRef] [Green Version]
- Wójcikowski, M.; Ballester, P.J.; Siedlecki, P. Performance of machine-learning scoring functions in structure-based virtual screening. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Li, Y.; Cheng, T.; Liu, Z.; Wang, R. Evaluation of the performance of four molecular docking programs on a diverse set of protein-ligand complexes. J. Comput. Chem. 2010, 31, 2109–2125. [Google Scholar] [CrossRef] [PubMed]
- Hargrove, T.Y.; Friggeri, L.; Wawrzak, Z.; Qi, A.; Hoekstra, W.J.; Schotzinger, R.J.; York, J.D.; Guengerich, F.P.; Lepesheva, G.I. Structural analyses of Candida albicans sterol 14α-demethylasecomplexed with azole drugs address the molecular basis of azole-mediated inhibition of fungal sterol biosynthesis. J. Biol. Chem. 2017, 292, 6728–6743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, B.R.; Kim, H.Y.; Choi, I.; Kim, J.B.; Jin, C.H.; Han, A.R. DPP-IV Inhibitory Potentials of Flavonol Glycosides Isolated from the Seeds of Lens culinaris: In Vitro and Molecular Docking Analyses. Molecules 2018, 23, 1998. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [Green Version]
- Gfeller, D.; Michielin, O.; Zoete, V. Shaping the interaction landscape of bioactive molecules. Bioinformatics 2013, 29, 3073–3079. [Google Scholar] [CrossRef]
- Schrödinger Release 2015-1: Maestro; Schrödinger, LLC.: New York, NY, USA, 2015.
- Forli, S.; Huey, R.; Pique, M.E.; Sanner, M.F.; Goodsell, D.S.; Olson, A.J. Computational protein–ligand docking and virtual drug screening with the AutoDock suite. Nat. Protoc. 2016, 11, 905–919. [Google Scholar] [CrossRef] [Green Version]





| Groups | Glucose Level (mg/dL) | Total Antioxidant Capacity (mM/L) | α-Amylase Level (U/L) |
|---|---|---|---|
| Negative control | 119.1 ± 5.83 | 4.96 ± 0.04 | 104.5 ± 1.569 |
| Positive control (STZ 45 mg/kg) | 408.3 ± 36.32 a | 4.65 ± 0.01 a | 317.4 ± 12.86 a |
| Glimepiride (0.1 mg/kg) | 121.5 ± 11.31 b | 5.65 ± 0.07 a,b | 140.2 ± 10.6 a,b |
| Compound 4(0.004 mg/kg) | 122.4 ± 3.2 b | 4.69 ± 0.004 a | 117.6 ± 1.51 a,b |
| Compound 5(0.006 mg/kg) | 116.5 ± 7.2 b | 4.76 ± 0.01 a | 78.41 ± 1.04 a,b |
| Groups | Total Cholesterol (TC) Level (mg/dL) | High Density Lipoprotein Cholesterol (HDL-C) Level (mg/dL) |
|---|---|---|
| Negative control | 238.7 ± 3.92 | 385 ± 4.35 |
| Positive control (STZ 45 mg/kg) | 241.26 ± 7.97 | 200.75 ± 5.5 a |
| Glimepiride (0.1 mg/kg) | 237.47 ± 2.56 | 341.83 ± 0.34 a,b |
| Compound 4(0.004 mg/kg) | 244.6 ± 3.87 | 511.78 ± 24.21 a,b |
| Compound 5 (0.006 mg/kg) | 244.27 ± 3.03 | 491.98 ± 17.37 a,b |
| Inhibition Zone Diameter mm/mg | ||||
|---|---|---|---|---|
| Microorganism | Gram Strain | Compound 4 | Compound 5 | Reference Antibiotic |
| Bacillus cereus | positive | 10 | 13 | 30 |
| Escherichia coli | negative | 8 | 10 | 15 |
| Pseudomonas aeruginosa | negative | 10 | 12 | 20 |
| Staphylococcus aureus | positive | 10 | 10 | 30 |
| Salmonella typhimurium | negative | 9 | 10 | 15 |
| Candida albicans | yeast | 19 | 25 | 35 |
| Rhizopus oligosporus | fungus | 12 | 15 | 25 |
| Mucormiehei | fungus | 10 | 10 | 28 |
| Aspergillus niger | fungus | 10 | 10 | 25 |
| Microorganism | Clear Zone of Compound 5 Concentration (µL) | |||
|---|---|---|---|---|
| 5 µL | 10 µL | 15 µL | 20 µL | |
| E.coli | 10 | 10 | 10 | 10 |
| Salmonella typhimurium | 10 | 10 | 10 | 10 |
| Aspergillus niger | 10 | 15 | 17 | 15 |
| Compound | AMPC | CYP51 | DPP-IV |
|---|---|---|---|
| 4 | −7.7 | −7.3 | −8.2 |
| 5 | −7.2 | −7.2 | −8.2 |
| Native ligand | −6.5 (3-[(4-chloroanilino)sulfonyl] thiophene-2-carboxylic acid) | −10.7 (VT-1161) | −8.0 (Sitagliptin) |
| Compound | AMPC | CYP51 | DPP-IV |
|---|---|---|---|
| 4 | Hydrogen bonds (Asp123, Ser212, Tyr221) π-Stacking T(Tyr221) | Hydrogen bonds (Glu444, Ser453, Arg469) Salt bridge (Glu473) with tertamine group | Hydrogen bonds (Glu347, Ser376) Salt bridge with tertamine group (Glu378) |
| 5 | Hydrogen bonds (Ala194, Arg296, Asn346) | Hydrophobic (Leu461), Hydrogen bonds (Ser438), Ser453, Gly455 | Hydrogen bonds (Ser376, Glu378) Salt bridge (Glu378) |
| Native ligand | [3-[(4-chloroanilino)sulfonyl]thiophene-2-carboxylic acid] Hydrophobic (Tyr221), Hydrogen bonds (Ser64, Lys76, Asn152, Ala318), Water bridge (Asn346) | Composite ligand consists of Heme and (VT-1161) Hydrophobic interaction (Tyr118, Phe126, Ile304, Thr311, Pro375, Leu376, Phe380, Phe463, Ile471, Phe475, Ala476),Hydrogen bonds (Tyr118, tyr132,His377, His468) Halogen bond (Tyr132) Salt bridges (between carboxylate group and Lys143, Arg381, His468) Heme group forms metal complex (Cys470) | (Sitagliptin) Hydrophobic (Val711),Hydrogen bonds (Glu205,Glu206,Tyr662), Water bridge (Ser209), π-Stacking P(Phe357), P(Tyr662), T(Tyr666) Halogen bond (Ser209) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bassyouni, F.; Tarek, M.; Salama, A.; Ibrahim, B.; Salah El Dine, S.; Yassin, N.; Hassanein, A.; Moharam, M.; Abdel-Rehim, M. Promising Antidiabetic and Antimicrobial Agents Based on Fused Pyrimidine Derivatives: Molecular Modeling and Biological Evaluation with Histopathological Effect. Molecules 2021, 26, 2370. https://doi.org/10.3390/molecules26082370
Bassyouni F, Tarek M, Salama A, Ibrahim B, Salah El Dine S, Yassin N, Hassanein A, Moharam M, Abdel-Rehim M. Promising Antidiabetic and Antimicrobial Agents Based on Fused Pyrimidine Derivatives: Molecular Modeling and Biological Evaluation with Histopathological Effect. Molecules. 2021; 26(8):2370. https://doi.org/10.3390/molecules26082370
Chicago/Turabian StyleBassyouni, Fatma, Mohammad Tarek, Abeer Salama, Bassant Ibrahim, Sawsan Salah El Dine, Nemat Yassin, Amina Hassanein, Maysa Moharam, and Mohamed Abdel-Rehim. 2021. "Promising Antidiabetic and Antimicrobial Agents Based on Fused Pyrimidine Derivatives: Molecular Modeling and Biological Evaluation with Histopathological Effect" Molecules 26, no. 8: 2370. https://doi.org/10.3390/molecules26082370






