Microbiological Study in Petrol-Spiked Soil
Abstract
:1. Introduction
2. Results
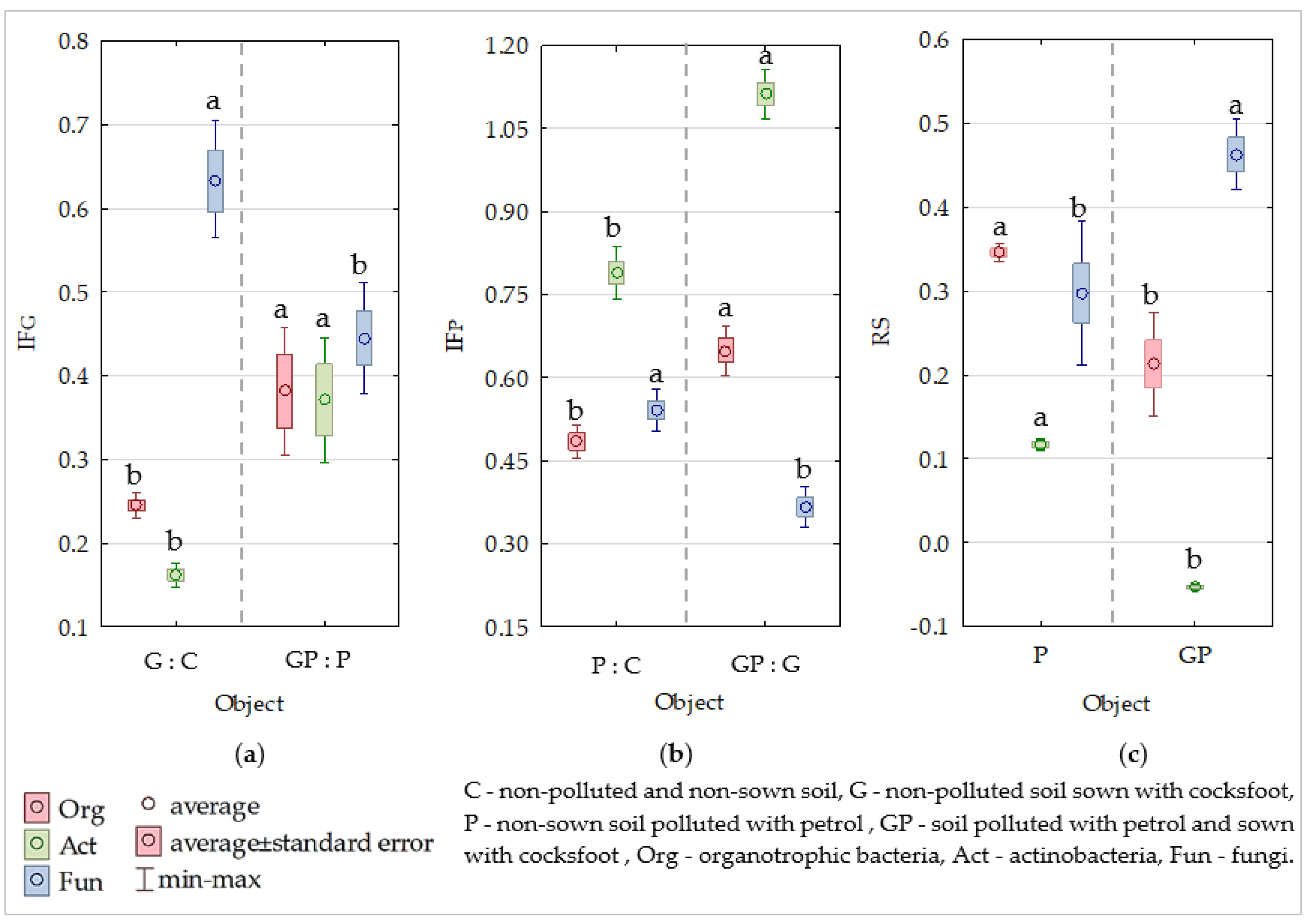
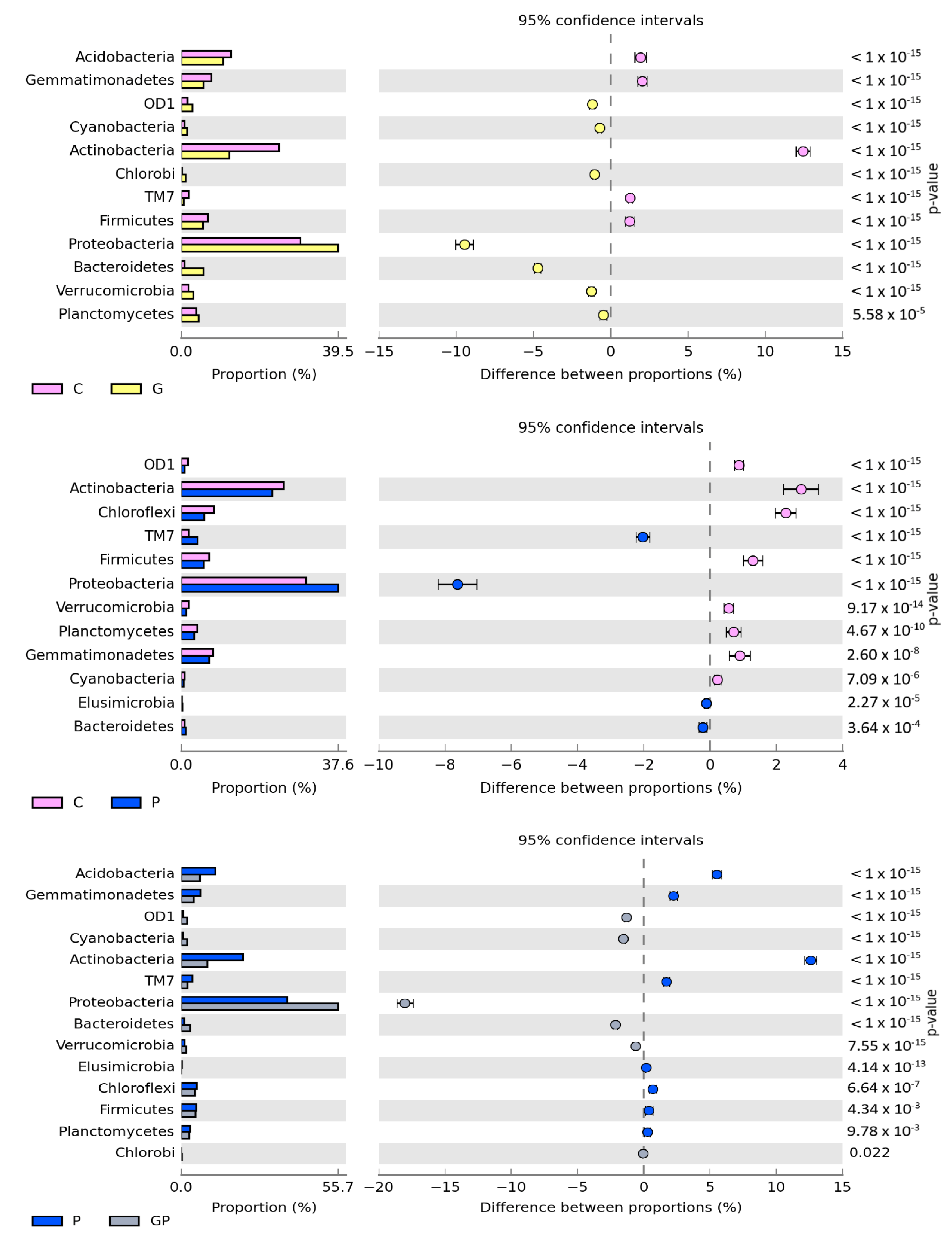
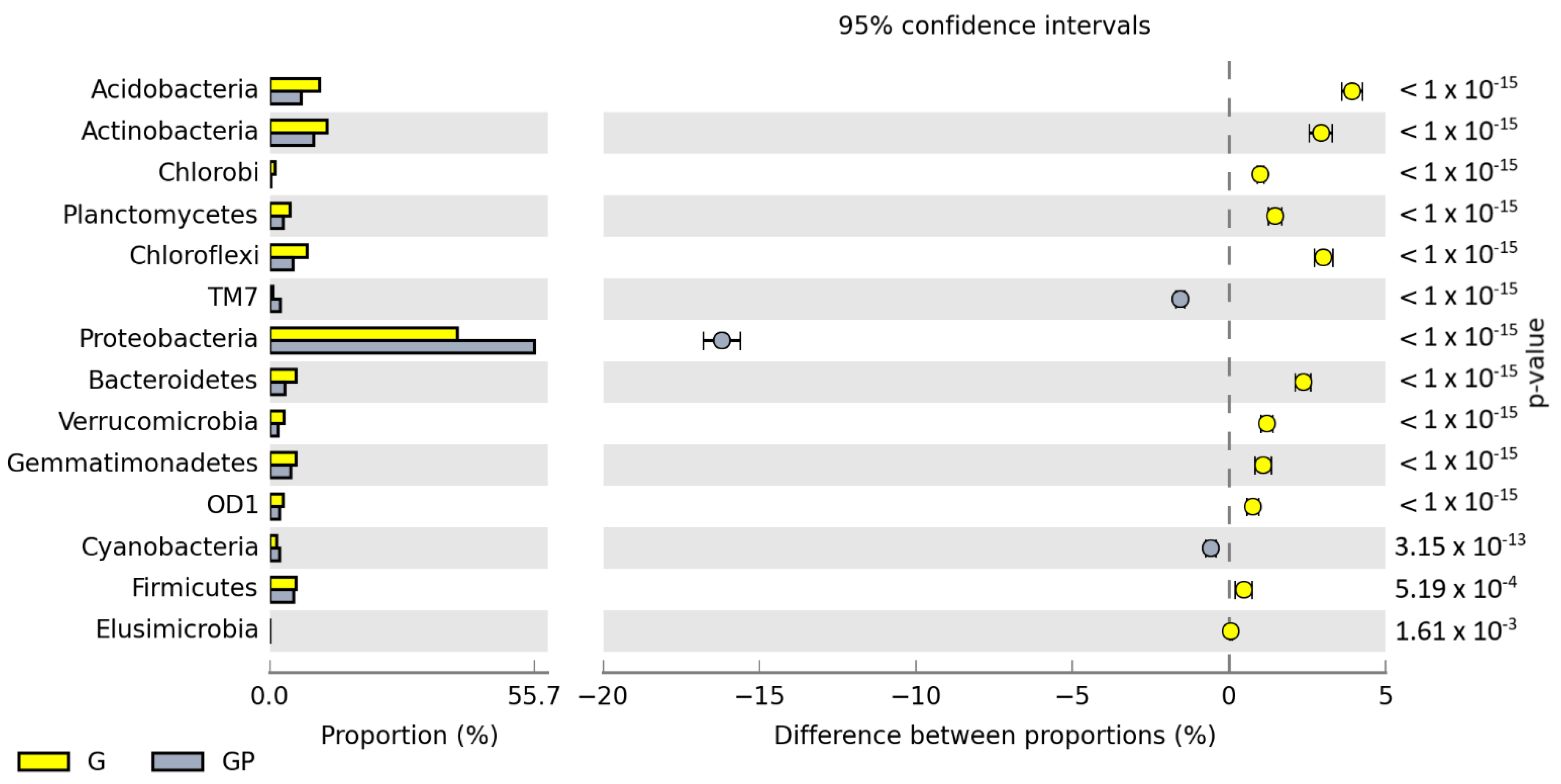
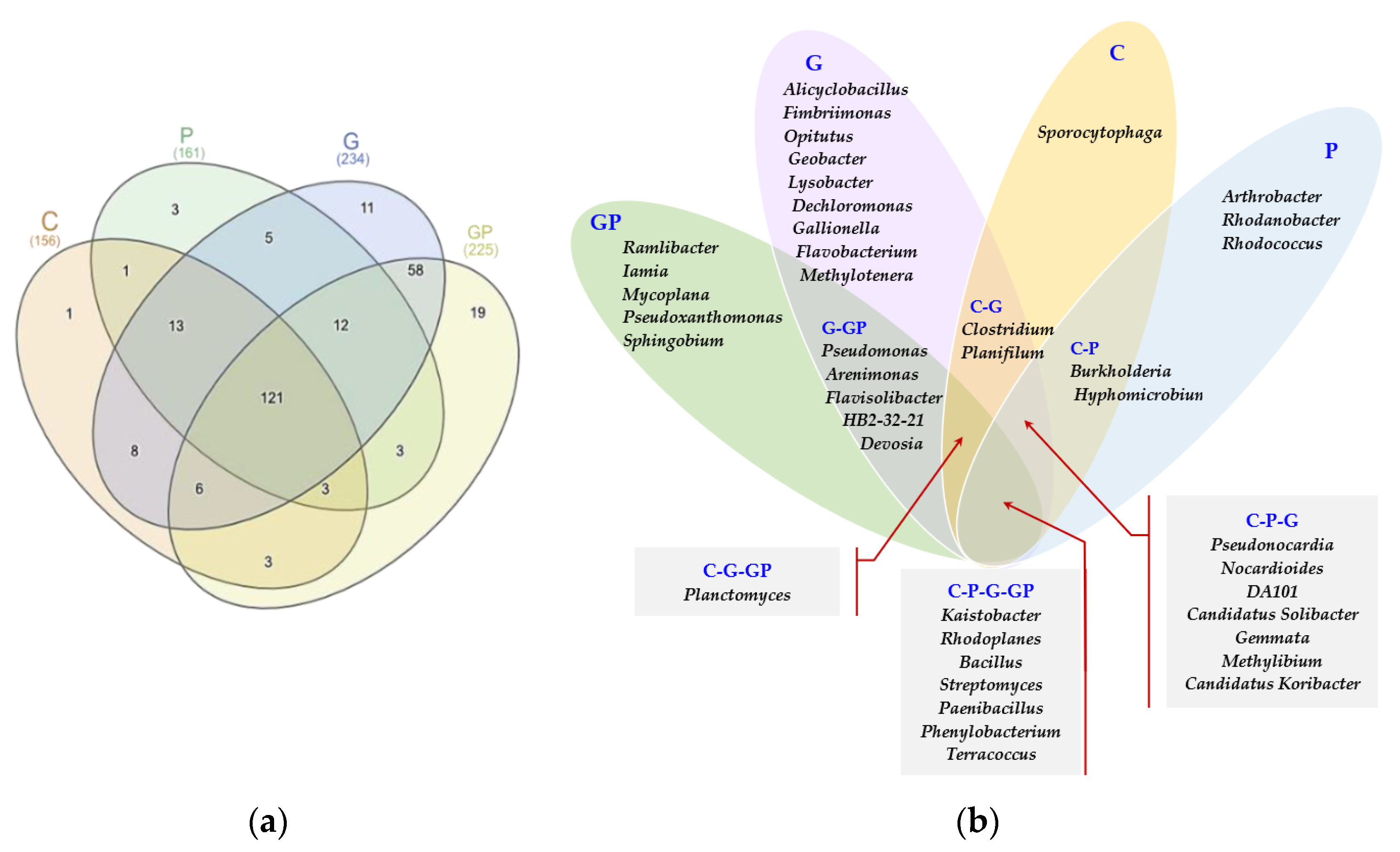
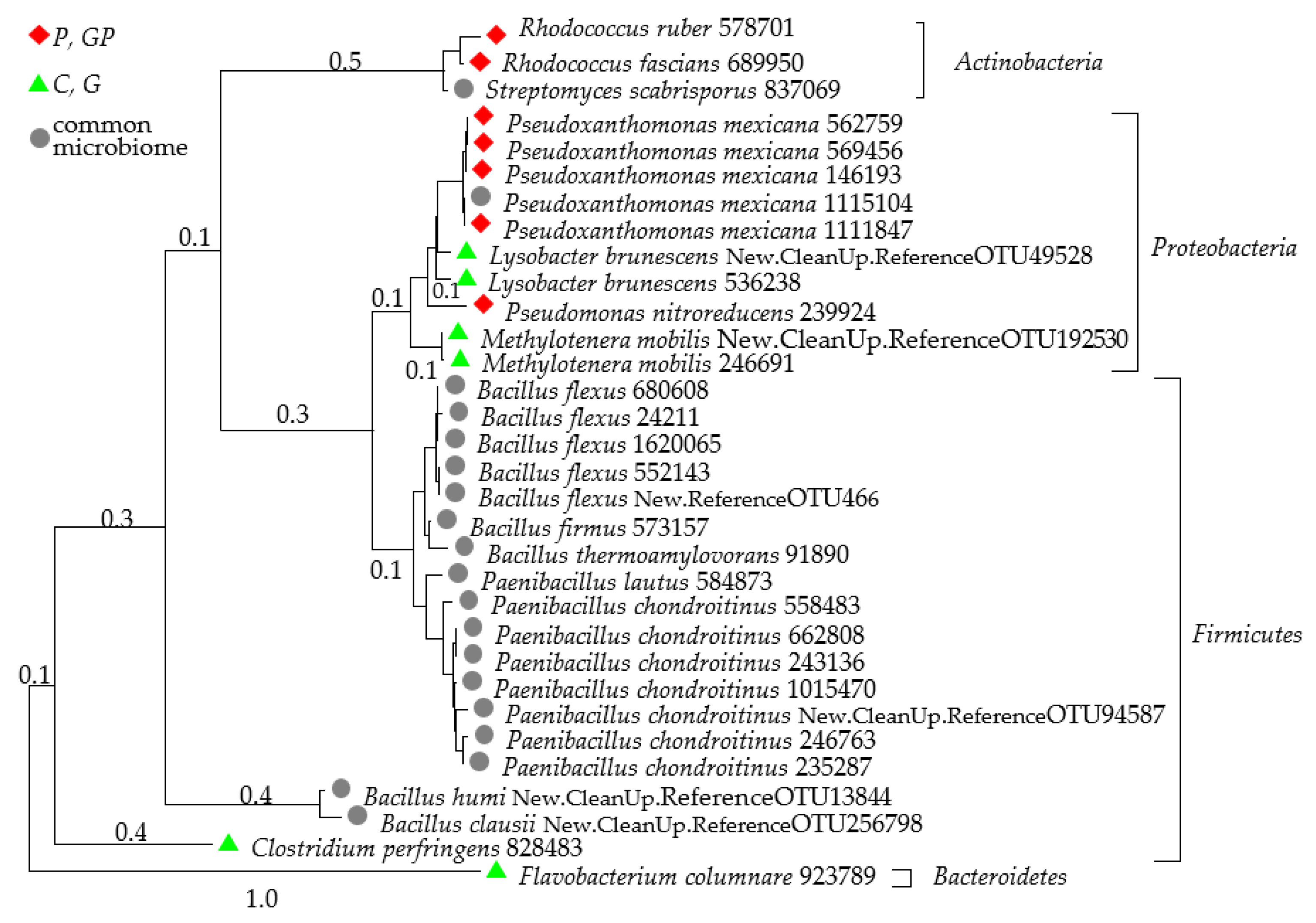
2.1. Microbiological Properties of Soil
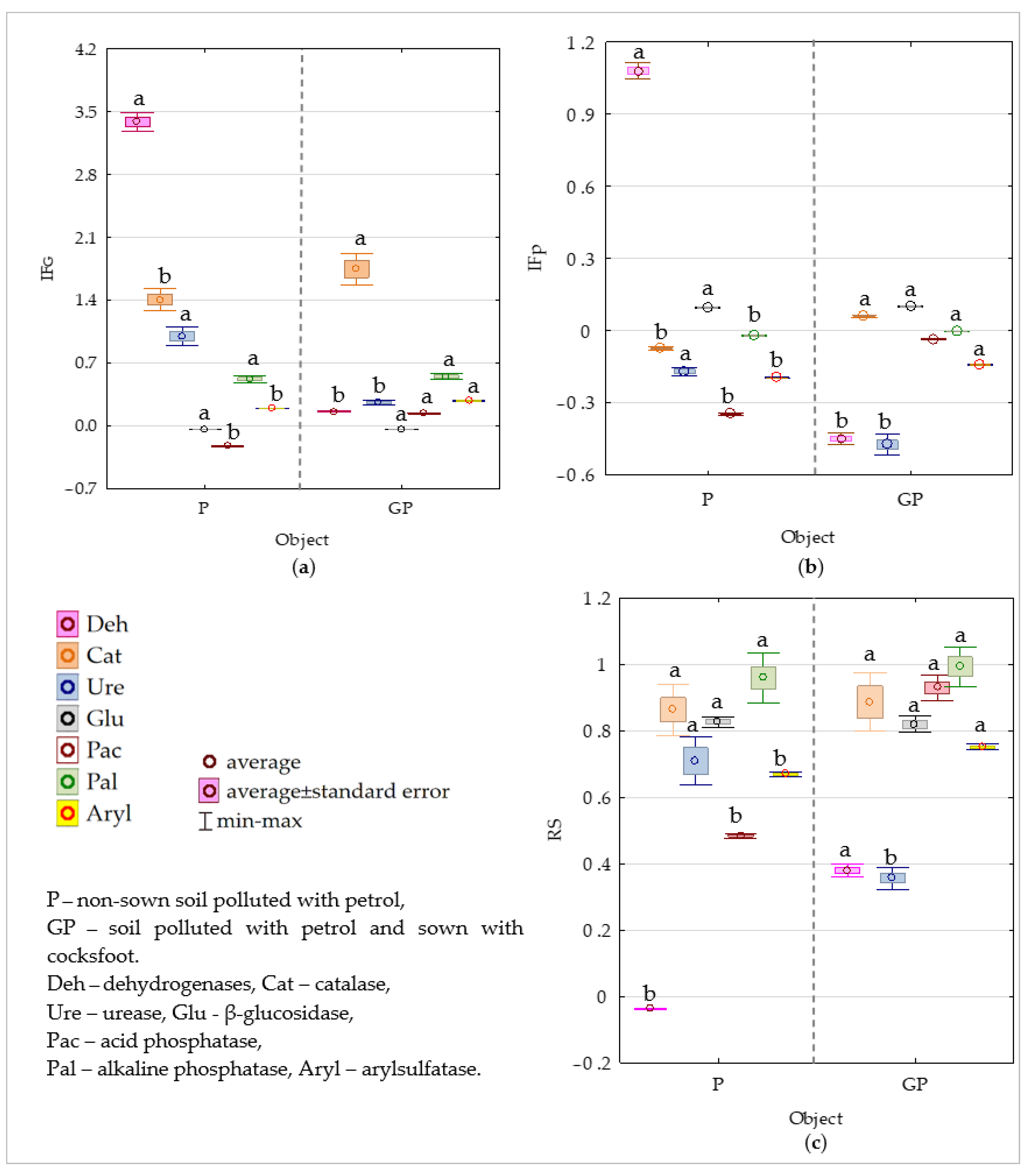
2.2. Biochemical Properties of Soil
2.3. Degradation of Petrol
3. Discussion
3.1. Response of Microorganisms and Enzymes to Soil Pollution with Petrol
3.2. The Role of Cocksfoot in Restoring the Biological Homeostasis of Petrol-Polluted Soil
4. Materials and Methods
4.1. Petrol
4.2. Field Studies
4.3. Greenhouse Experiment
4.4. Microbiological Analyses
4.4.1. Determination of Population Numbers of Microorganisms
4.4.2. DNA Isolation
4.4.3. 16S rRNA Gene Amplicon Sequencing
4.4.4. Bioinformatic Analysis
4.4.5. Biochemical, Chemical, and Physicochemical Analyses of Soil
4.4.6. Calculations and Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Erisman, J.W.; van Eekeren, N.; De Wit, J.; Koopmans, C.J.; Cuijpers, W.J.M.; Oerlemans, N.; Koks, B. Agriculture and biodiversity: A better balance benefits both. AIMS Agric. Food 2016, 1, 157–174. [Google Scholar] [CrossRef]
- Huera-Lucero, T.; Labrador-Moreno, J.; Blanco-Salas, J.; Ruiz-Téllez, T. A framework to incorporate biological soil quality indicators into assessing the sustainability of territories in the Ecuadorian Amazon. Sustainability 2020, 12, 3007. [Google Scholar] [CrossRef] [Green Version]
- Odukoya, J.; Lambert, R.; Sakrabani, R. Understanding the impacts of crude oil and its induced abiotic stresses on agrifood production: A Review. Horticulturae 2019, 5, 47. [Google Scholar] [CrossRef] [Green Version]
- Alengebawy, A.; Abdelkhalek, S.T.; Qureshi, S.R.; Wang, M.-Q. Heavy metals and pesticides toxicity in agricultural soil and plants: Ecological risks and human health implications. Toxics 2021, 9, 42. [Google Scholar] [CrossRef]
- Kumar, A.; Kumar, A.; da Silva Cabral Pinto, M.-M.; Chaturvedi, A.K.; Shabnam, A.A.; Subrahmanyam, G.; Mondal, R.; Gupta, D.K.; Malyan, S.K.; Kumar, S.S.; et al. Lead toxicity: Health hazards, influence on food chain, and sustainable remediation approaches. Int. J. Environ. Res. Public Health 2020, 17, 2179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garousin, H.; Pourbabaee, A.A.; Alikhani, H.A.; Yazdanfar, N. A combinational strategy mitigated old-aged petroleum contaminants: Ineffectiveness of biostimulation as a bioremediation technique. Front. Microbiol. 2021, 12. [Google Scholar] [CrossRef]
- Ławniczak, Ł.; Woźniak-Karczewska, M.; Loibner, A.P.; Heipieper, H.J.; Chrzanowski, Ł. Microbial degradation of hydrocarbons—basic principles for bioremediation: A review. Molecules 2020, 25, 856. [Google Scholar] [CrossRef] [Green Version]
- Varjani, S.; Upasani, V.N. Influence of abiotic factors, natural attenuation, bioaugmentation and nutrient supplementation on bioremediation of petroleum crude contaminated agricultural soil. J. Environ. Manag. 2019, 245, 358–366. [Google Scholar] [CrossRef]
- Hernández-Adame, N.M.; López-Miranda, J.; Martínez-Prado, M.A.; Cisneros-de la Cueva, S.; Rojas-Contreras, J.A.; Medrano-Roldán, H. Increase in total petroleum hydrocarbons removal rate in contaminated mining soil through bioaugmentation with autochthonous fungi during the slow bioremediation stage. Water Air Soil Pollut. 2021, 232, 95. [Google Scholar] [CrossRef]
- Jeyasundar, P.G.S.A.; Ali, A.; Azeem, M.; Li, Y.; Guo, D.; Sikdar, A.; Abdelrahman, H.; Kwon, E.; Antoniadis, V.; Mani, V.M.; et al. Green remediation of toxic metals contaminated mining soil using bacterial consortium and Brassica juncea. Environ. Pollut. 2021, 277, 116789. [Google Scholar] [CrossRef]
- Lipińska, A.; Wyszkowska, J.; Kucharski, J. Microbiological and biochemical activity in soil contaminated with pyrene subjected to bioaugmentation. Water Air Soil Pollut. 2021, 232, 45. [Google Scholar] [CrossRef]
- Varjani, S.J.; Gnansounou, E.; Pandey, A. Comprehensive review on toxicity of persistent organic pollutants from petroleum refinery waste and their degradation by microorganisms. Chemosphere 2017, 188, 280–291. [Google Scholar] [CrossRef]
- Blagodatskaya, E.; Kuzyakov, Y. Active microorganisms in soil: Critical review of estimation criteria and approaches. Soil Biol. Biochem. 2013, 67, 192–211. [Google Scholar] [CrossRef]
- Vestergaard, G.; Schulz, S.; Schöler, A.; Schloter, M. Making Big Data Smart—How to use metagenomics to understand soil quality. Biol. Fertil. Soils 2017, 53, 479–484. [Google Scholar] [CrossRef]
- Nesme, J.; Achouak, W.; Agathos, S.N.; Bailey, M.; Baldrian, P.; Brunel, D.; Frostegård, Å.; Heulin, T.; Jansson, J.K.; Jurkevitch, E.; et al. Back to the future of soil metagenomics. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef]
- Amorim, H.C.S.; Ashworth, A.J.; Wienhold, B.J.; Savin, M.C.; Allen, F.L.; Saxton, A.M.; Owens, P.R.; Curi, N. Soil quality indices based on long-term conservation cropping systems management. Agrosyst. Geosci. Environ. 2020, 3, e20036. [Google Scholar] [CrossRef] [Green Version]
- Andrea, F.; Bini, C.; Amaducci, S. Soil and ecosystem services: Current knowledge and evidences from Italian case studies. Appl. Soil Ecol. 2018, 123, 693–698. [Google Scholar] [CrossRef]
- Cardoso, E.J.B.N.; Vasconcellos, R.L.F.; Bini, D.; Miyauchi, M.Y.H.; dos Santos, C.A.; Alves, P.R.L.; de Paula, A.M.; Nakatani, A.S.; de Moraes Pereira, J.; Nogueira, M.A. Soil health: Looking for suitable indicators. What should be considered to assess the effects of use and management on soil health? Sci. Agric. 2013, 70, 274–289. [Google Scholar] [CrossRef] [Green Version]
- Juhos, K.; Czigány, S.; Madarász, B.; Ladányi, M. Interpretation of soil quality indicators for land suitability assessment—A multivariate approach for Central European arable soils. Ecol. Indic. 2019, 99, 261–272. [Google Scholar] [CrossRef]
- Cederlund, H.; Wessén, E.; Enwall, K.; Jones, C.; Juhanson, J.; Pell, M.; Philippot, L.; Hallin, S. Soil carbon quality and nitrogen fertilization structure bacterial communities with predictable responses of major bacterial phyla. Appl. Soil Ecol. 2014, 84, 62–68. [Google Scholar] [CrossRef]
- Deru, J.G.C.; Bloem, J.; de Goede, R.; Keidel, H.; Kloen, H.; Rutgers, M.; van den Akker, J.; Brussaard, L.; van Eekeren, N. Soil ecology and ecosystem services of dairy and semi-natural grasslands on peat. Appl. Soil Ecol. 2018, 125, 26–34. [Google Scholar] [CrossRef]
- Nelson, M.B.; Martiny, A.C.; Martiny, J.B.H. Global biogeography of microbial nitrogen-cycling traits in soil. Proc. Natl. Acad. Sci. USA 2016, 113, 8033–8040. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mori, T.; Wang, S.; Wang, Z.; Wang, C.; Mo, H.; Mo, J.; Lu, X. Testing potassium limitation on soil microbial activity in a sub-tropical forest. J. For. Res. 2019, 30, 2341–2347. [Google Scholar] [CrossRef]
- Pereira, D.G.C.; Santana, I.A.; Megda, M.M.; Megda, M.X.V.; Pereira, D.G.C.; Santana, I.A.; Megda, M.M.; Megda, M.X.V. Potassium chloride: Impacts on soil microbial activity and nitrogen mineralization. Ciência Rural 2019, 49. [Google Scholar] [CrossRef]
- Rousk, J.; Bååth, E.; Brookes, P.C.; Lauber, C.L.; Lozupone, C.; Caporaso, J.G.; Knight, R.; Fierer, N. Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 2010, 4, 1340–1351. [Google Scholar] [CrossRef]
- Shen, C.; Xiong, J.; Zhang, H.; Feng, Y.; Lin, X.; Li, X.; Liang, W.; Chu, H. Soil pH drives the spatial distribution of bacterial communities along elevation on changbai mountain. Soil Biol. Biochem. 2013, 57, 204–211. [Google Scholar] [CrossRef]
- Biological Characterization ISO Standards. Available online: https://www.iso.org/committee/54366/x/catalogue/completed (accessed on 23 February 2021).
- Bünemann, E.K.; Bongiorno, G.; Bai, Z.; Creamer, R.E.; De Deyn, G.; de Goede, R.; Fleskens, L.; Geissen, V.; Kuyper, T.W.; Mäder, P.; et al. Soil quality—A critical review. Soil Biol. Biochem. 2018, 120, 105–125. [Google Scholar] [CrossRef]
- Perez-Mon, C.; Frey, B.; Frossard, A. Functional and structural responses of arctic and alpine soil prokaryotic and fungal communities under freeze-thaw cycles of different frequencies. Front. Microbiol. 2020, 11. [Google Scholar] [CrossRef]
- Schloter, M.; Nannipieri, P.; Sørensen, S.J.; van Elsas, J.D. Microbial indicators for soil quality. Biol. Fertil. Soils 2018, 54, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Gunina, A.; Dippold, M.A.; Glaser, B.; Kuzyakov, Y. Fate of low molecular weight organic substances in an arable soil: From microbial uptake to utilisation and stabilisation. Soil Biol. Biochem. 2014, 77, 304–313. [Google Scholar] [CrossRef]
- Praeg, N.; Pauli, H.; Illmer, P. Microbial diversity in bulk and rhizosphere soil of ranunculus glacialis along a high-alpine altitudinal gradient. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef]
- Chaparro, J.M.; Sheflin, A.M.; Manter, D.K.; Vivanco, J.M. Manipulating the soil microbiome to increase soil health and plant fertility. Biol. Fertil. Soils 2012, 48, 489–499. [Google Scholar] [CrossRef]
- Huang, C.; Han, X.; Yang, Z.; Chen, Y.; Rengel, Z. Sowing methods influence soil bacterial diversity and community composition in a winter wheat-summer maize rotation system on the Loess Plateau. Front. Microbiol. 2020, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, Y.; Bing, H.; Fang, L.; Wu, Y.; Yu, J.; Shen, G.; Jiang, M.; Wang, X.; Zhang, X. Diversity patterns of the rhizosphere and bulk soil microbial communities along an altitudinal gradient in an alpine ecosystem of the Eastern Tibetan Plateau. Geoderma 2019, 338, 118–127. [Google Scholar] [CrossRef]
- Dos Santos, J.J.; Maranho, L.T. Rhizospheric microorganisms as a solution for the recovery of soils contaminated by petroleum: A review. J. Environ. Manag. 2018, 210, 104–113. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Adams, C.A.; Yang, W.; Sun, Y.; Shi, Z. Benefits of arbuscular mycorrhizal fungi in reducing organic contaminant residues in crops: Implications for cleaner agricultural production. Crit. Rev. Environ. Sci. Technol. 2020, 50, 1580–1612. [Google Scholar] [CrossRef]
- Berendsen, R.L.; Pieterse, C.M.J.; Bakker, P.A.H.M. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012, 17, 478–486. [Google Scholar] [CrossRef]
- Gumiere, T.; Rousseau, A.N.; da Costa, D.P.; Cassetari, A.; Cotta, S.R.; Andreote, F.D.; Gumiere, S.J.; Pavinato, P.S. Phosphorus source driving the soil microbial interactions and improving sugarcane development. Sci. Rep. 2019, 9, 4400. [Google Scholar] [CrossRef]
- Oliverio, A.M.; Bissett, A.; McGuire, K.; Saltonstall, K.; Turner, B.L.; Fierer, N. The role of phosphorus limitation in shaping soil bacterial communities and their metabolic capabilities. mBio 2020, 11. [Google Scholar] [CrossRef]
- Bidja Abena, M.T.; Li, T.; Shah, M.N.; Zhong, W. Biodegradation of total petroleum hydrocarbons (TPH) in highly contaminated soils by natural attenuation and bioaugmentation. Chemosphere 2019, 234, 864–874. [Google Scholar] [CrossRef]
- Camacho-Montealegre, C.M.; Rodrigues, E.M.; Tótola, M.R. Microbial diversity and bioremediation of rhizospheric soils from Trindade Island—Brazil. J. Environ. Manag. 2019, 236, 358–364. [Google Scholar] [CrossRef] [PubMed]
- Mafiana, M.O.; Kang, X.-H.; Leng, Y.; He, L.-F.; Li, S.-W. Petroleum contamination significantly changes soil microbial communities in three oilfield locations in Delta State, Nigeria. Environ. Sci. Pollut. Res. Int. 2021. [Google Scholar] [CrossRef] [PubMed]
- Mendes, L.W.; Tsai, S.M.; Navarrete, A.A.; de Hollander, M.; van Veen, J.A.; Kuramae, E.E. Soil-borne microbiome: Linking diversity to function. Microb. Ecol. 2015, 70, 255–265. [Google Scholar] [CrossRef] [PubMed]
- Allamin, I.A.; Halmi, M.I.E.; Yasid, N.A.; Ahmad, S.A.; Abdullah, S.R.S.; Shukor, Y. Rhizodegradation of petroleum oily sludge-contaminated soil using cajanus cajan increases the diversity of soil microbial community. Sci. Rep. 2020, 10, 4094. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bouchez, T.; Blieux, A.L.; Dequiedt, S.; Domaizon, I.; Dufresne, A.; Ferreira, S.; Godon, J.J.; Hellal, J.; Joulian, C.; Quaiser, A.; et al. Molecular microbiology methods for environmental diagnosis. Environ. Chem. Lett. 2016, 14, 423–441. [Google Scholar] [CrossRef]
- Hartmann, M.; Frey, B.; Mayer, J.; Mäder, P.; Widmer, F. Distinct soil microbial diversity under long-term organic and conventional farming. ISME 2015, 9, 1177–1194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borowik, A.; Wyszkowska, J.; Oszust, K. Changes in the functional diversity of bacterial communities in soil contaminated with diesel oil. J. Elem. 2018, 23, 1099–1117. [Google Scholar] [CrossRef]
- Stone, D.; Ritz, K.; Griffiths, B.G.; Orgiazzi, A.; Creamer, R.E. Selection of biological indicators appropriate for European soil monitoring. Appl. Soil Ecol. 2016, 97, 12–22. [Google Scholar] [CrossRef]
- Lynch, M.D.J.; Neufeld, J.D. Ecology and exploration of the rare biosphere. Nat. Rev. Microbiol. 2015, 13, 217–229. [Google Scholar] [CrossRef]
- Kandziora-Ciupa, M.; Nadgórska-Socha, A.; Barczyk, G. The influence of heavy metals on biological soil quality assessments in the Vaccinium myrtillus L. rhizosphere under different field conditions. Ecotoxicology 2021, 30, 292–310. [Google Scholar] [CrossRef]
- Zhu, H.; Teng, Y.; Wang, X.; Zhao, L.; Ren, W.; Luo, Y.; Christie, P. Changes in clover rhizosphere microbial community and diazotrophs in mercury-contaminated soils. Sci. Total Environ. 2021, 767, 145473. [Google Scholar] [CrossRef] [PubMed]
- Ite, A.E.; Ibok, U.J. Role of plants and microbes in bioremediation of petroleum hydrocarbons contaminated soils. Int. J. Environ. Bioremediat. Biodegrad. 2019, 7, 1–19. [Google Scholar] [CrossRef]
- Shen, H.; Huang, Y.; Wang, R.; Zhu, D.; Li, W.; Shen, G.; Wang, B.; Zhang, Y.; Chen, Y.; Lu, Y.; et al. Global atmospheric emissions of polycyclic aromatic hydrocarbons from 1960 to 2008 and future predictions. Environ Sci Technol. 2013, 47, 6415–6424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zaborowska, M.; Wyszkowska, J.; Borowik, A. Soil microbiome response to contamination with bisphenol A, bisphenol F and bisphenol S. Int. J. Mol. Sci. 2020, 21, 3529. [Google Scholar] [CrossRef] [PubMed]
- Ali, N.; Dashti, N.; Khanafer, M.; Al-Awadhi, H.; Radwan, S. Bioremediation of soils saturated with spilled crude oil. Sci. Rep. 2020, 10. [Google Scholar] [CrossRef] [Green Version]
- Shen, D.; Yuan, X.; Han, Y.; Mo, R.; Zhong, D.; Tang, F.; Liu, Y. Evaluation of Polycyclic Aromatic Hydrocarbons (PAHs) in bamboo shoots from soil. Bull. Environ. Contam. Toxicol. 2021. [Google Scholar] [CrossRef]
- Galitskaya, P.; Biktasheva, L.; Blagodatsky, S.; Selivanovskaya, S. Response of bacterial and fungal communities to high petroleum pollution in different soils. Sci. Rep. 2021, 11, 164. [Google Scholar] [CrossRef]
- Borowik, A.; Wyszkowska, J.; Kucharski, M.; Kucharski, J. Implications of soil pollution with diesel oil and BP petroleum with active technology for soil health. Int. J. Environ. Res. Public Health 2019, 16, 2474. [Google Scholar] [CrossRef] [Green Version]
- Nebeská, D.; Auer Malinská, H.; Erol, A.; Pidlisnyuk, V.; Kuráň, P.; Medžová, A.; Smaha, M.; Trögl, J. Stress response of miscanthus plants and soil microbial communities: A case study in metals and hydrocarbons contaminated soils. Appl. Sci. 2021, 11, 1866. [Google Scholar] [CrossRef]
- Benedek, T.; Vajna, B.; Táncsics, A.; Márialigeti, K.; Lányi, S.; Máthé, I. Remarkable impact of PAHs and TPHs on the richness and diversity of bacterial species in surface soils exposed to long-term hydrocarbon pollution. World J. Microbiol. Biotechnol. 2013, 29, 1989–2002. [Google Scholar] [CrossRef]
- Siles, J.A.; Margesin, R. Insights into microbial communities mediating the bioremediation of hydrocarbon-contaminated soil from an Alpine former military site. Appl. Microbiol. Biotechnol. 2018, 102, 4409–4421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhong, Z.; Zhang, X.; Wang, X.; Fu, S.; Wu, S.; Lu, X.; Ren, C.; Han, X.; Yang, G. Soil bacteria and fungi respond differently to plant diversity and plant family composition during the secondary succession of abandoned farmland on the Loess Plateau, China. Plant Soil 2020, 448, 183–200. [Google Scholar] [CrossRef]
- Tejeda-Agredano, M.C.; Gallego, S.; Vila, J.; Grifoll, M.; Ortega-Calvo, J.J.; Cantos, M. Influence of the sunflower rhizosphere on the biodegradation of PAHs in soil. Soil Biol. Biochem. 2013, 57, 830–840. [Google Scholar] [CrossRef] [Green Version]
- Kumar, V.; AlMomin, S.; Al-Aqeel, H.; Al-Salameen, F.; Nair, S.; Shajan, A. Metagenomic analysis of rhizosphere microflora of oil-contaminated soil planted with barley and alfalfa. PLoS ONE 2018, 13, e0202127. [Google Scholar] [CrossRef]
- De la Cueva, S.C.; Rodríguez, C.H.; Cruz, N.O.S.; Contreras, J.A.R.; Miranda, J.L. Changes in bacterial populations during bioremediation of soil contaminated with petroleum hydrocarbons. Water Air Soil Pollut. 2016, 227, 91. [Google Scholar] [CrossRef]
- Zhang, B.; Zhang, L.; Zhang, X. Bioremediation of petroleum hydrocarbon-contaminated soil by petroleum-degrading bacteria immobilized on biochar. RSC Adv. 2019, 9, 35304–35311. [Google Scholar] [CrossRef] [Green Version]
- Bhowmik, A.; Kukal, S.S.; Saha, D.; Sharma, H.; Kalia, A.; Sharma, S. Potential indicators of soil health degradation in different land use-based ecosystems in the Shiwaliks of Northwestern India. Sustainability 2019, 11, 3908. [Google Scholar] [CrossRef] [Green Version]
- Galitskaya, P.; Biktasheva, L.; Kuryntseva, P.; Selivanovskaya, S. Response of soil bacterial communities to high petroleum content in the absence of remediation procedures. Environ. Sci. Pollut. Res. 2021, 28, 9610–9627. [Google Scholar] [CrossRef]
- Borowik, A.; Wyszkowska, J.; Wyszkowski, M. Resistance of aerobic microorganisms and soil enzyme response to soil contamination with Ekodiesel Ultra Fuel. Environ. Sci. Pollut. Res. 2017, 24, 24346–24363. [Google Scholar] [CrossRef] [Green Version]
- Ramadass, K.; Megharaj, M.; Venkateswarlu, K.; Naidu, R. Ecotoxicity of measured concentrations of soil-applied diesel: Effects on earthworm survival, dehydrogenase, urease and nitrification activities. Appl. Soil Ecol. 2017, 119, 1–7. [Google Scholar] [CrossRef]
- Novakovskiy, A.B.; Kanev, V.A.; Markarova, M.Y. Long-term dynamics of plant communities after biological remediation of oil-contaminated soils in Far North. Sci. Rep. 2021, 11, 4888. [Google Scholar] [CrossRef]
- Alrumman, S.A.; Standing, D.B.; Paton, G.I. Effects of hydrocarbon contamination on soil microbial community and enzyme activity. J. King Saud Univ. Sci. 2015, 27, 31–41. [Google Scholar] [CrossRef] [Green Version]
- Dindar, E.; Topaç Şağban, F.O.; Başkaya, H.S. Variations of soil enzyme activities in petroleum-hydrocarbon contaminated soil. Int. Biodeter. Biodegr. 2015, 105, 268–275. [Google Scholar] [CrossRef]
- Curyło, K.; Telesiński, A.; Jarnuszewski, G.; Krzyśko-Łupicka, T.; Cybulska, K. Analysis of chemical and biochemical parameters of petrol-contaminated soil after biostimulation with an enzyme reagent. Processes 2020, 8, 949. [Google Scholar] [CrossRef]
- Kaczyńska, G.; Borowik, A.; Wyszkowska, J. Soil dehydrogenases as an indicator of contamination of the environment with petroleum products. Water Air Soil Pollut. 2015, 226, 372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aravindh, S.; Chinnadurai, C.; Balachandar, D. Development of a soil biological quality index for soils of semi-arid tropics. Soil 2020, 6, 483–497. [Google Scholar] [CrossRef]
- De la Paz Jimenez, M.; de la Horra, A.; Pruzzo, L.; Palma, M. Soil quality: A new index based on microbiological and biochemical parameters. Biol. Fertil. Soils 2002. [Google Scholar] [CrossRef]
- Klimkowicz-Pawlas, A.; Ukalska-Jaruga, A.; Smreczak, B. Soil quality index for agricultural areas under different levels of anthropopressure. Int. Agrophys. 2019, 33, 455–462. [Google Scholar] [CrossRef]
- Wyszkowska, J.; Borowik, A.; Kucharski, M.; Kucharski, J. Effect of cadmium, copper and zinc on plants, soil microorganisms and soil enzymes. J. Elem. 2013, 18, 769–796. [Google Scholar] [CrossRef]
- Chakravarty, P.; Deka, H. Enzymatic defense of Cyperus brevifolius in hydrocarbons stress environment and changes in soil properties. Sci. Rep. 2021, 11, 718. [Google Scholar] [CrossRef]
- Gao, Y.; Wang, J.; Xu, J.; Kong, X.; Zhao, L.; Zeng, D.-H. Assessing the quality of oil-contaminated saline soil using two composite indices. Ecol. Indic. 2013, 24, 105–112. [Google Scholar] [CrossRef]
- Gospodarek, J.; Rusin, M.; Barczyk, G.; Nadgórska-Socha, A. The effect of petroleum-derived substances and their bioremediation on soil enzymatic activity and soil invertebrates. Agronomy 2021, 11, 80. [Google Scholar] [CrossRef]
- Zuzolo, D.; Guarino, C.; Tartaglia, M.; Sciarrillo, R. Plant-soil-microbiota combination for the removal of total petroleum hydrocarbons (TPH): An in-field experiment. Front. Microbiol. 2021, 11. [Google Scholar] [CrossRef]
- Al-Sayegh, A.; Al-Wahaibi, Y.; Joshi, S.; Al-Bahry, S.; Elshafie, A.; Al-Bemani, A. Bioremediation of heavy crude oil contamination. Open Biotechnol. J. 2016, 10. [Google Scholar] [CrossRef]
- Correa-García, S.; Rheault, K.; Tremblay, J.; Séguin, A.; Yergeau, E. Soil characteristics constrain the response of microbial communities and associated hydrocarbon degradation genes during phytoremediation. Appl. Environ. Microbiol. 2021, 87. [Google Scholar] [CrossRef] [PubMed]
- Hoang, S.A.; Lamb, D.; Seshadri, B.; Sarkar, B.; Cheng, Y.; Wang, L.; Bolan, N.S. Petroleum hydrocarbon rhizoremediation and soil microbial activity improvement via cluster root formation by wild proteaceae plant species. Chemosphere 2021, 275, 130135. [Google Scholar] [CrossRef]
- BP, p.l.c. Website. Available online: https://www.bp.com/ (accessed on 23 February 2021).
- BP’s Fuels Characteristics Cards. Available online: https://www.bp.com/pl_pl/poland/home/produkty_uslugi/hurt_paliw.html (accessed on 23 February 2021).
- Raman, J.K.; Alves, C.M.; Gnansounou, E. A review on moringa tree and vetiver grass—Potential biorefinery feedstocks. Bioresour. Technol. 2018, 249, 1044–1051. [Google Scholar] [CrossRef]
- Visconti, D.; Álvarez-Robles, M.J.; Fiorentino, N.; Fagnano, M.; Clemente, R. Use of Brassica juncea and Dactylis glomerata for the phytostabilization of mine soils amended with compost or biochar. Chemosphere 2020, 260, 127661. [Google Scholar] [CrossRef]
- Yang, Z.; Nie, G.; Feng, G.; Han, J.; Huang, L.; Zhang, X. Genome-wide identification, characterization, and expression analysis of the NAC transcription factor family in orchardgrass (Dactylis glomerata L.). BMC Genom. 2021, 22, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Gajić, G.; Djurdjević, L.; Kostić, O.; Jarić, S.; Stevanović, B.; Mitrović, M.; Pavlović, P. Phytoremediation potential, photosynthetic and antioxidant response to arsenic-induced stress of Dactylis glomerata L. sown on fly ash deposits. Plants 2020, 9, 657. [Google Scholar] [CrossRef]
- Li, S.; Nie, Z.; Zhang, D. Competition between Cocksfoot (Dactylis Glomerata L.) and companion species: Evidence for allelopathy. Field Crops Res. 2016, 196, 452–462. [Google Scholar] [CrossRef]
- Sanderson, M.A.; Skinner, R.H.; Elwinger, G.F. Seedling development and field performance of prairiegrass, grazing bromegrass, and orchardgrass. Crop Sci. 2002, 42, 224–230. [Google Scholar] [CrossRef] [PubMed]
- Gałązka, A.; Gałązka, R. Phytoremediation of polycyclic aromatic hydrocarbons in soils artificially polluted using plant-associated-endophytic bacteria and Dactylis glomerata as the bioremediation plant. Pol. J. Microbiol. 2015, 64, 241–252. [Google Scholar] [CrossRef]
- Wyszkowska, J.; Borowik, A.; Olszewski, J.; Kucharski, J. Soil bacterial community and soil enzyme activity depending on the cultivation of Triticum aestivum, Brassica napus, and Pisum sativum ssp. arvense. Diversity 2019, 11, 246. [Google Scholar] [CrossRef] [Green Version]
- National Center for Biotechnology Information. Available online: https://www.ncbi.nlm.nih.gov/ (accessed on 22 March 2021).
- ISO 18287 Soil Quality—Determination of Polycyclic Aromatic Hydrocarbons (PAH)—Gas Chromatographic Method with Mass Spectrometric Detection (GC-MS); International Organization for Standardization: Geneva, Switzerland, 2006.
- EN ISO 16703 Soil Quality—Determination of Content of Hydrocarbon in the Range c10 to c40 by Gas Chromatography; International Organization for Standardization: Geneva, Switzerland, 2004.
- EN ISO 22155 Soil Quality—Gas Chromatographic Determination of Volatile Aromatic and Halogenated Hydrocarbons and Selected Ethers—Static Headspace Method; International Organization for Standardization: Geneva, Switzerland, 2016.
- De Leij, F.A.A.M.; Whipps, J.M.; Lynch, J.M. The use of colony development for the characterization of bacterial communities in soil and on roots. Microb. Ecol. 1993, 27, 81–97. [Google Scholar] [CrossRef] [PubMed]
- Orwin, K.H.; Wardle, D.A. New indices for quantifying the resistance and resilience of soil biota to exogenous disturbances. Soil Biol. Biochem. 2004, 36, 1907–1912. [Google Scholar] [CrossRef]
- TIBCO Software Inc. Statistica (Data Analysis Software System), version 13.3; TIBCO Software Inc.: Palo Alto, CA, USA, 2017. [Google Scholar]
- Parks, D.H.; Tyson, G.W.; Hugenholtz, P.; Beiko, R.G. STAMP: Statistical Analysis of Taxonomic and Functional Profiles. Bioinformatics 2014, 30, 3123–3124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- RStudio Team. RStudio: Integrated Development for R; RStudio, Inc.: Boston, MA, USA, 2019; Available online: http://www.rstudio.com/ (accessed on 23 February 2020).
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019; Available online: https://www.r-project.org/ (accessed on 23 February 2020).
- Warnes, G.R.; Bolker, B.; Bonebakker, L.; Gentleman, R.; Huber, W.; Liaw, A.; Lumley, T.; Maechler, M.; Magnusson, M.; Moeller, S.; et al. Gplots: Various R Programming Tools for Plotting Data; R Package Version 2.17.0; 2020; Available online: https://CRAN.R-project.org/package=gplots (accessed on 23 February 2020).
- Heberle, H.; Meirelles, G.V.; da Silva, F.R.; Telles, G.P.; Minghim, R. InteractiVenn: A web-based tool for the analysis of sets through Venn diagrams. BMC Bioinform. 2015, 16, 169. [Google Scholar] [CrossRef] [PubMed]




| Object | C6–C12 | C12–C35 | Ben | EtB | Tol | Xyl | Nap |
| P | 99.53 ± 1.06 a | 71.41 ± 0.70 b | 95.81 ± 0.09 b | 99.83 ± 0.09 a | 99.86 ± 0.15 a | 99.87 ± 0.10 a | 90.63 ± 0.09 b |
| GP | 99.76 ± 0.90 a | 76.56 ± 0.69 a | 98.11 ± 0.19 a | 99.95 ± 0.13 a | 99.96 ± 0.20 a | 99.96 ± 0.13 a | 99.84 ± 0.09 a |
| Object | Ant | Chr | BaA | BaP | BbF | BkF | IP |
| P | 59.09 ± 0.14 b | 18.33 ± 0.10 b | 40.91 ± 0.09 b | 35.00 ± 0.07 b | 16.92 ± 0.04 b | 13.33 ± 0.07 b | 40.00 ± 0.08 a |
| GP | 84.85 ± 0.11a | 41.67 ± 0.09 a | 54.55 ± 0.11 a | 50.00 ± 0.10 a | 30.77 ± 0.06 a | 33.33 ± 0.07 a | 40.00 ± 0.08 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Borowik, A.; Wyszkowska, J.; Kucharski, J. Microbiological Study in Petrol-Spiked Soil. Molecules 2021, 26, 2664. https://doi.org/10.3390/molecules26092664
Borowik A, Wyszkowska J, Kucharski J. Microbiological Study in Petrol-Spiked Soil. Molecules. 2021; 26(9):2664. https://doi.org/10.3390/molecules26092664
Chicago/Turabian StyleBorowik, Agata, Jadwiga Wyszkowska, and Jan Kucharski. 2021. "Microbiological Study in Petrol-Spiked Soil" Molecules 26, no. 9: 2664. https://doi.org/10.3390/molecules26092664





