Quantitative Label-Free Comparison of the Metabolic Protein Fraction in Old and Modern Italian Wheat Genotypes by a Shotgun Approach
Abstract
:1. Introduction
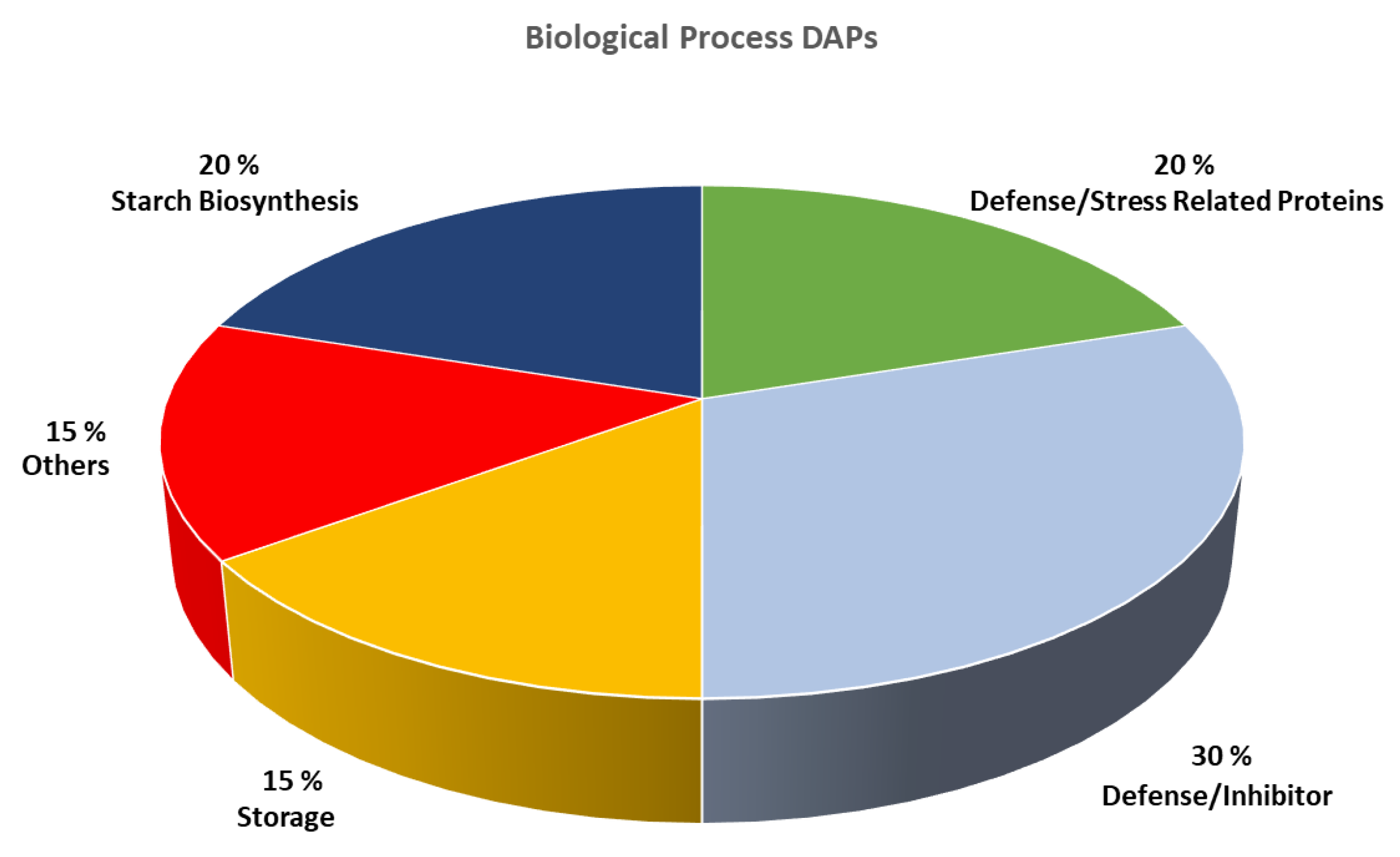
2. Results
Differentially Abundant Proteins Genotype-Related
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Samples Collection and Treatment
4.3. Mass Spectrometry Analysis
4.4. Database Search and Protein Identification
4.5. Label-Free Quantification
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| MS | Mass Spectrometry |
| CD | Celiac Deasease |
| IgE | Immunoglobulin E |
| NCWS | Non Celiac Wheat Sentivity |
| DAP | Differentially Abundant Proteins |
| WSCI | Subtilisin Chymotrypsin Inhibitor |
| ALPs | Avenin-Like Proteins |
| SUS1 | Sucrose Synthase 1 |
| SUS3 | Sucrose Synthase 3 |
| LOX | Linoleate Lipoxygenase |
| FA | Formic Acid |
| EDTA | Ethylenediaminetetraacetic acid |
| DTT | Dithiothreitol |
| IAA | Iodoacetamide |
| ACN | Acetonitrile |
| c-RAP | common Repository of Adventitious Proteins |
| PSMs | Peptide Spectral Matches |
| FDR | False Discovery Rate |
| RT | Retention Time |
References
- Cunsolo, V.; Foti, S.; Saletti, R. Mass spectrometry in the characterisation of cereal seed proteins. Eur. J. Mass Spectrom. 2004, 10, 359–370. [Google Scholar] [CrossRef]
- Cunsolo, V.; Muccilli, V.; Saletti, R.; Foti, S. Mass spectrometry in the proteome analysis of mature cereal kernels. Mass Spectrom. Rev. 2012, 31, 448–465. [Google Scholar] [CrossRef]
- Bromilow, S.; Gethings, L.A.; Buckley, M.; Bromley, M.; Shewry, P.R.; Langridge, J.I.; Mills, E.N.C. A curated gluten protein sequence database to support development of proteomics methods for determination of gluten in gluten-free foods. J. Proteom. 2017, 163, 67–75. [Google Scholar] [CrossRef]
- Foti, S.; Maccarrone, G.; Saletti, R.; Roepstorff, P.; Gilbert, S.; Tatham, A.; Shewry, P. Verification of the cDNA Deduced Sequence of Glutenin Subunit 1Dx5 and an Mr58 000 Repetitive Peptide by Matrix-Assisted Laser Desorption/Ionisation Mass Spectrometry (MALDI-MS). J. Cereal Sci. 2000, 3, 173–183. [Google Scholar] [CrossRef]
- Cunsolo, V.; Foti, S.; Saletti, R.; Gilbert, S.; Tatham, A.S.; Shewry, P.R. Investigation and correction of the gene-derived sequence of glutenin subunit 1Dx2 by matrix-assisted laser desorption/ionisation mass spectrometry. Rapid Commun. Mass Spectrom. 2002, 16, 1911–1918. [Google Scholar] [CrossRef]
- Cunsolo, V.; Foti, S.; Saletti, R.; Gilbert, S.; Tatham, A.S.; Shewry, P.R. Structural studies of glutenin subunits 1Dy10 and 1Dy12 by matrix-assisted laser desorption/ionisation mass spectrometry and high-performance liquid chromatography/electrospray ionisation mass spectrometry. Rapid Commun. Mass Spectrom. 2003, 17, 442–454. [Google Scholar] [CrossRef]
- Cunsolo, V.; Foti, S.; Saletti, R.; Gilbert, S.; Tatham, A.S.; Shewry, P.R. Structural studies of the allelic wheat glutenin subunits 1Bx7 and 1Bx20 by matrix-assisted laser desorption/ionization mass spectrometry and high-performance liquid chromatography/electrospray ionization mass spectrometry. J. Mass Spectrom. 2004, 39, 66–78. [Google Scholar] [CrossRef]
- Mamone, G.; De Caro, S.; Di Luccia, A.; Addeo, F.; Ferranti, P. Proteomic-based analytical approach for the characterization of glutenin subunits in durum wheat. J. Mass Spectrom. 2009, 44, 1709–1723. [Google Scholar] [CrossRef]
- Labuschagne, M.; Masci, S.; Tundo, S.; Muccilli, V.; Saletti, R.; van Biljon, A. Proteomic Analysis of Proteins Responsive to Drought and Low Temperature Stress in a Hard Red Spring Wheat Cultivar. Molecules 2020, 25, 1366. [Google Scholar] [CrossRef] [Green Version]
- Muccilli, V.; Cunsolo, V.; Saletti, R.; Foti, S.; Margiotta, B.; Scossa, F.; Masci, S.; Lafiandra, D. Characterisation of a specific class of typical low molecular weight glutenin subunits of durum wheat by a proteomic approach. J. Cereal Sci. 2010, 51, 134–139. [Google Scholar] [CrossRef] [Green Version]
- Sestili, F.; Paoletti, F.; Botticella, E.; Masci, S.; Saletti, R.; Muccilli, V.; Lafiandra, D. Comparative proteomic analysis of kernel proteins of two high amylose transgenic durum wheat lines obtained by biolistic and Agrobacterium-mediated transformations. J. Cereal Sci. 2013, 58, 15–22. [Google Scholar] [CrossRef]
- Sestili, F.; Pagliarello, R.; Zega, A.; Saletti, R.; Pucci, A.; Botticella, E.; Masci, S.; Tundo, S.; Moscetti, I.; Foti, S.; et al. Enhancing grain size in durum wheat using RNAi to knockdown GW2 genes. Theor. Appl. Genet. 2019, 132, 419–429. [Google Scholar] [CrossRef]
- Vici, G.; Belli, L.; Biondi, M.; Polzonetti, V. Gluten free diet and nutrient deficiencies: A review. Clin. Nutr. 2016, 35, 1236–1241. [Google Scholar] [CrossRef]
- Sapone, A.; Bai, J.C.; Ciacci, C.; Dolinsek, J.; Green, P.H.R.; Hadjivassiliou, M.; Kaukinen, K.; Rostami, K.; Sanders, D.S.; Schumann, M.; et al. Spectrum of gluten-related disorders: Consensus on new nomenclature and classification. BMC Med. 2012, 10, 13. [Google Scholar] [CrossRef] [Green Version]
- McAllister, B.P.; Clarke, K.; Williams, E. A Comprehensive Review of Celiac Disease/Gluten-Sensitive Enteropathies. Clin. Rev. Allergy Immunol. 2018, 39–41. [Google Scholar] [CrossRef]
- Potter, M.D.E.; Walker, M.M.; Talley, N.J. Non-coeliac gluten or wheat sensitivity: Emerging disease or misdiagnosis? Med. J. Aust. 2017, 207, 211–215. [Google Scholar] [CrossRef]
- Boukid, F.; Folloni, S.; Sforza, S.; Vittadini, E.; Prandi, B. Current Trends in Ancient Grains-Based Foodstuffs: Insights into Nutritional Aspects and Technological Applications. Compr. Rev. Food Sci. Food Saf. 2018, 17, 123–136. [Google Scholar] [CrossRef] [Green Version]
- Khush, G.S. Green revolution: The way forward. Nat. Rev. Genet. 2001, 2, 815–822. [Google Scholar] [CrossRef]
- Shewry, P.R. Do ancient types of wheat have health benefits compared with modern bread wheat? J. Cereal Sci. 2018, 79, 469–476. [Google Scholar] [CrossRef]
- De Santis, M.A.; Giuliani, M.M.; Giuzio, L.; De Vita, P.; Lovegrove, A.; Shewry, P.R.; Flagella, Z. Differences in gluten protein composition between old and modern durum wheat genotypes in relation to 20th century breeding in Italy. Eur. J. Agron. 2017, 87, 19–29. [Google Scholar] [CrossRef]
- Prandi, B.; Tedeschi, T.; Folloni, S.; Galaverna, G.; Sforza, S. Peptides from gluten digestion: A comparison between old and modern wheat varieties. Food Res. Int. 2017, 91, 92–102. [Google Scholar] [CrossRef]
- De Santis, M.A.; Cunsolo, V.; Giuliani, M.M.; Di Francesco, A.; Saletti, R.; Foti, S.; Flagella, Z. Gluten proteome comparison among durum wheat genotypes with different release date. J. Cereal Sci. 2020, 96, 103092. [Google Scholar] [CrossRef]
- Di Francesco, A.; Saletti, R.; Cunsolo, V.; Svensson, B.; Muccilli, V.; De Vita, P.; Foti, S. Qualitative proteomic comparison of metabolic and CM-like protein fractions in old and modern wheat Italian genotypes by a shotgun approach. J. Proteom. 2020, 211, 103530. [Google Scholar] [CrossRef]
- Dupont, F.M.; Altenbach, S.B. Molecular and biochemical impacts of environmental factors on wheat grain development and protein synthesis. J. Cereal Sci. 2003, 38, 133–146. [Google Scholar] [CrossRef]
- Boggini, G.; Palumbo, M.; Calcagno, F. Characterization and utilization of sicilian landraces of durum wheat in breeding programs. In Wheat Genetic Resources: Meeting Diverse Needs; John Wiley & Sons: Hoboken, NJ, USA, 1990; pp. 223–234. [Google Scholar]
- Taranto, F.; D’Agostino, N.; Rodriguez, M.; Pavan, S.; Minervini, A.P.; Pecchioni, N.; Papa, R.; De Vita, P. Whole Genome Scan Reveals Molecular Signatures of Divergence and Selection Related to Important Traits in Durum Wheat Germplasm. Front. Genet. 2020, 11. [Google Scholar] [CrossRef] [Green Version]
- Muccilli, V.; Lo Bianco, M.; Cunsolo, V.; Saletti, R.; Gallo, G.; Foti, S. High Molecular Weight Gluten in Subunits in Some Durum Wheat Cultivars Investigated by Means of Mass Spectrometric Techniques. J. Agric. Food Chem. 2011, 59, 12226–12237. [Google Scholar] [CrossRef]
- Taranto, G.H.; Kvile, K.O.; Pitcher, T.J.; Morato, T. An Ecosystem Evaluation Framework for Global Seamount Conservation and Management. PLoS ONE 2012, 7, e42950. [Google Scholar] [CrossRef] [Green Version]
- Giancaspro, A.; Giove, S.L.; Zito, D.; Blanco, A.; Gadaleta, A. Mapping QTLs for Fusarium Head Blight Resistance in an Interspecific Wheat Population. Front. Plant Sci. 2016, 7, 1381. [Google Scholar] [CrossRef]
- Venora, G.; Blangiforti, S.; Castiglione, M.R.; Pignone, D.; Losavio, F.; Cremonini, R. Chromatin organisation and computer aided karyotyping of Triticum durum Desf. cv. Timilia. Caryologia 2002, 55, 91–98. [Google Scholar] [CrossRef] [Green Version]
- Bonman, J.M.; Bockelman, H.E.; Jin, Y.; Hijmans, R.J.; Gironella, A.I.N. Geographic distribution of stem rust resistance in wheat landraces. Crop. Sci. 2007, 47, 1955–1963. [Google Scholar] [CrossRef] [Green Version]
- Ouaja, M.; Aouini, L.; Bahri, B.; Ferjaoui, S.; Medini, M.; Marcel, T.C.; Hamza, S. Identification of valuable sources of resistance to Zymoseptoria tritici in the Tunisian durum wheat landraces. Eur. J. Plant Pathol. 2020, 156, 647–661. [Google Scholar] [CrossRef]
- Beres, B.L.; Rahmani, E.; Clarke, J.M.; Grassini, P.; Pozniak, C.J.; Geddes, C.M.; Porker, K.D.; May, W.E.; Ransom, J.K. A Systematic Review of Durum Wheat: Enhancing Production Systems by Exploring Genotype, Environment, and Management (G × E × M) Synergies. Front. Plant Sci. 2020, 11, 568657. [Google Scholar] [CrossRef]
- Benbow, H.R.; Jermiin, L.S.; Doohan, F.M. Serpins: Genome-Wide Characterisation and Expression Analysis of the Serine Protease Inhibitor Family in Triticum aestivum. G3 Genes Genomes Genet. 2019, 9, 2709–2722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, H.; He, M.; Zhu, C.; Yuan, L.L.; Dong, L.W.; Bian, Y.W.; Zhang, W.Y.; Yan, Y.M. Distinct metabolic changes between wheat embryo and endosperm during grain development revealed by 2D-DIGE-based integrative proteome analysis. Proteomics 2016, 16, 1515–1536. [Google Scholar] [CrossRef] [PubMed]
- Altenbach, S.B.; Chang, H.-C.; Simon-Buss, A. Deciphering the immunogenic potential of wheat flour: A reference map of the salt-soluble proteome from the US wheat Butte 86. Proteome Sci. 2020, 18, 8. [Google Scholar] [CrossRef] [PubMed]
- Sotkovsky, P.; Hubalek, M.; Hernychova, L.; Novak, P.; Havranova, M.; Setinova, I.; Kitanovicova, A.; Fuchs, M.; Stulik, J.; Tuckova, L. Proteomic analysis of wheat proteins recognized by IgE antibodies of allergic patients. Proteomics 2008, 8, 1677–1691. [Google Scholar] [CrossRef]
- Salcedo, G.; Quirce, S.; Diaz-Perales, A. Wheat Allergens Associated With Baker’s Asthma. J. Investig. Allergol. Clin. Immunol. 2011, 21, 81–92. [Google Scholar]
- Sander, I.; Rozynek, P.; Rihs, H.P.; van Kampen, V.; Chew, F.T.; Lee, W.S.; Kotschy-Lang, N.; Merget, R.; Bruening, T.; Raulf-Heimsoth, M. Multiple wheat flour allergens and cross-reactive carbohydrate determinants bind IgE in baker’s asthma. Allergy 2011, 66, 1208–1215. [Google Scholar] [CrossRef]
- Akagawa, M.; Handoyo, T.; Ishii, T.; Kumazawa, S.; Morita, N.; Suyama, K. Proteomic analysis of wheat flour allergens. J. Agric. Food Chem. 2007, 55, 6863–6870. [Google Scholar] [CrossRef] [PubMed]
- Salcedo, G.; Sánchez-Monge, R.; García-Casado, G.; Armentia, A.; Gomez, L.; Barber, D. The Cereal α-Amylase/Trypsin Inhibitor Family Associated with Bakers’ Asthma and Food Allergy, 2004 ed.; Mills, E.N.C., Shewry, P.R., Eds.; Plant Food Allergens: Oxford, UK, 2004; Volume 5, pp. 70–87. [Google Scholar]
- Xu, E.S.; Chen, M.M.; He, H.; Zhan, C.F.; Cheng, Y.H.; Zhang, H.S.; Wang, Z.F. Proteomic Analysis Reveals Proteins Involved in Seed Imbibition under Salt Stress in Rice. Front. Plant Sci. 2017, 7, 1070. [Google Scholar] [CrossRef] [Green Version]
- Khan, A.; Tabassum, B.; Aaliya, K.; Tariq, M.; Nasir, I.A.; Hassan, S.; Ismail, T.; Ali, N.; Ponya, Z. The effectiveness of recombinat chitinase obtained from barley (Hordeum vulgare L.) against potato pathogens. Appl. Ecol. Environ. Res. 2019, 17, 4147–4157. [Google Scholar] [CrossRef]
- Hou, A.Y.; Kakar, R.K.; Neeck, S.; Azarbarzin, A.A.; Kummerow, C.D.; Kojima, M.; Oki, R.; Nakamura, K.; Iguchi, T. The global precipitation measurement mission. Bull. Am. Meteorol. Soc. 2014, 95, 701–722. [Google Scholar] [CrossRef]
- Chen, X.Y.; Cao, X.Y.; Zhang, Y.J.; Islam, S.; Zhang, J.J.; Yang, R.C.; Liu, J.J.; Li, G.Y.; Appels, R.; Keeble-Gagnere, G.; et al. Genetic characterization of cysteine-rich type-b avenin-like protein coding genes in common wheat. Sci. Rep. 2016, 6, 30692. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.Q.; Li, M.; Guan, Y.B.; Li, L.; Sun, F.S.; Han, J.P.; Chang, J.L.; Chen, M.J.; Yang, G.X.; Wang, Y.S.; et al. Effects of an Additional Cysteine Residue of Avenin-like b Protein by Site-Directed Mutagenesis on Dough Properties in Wheat (Triticum aestivum L.). J. Agric. Food Chem. 2019, 67, 8559–8572. [Google Scholar] [CrossRef]
- Ma, F.Y.; Li, M.; Li, T.T.; Liu, W.; Liu, Y.Y.; Li, Y.; Hu, W.; Zheng, Q.; Wang, Y.Q.; Li, K.X.; et al. Overexpression of Avenin-Like b Proteins in Bread Wheat (Triticum aestivum L.) Improves Dough Mixing Properties by Their Incorporation into Glutenin Polymers. PLoS ONE 2013, 8, e66758. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Hu, X.; Islam, S.; She, M.; Peng, Y.; Yu, Z.; Wylie, S.; Juhasz, A.; Dowla, M.; Yang, R.; et al. New insights into the evolution of wheat avenin-like proteins in wild emmer wheat (Triticum dicoccoides). Proc. Natl. Acad. Sci. USA 2018, 115, 13312–13317. [Google Scholar] [CrossRef] [Green Version]
- Siedow, J.N. Plant lipoxygenase—Structure and function. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1991, 42, 145–188. [Google Scholar] [CrossRef]
- Verlotta, A.; De Simone, V.; Mastrangelo, A.M.; Cattivelli, L.; Papa, R.; Trono, D. Insight into durum wheat Lpx-B1: A small gene family coding for the lipoxygenase responsible for carotenoid bleaching in mature grains. BMC Plant Biol. 2010, 10, 263. [Google Scholar] [CrossRef] [Green Version]
- Lopez-Ribera, I.; La Paz, J.L.; Repiso, C.; Garcia, N.; Miquel, M.; Hernandez, M.L.; Martinez-Rivas, J.M.; Vicient, C.M. The Evolutionary Conserved Oil Body Associated Protein OBAP1 Participates in the Regulation of Oil Body Size. Plant Physiol. 2014, 164, 1237–1249. [Google Scholar] [CrossRef] [Green Version]
- Lin, H.; He, L.; Ma, B. A combinatorial approach to the peptide feature matching problem for label-free quantification. Bioinformatics 2013, 29, 1768–1775. [Google Scholar] [CrossRef] [Green Version]
- Calò, D.G.; Scotto, F.; Ravaglia, S. Contenuto proteico del grano e variabili agronomiche: Un’analisi statistica. Statistica 2002, 62, 501–513. [Google Scholar] [CrossRef]
- Bose, U.; Juhasz, A.; Broadbent, J.A.; Byrne, K.; Howitt, C.A.; Colgrave, M.L. Identification and Quantitation of Amylase Trypsin Inhibitors Across Cultivars Representing the Diversity of Bread Wheat. J. Proteome Res. 2020, 19, 2136–2148. [Google Scholar] [CrossRef] [PubMed]
- Camerlengo, F.; Frittelli, A.; Sparks, C.; Doherty, A.; Martignago, D.; Larre, C.; Lupi, R.; Sestili, F.; Masci, S. CRISPR-Cas9 Multiplex Editing of the alpha-Amylase/Trypsin Inhibitor Genes to Reduce Allergen Proteins in Durum Wheat. Front. Sustain. Food Syst. 2020, 4, 104. [Google Scholar] [CrossRef]
- Kalunke, R.M.; Tundo, S.; Sestili, F.; Camerlengo, F.; Lafiandra, D.; Lupi, R.; Larre, C.; Denery-Papini, S.; Islam, S.; Ma, W.; et al. Reduction of Allergenic Potential in Bread Wheat RNAi Transgenic Lines Silenced for CM3, CM16 and 0.28 ATI Genes. Int. J. Mol. Sci. 2020, 21, 5817. [Google Scholar] [CrossRef] [PubMed]
- Sievers, S.; Rohrbach, A.; Beyer, K. Wheat-induced food allergy in childhood: Ancient grains seem no way out. Eur. J. Nutr. 2019, 59, 2693–2707. [Google Scholar] [CrossRef] [PubMed]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef] [PubMed]
| Proteins | Gene Code | Growing Season 2010/2011 | Growing Season 2011/2012 | Fold Change | |
|---|---|---|---|---|---|
| Fold Change | Fold Change | ||||
| Russello vs. Simeto |  | 64 | |||
| Avenin-like b1 | AVLB1 | 0.12 | 0.12 | ||
| Oil body-associated protein 1A | OBP1A | 4.75 | 9.73 | ||
| Serpin-Z1C | SPZ1C * | 0.4 | 0.21 | ||
| Timilia vs. Simeto | |||||
| 26 kDa endochitinase 2 | CHI2 | 0.44 | 0.41 | ||
| a-amylase inhibitor 0.28 | IAA2 | 64 | 64 | ||
| Antifungal protein R (Fragment) | THHR | 0.27 | 0.32 | ||
| Avenin-like b1 | AVLB1 * | 3.33 | 2.5 | ||
| L-ascorbate peroxidase 2 cytosolic | APX2 * | 5.15 | 2.32 | ||
| Oil body-associated protein 1A | OBP1A | 4.6 | 9.09 | ||
| Subtilisin-chymotrypsin inhibitor WSCI | ICIW | 6.61 | 4.67 | 1 | |
| Sucrose synthase 1 | SUS1 | 6.31 | 3.95 | ||
| Sucrose synthase 3 | SUS3 | 3.13 | 2.1 | ||
| Timilia vs. Russello | |||||
| 26 kD aendochitinase 2 | CHI2 | 0.27 | 0.33 | ||
| a-amylase inhibitor WDAI-3 (Fragment) | IAA3 * | 0.39 | 0.39 | ||
| Avenin-like b1 | AVLB1 * | 38.49 | 25.39 | ||
| Linoleate 9S-lipoxygenase 1 | LOX1 | 2.42 | 2.28 | ||
| Serpin-Z2A | SPZ2A | 0.02 | 0.02 | ||
| Subtilisin-chymotrypsin inhibitor WSCI | ICIW | 6.74 | 5.81 | ||
| Sucrose synthase 1 | SUS1 | 3.36 | 2.05 | ||
| Sucrose synthase 3 | SUS3 | 3.11 | 2.3 | ||
| UniProt Acc. No. | Gene Code | Organism | Protein Description | Growing Season 2010/2011 | Growing Season 2011/2012 | Intra-Genotype DAPs * | Biological Process | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fold Change | p-Value | Significance | Peptides | Unique Peptides | Fold Change | p-Value | Significance | Peptides | Unique Peptides | ||||||
| Russello vs. Simeto | |||||||||||||||
| Q2A783 | AVLB1 | Wheat | Avenin-like b1 | 0.12 | 4.84 × 10−11 | 69.40 | 7 | 6 | 0.12 | 3.03 × 10−8 | 109.29 | 7 | 6 | Storage | |
| B4FFZ9 | OBP1A | Maize | Oil body-associated protein 1A | 4.75 | 7.00 × 10−5 | 64.98 | 5 | 5 | 9.73 | 3.64 × 10−14 | 88.47 | 4 | 4 | Others | |
| Q9ST58 | SPZ1C | Wheat | Serpin-Z1C | 0.40 | 1.55 × 10−7 | 48.73 | 12 | 5 | 0.21 | 1.09 × 10−8 | 105.97 | 11 | 3 | Y (Russello) | Defense/Inhibitor |
| Timilia vs. Simeto | |||||||||||||||
| P23951 | CHI2 | Barley | 26 kDa endochitinase 2 | 0.44 | 9.95 × 10−9 | 79.42 | 12 | 7 | 0.41 | 3.01 × 10−5 | 61.90 | 12 | 6 | Defense/Stress Related Proteins | |
| P01083 | IAA2 | Wheat | α-amylase inhibitor 0.28 | 64.00 | 3.55 × 10−15 | 68.66 | 4 | 3 | 64.00 | 1.13 × 10−10 | 101.12 | 6 | 5 | Defense/Inhibitor | |
| P33044 | THHR | Barley | Antifungal protein R (Fragment) | 0.27 | 4.72 × 10−18 | 101.88 | 2 | 2 | 0.32 | 8.77 × 10−7 | 69.64 | 2 | 2 | Defense/Stress Related Proteins | |
| Q2A783 | AVLB1 | Wheat | Avenin-like b1 | 3.33 | 3.82 × 10−9 | 95.07 | 21 | 21 | 2.50 | 7.97 × 10−6 | 55.45 | 21 | 21 | Y (Timilia) | Storage |
| Q9FE01 | APX2 | Rice | L-ascorbate peroxidase 2 cytosolic | 5.15 | 4.43 × 10−8 | 87.16 | 2 | 2 | 2.32 | 2.13 × 10−5 | 51.24 | 2 | 2 | Y (Timilia) | Defense/Stress Related Proteins |
| B4FFZ9 | OBP1A | Maize | Oil body-associated protein 1A | 4.60 | 4.17 × 10−9 | 97.23 | 5 | 5 | 9.09 | 5.73 × 10−9 | 106.51 | 4 | 4 | Others | |
| P82977 | ICIW | Wheat | Subtilisin-chymotrypsin inhibitor WSCI | 6.61 | 7.87 × 10−9 | 36.55 | 2 | 2 | 4.67 | 6.87 × 10−8 | 84.71 | 3 | 3 | Defense/ Inhibitor | |
| P31922 | SUS1 | Barley | Sucrose synthase 1 | 6.31 | 1.37 × 10−7 | 94.78 | 10 | 2 | 3.95 | 9.51 × 10−7 | 51.48 | 11 | 3 | Starch biosynthesis | |
| Q43009 | SUS3 | Rice | Sucrosesynthase 3 | 3.13 | 3.21 × 10−7 | 77.26 | 7 | 2 | 2.10 | 9.56 × 10−5 | 42.12 | 10 | 2 | Starch biosynthesis | |
| Timilia vs. Russello | |||||||||||||||
| P23951 | CHI2 | Barley | 26 kD aendochitinase 2 | 0.27 | 3.29 × 10−4 | 53.12 | 17 | 10 | 0.33 | 4.27 × 10−8 | 87.68 | 12 | 6 | Defense/Stress Related Proteins | |
| P10846 | IAA3 | Wheat | α-amylase inhibitor WDAI-3 (Fragment) | 0.39 | 1.45 × 10−3 | 36.20 | 9 | 4 | 0.39 | 3.91 × 10−6 | 33.03 | 8 | 3 | Y (Timilia) | Defense/Inhibitor |
| Q2A783 | AVLB1 | Wheat | Avenin-like b1 | 38.49 | 2.37 × 10−11 | 99.23 | 8 | 6 | 25.39 | 1.84 × 10−8 | 119.91 | 8 | 6 | Y (Timilia) | Storage |
| P29114 | LOX1 | Barley | Linoleate 9S-lipoxygenase 1 | 2.42 | 1.31 × 10−6 | 42.63 | 8 | 8 | 2.28 | 2.30 × 10−6 | 55.34 | 9 | 9 | Others | |
| Q9ST57 | SPZ2A | Wheat | Serpin-Z2A | 0.02 | 4.04 × 10−6 | 96.72 | 7 | 3 | 0.02 | 3.47 × 10−13 | 94.42 | 5 | 2 | Defense/Inhibitor | |
| P82977 | ICIW | Wheat | Subtilisin-chymotrypsin inhibitor WSCI | 6.74 | 4.00 × 10−9 | 79.02 | 2 | 2 | 5.81 | 2.35 × 10−8 | 96.24 | 3 | 3 | Defense/Inhibitor | |
| P31922 | SUS1 | Barley | Sucrose synthase 1 | 3.36 | 3.11 × 10−7 | 68.34 | 13 | 3 | 2.05 | 2.74 × 10−5 | 45.72 | 12 | 4 | Starch biosynthesis | |
| Q43009 | SUS3 | Rice | Sucrose synthase 3 | 3.11 | 6.36 × 10−7 | 58.15 | 7 | 2 | 2.30 | 1.03 × 10−5 | 50.21 | 7 | 2 | Starch biosynthesis | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di Francesco, A.; Cunsolo, V.; Saletti, R.; Svensson, B.; Muccilli, V.; De Vita, P.; Foti, S. Quantitative Label-Free Comparison of the Metabolic Protein Fraction in Old and Modern Italian Wheat Genotypes by a Shotgun Approach. Molecules 2021, 26, 2596. https://doi.org/10.3390/molecules26092596
Di Francesco A, Cunsolo V, Saletti R, Svensson B, Muccilli V, De Vita P, Foti S. Quantitative Label-Free Comparison of the Metabolic Protein Fraction in Old and Modern Italian Wheat Genotypes by a Shotgun Approach. Molecules. 2021; 26(9):2596. https://doi.org/10.3390/molecules26092596
Chicago/Turabian StyleDi Francesco, Antonella, Vincenzo Cunsolo, Rosaria Saletti, Birte Svensson, Vera Muccilli, Pasquale De Vita, and Salvatore Foti. 2021. "Quantitative Label-Free Comparison of the Metabolic Protein Fraction in Old and Modern Italian Wheat Genotypes by a Shotgun Approach" Molecules 26, no. 9: 2596. https://doi.org/10.3390/molecules26092596






