Pure Ion Chromatograms Combined with Advanced Machine Learning Methods Improve Accuracy of Discriminant Models in LC–MS-Based Untargeted Metabolomics
Abstract
:1. Introduction
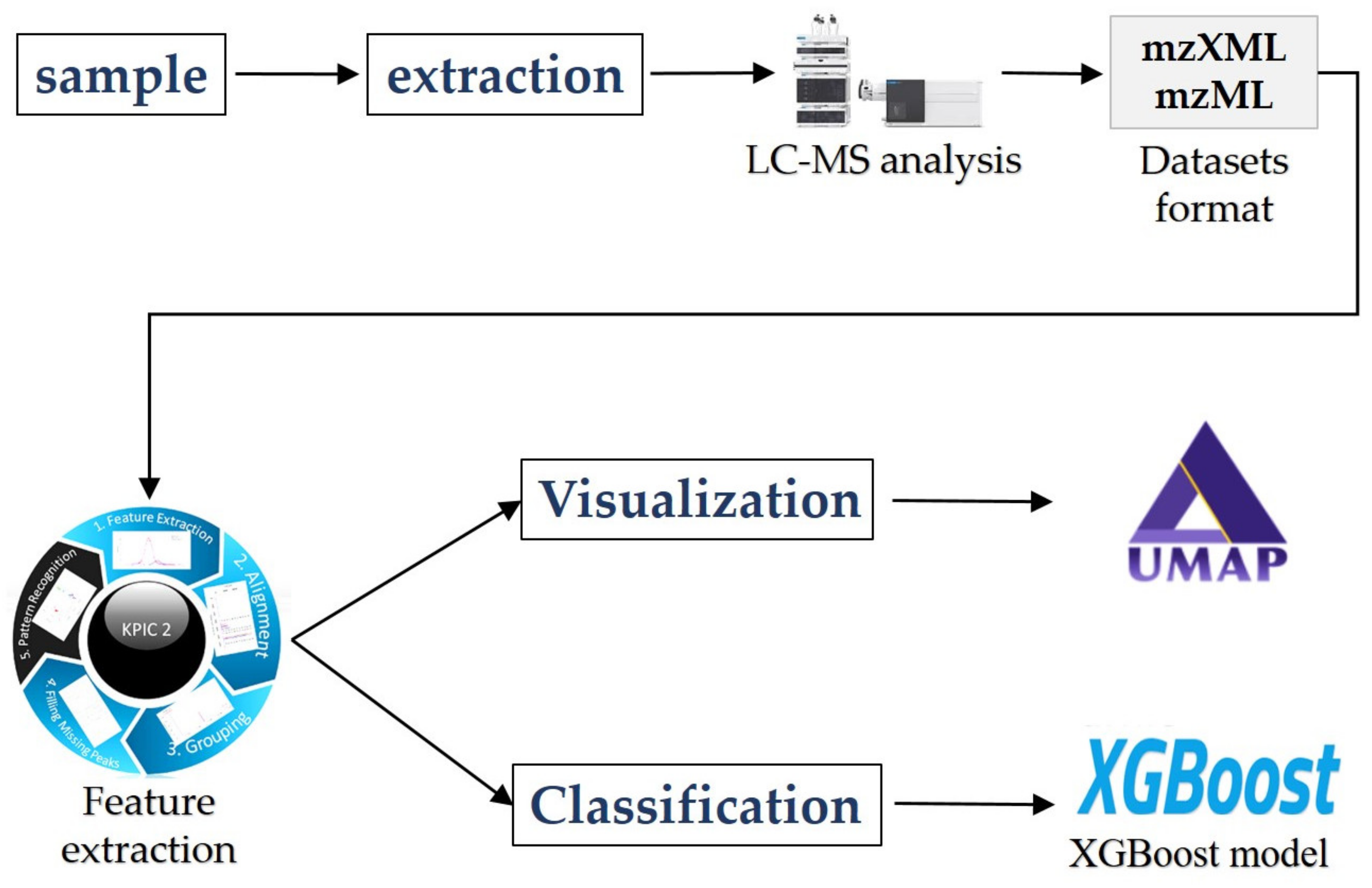
2. Materials and Methods
2.1. Theory of KPIC2
2.2. Theory of Advanced Machine Learning Methods
2.2.1. Visualization Methods
2.2.2. XGBoost
2.3. Datasets
2.3.1. Liquor Dataset
2.3.2. LCC Dataset
2.4. Comparison of KPIC2 and XCMS
2.5. Pattern Recognition
2.6. Evaluation Criteria
3. Results and Discussion
3.1. Comparison of XGBoost Performance Based on KPIC2 and XCMS
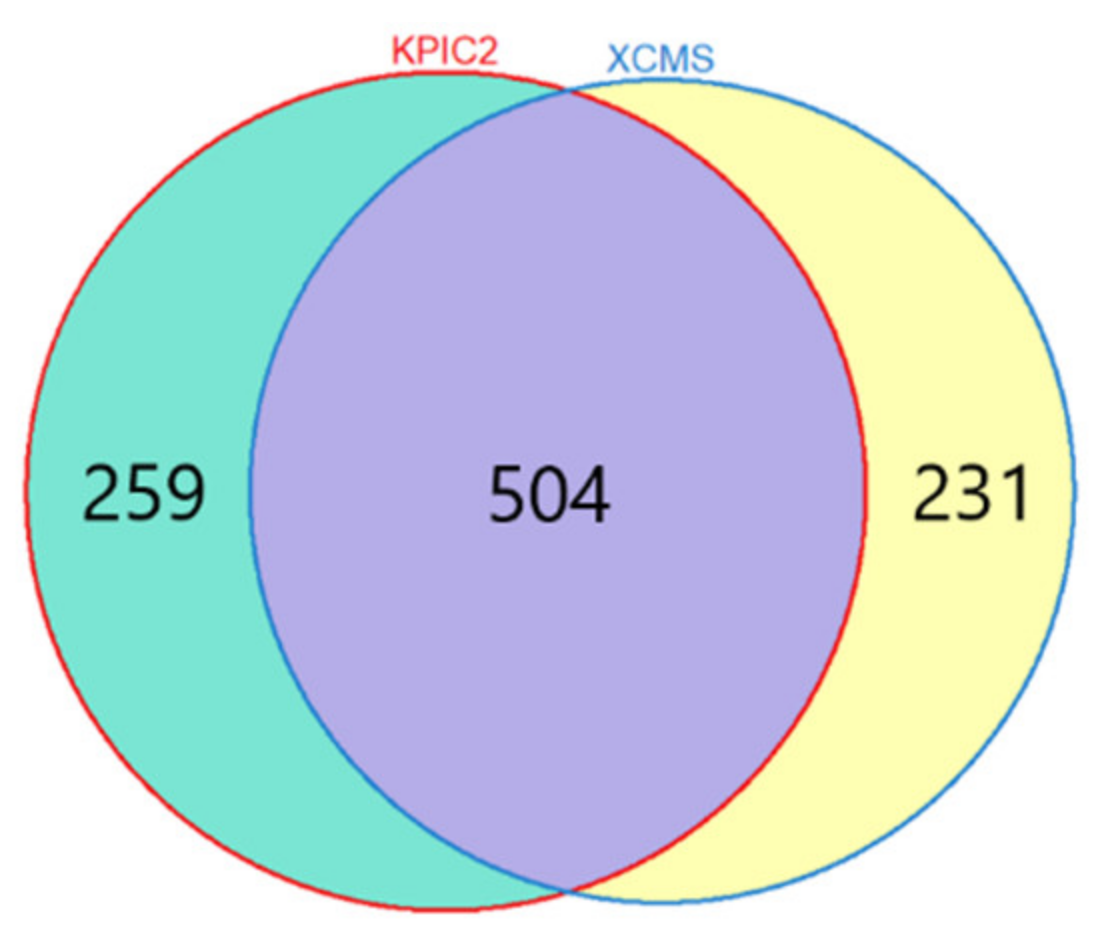
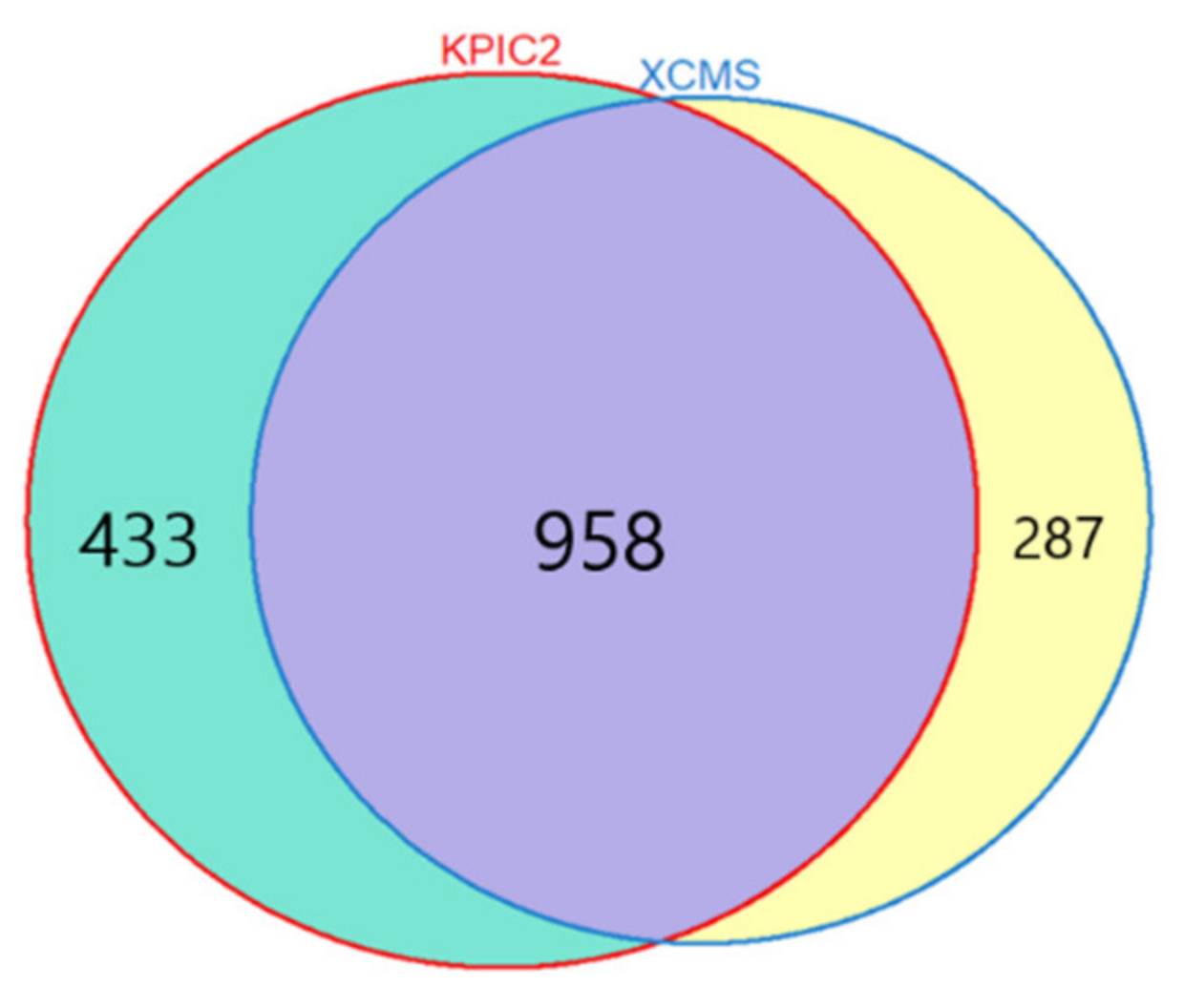
3.1.1. Results of Feature Extraction
Liquor Dataset
LCC Dataset
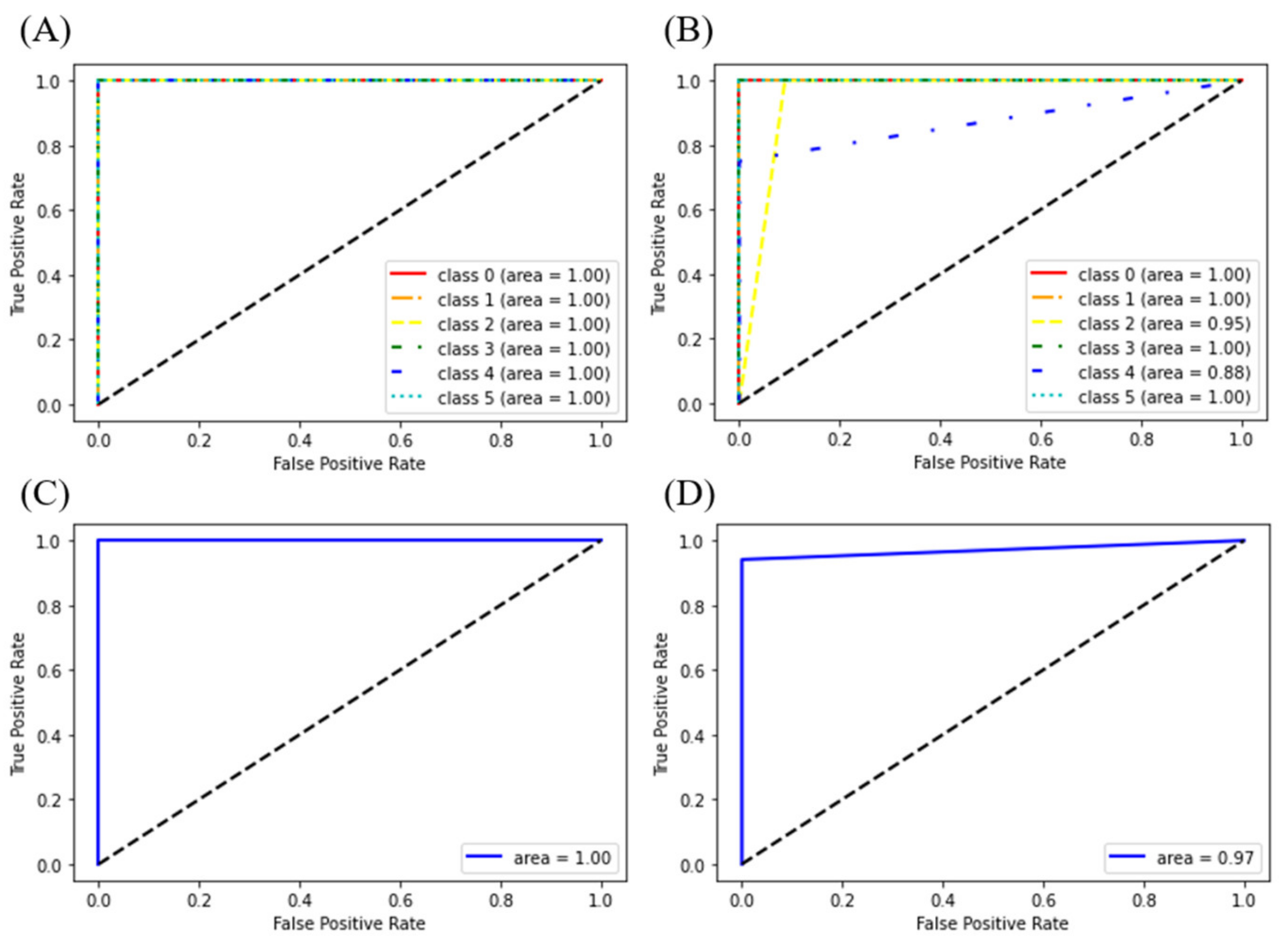
3.1.2. Discriminant Models for Liquor and LCC Datasets
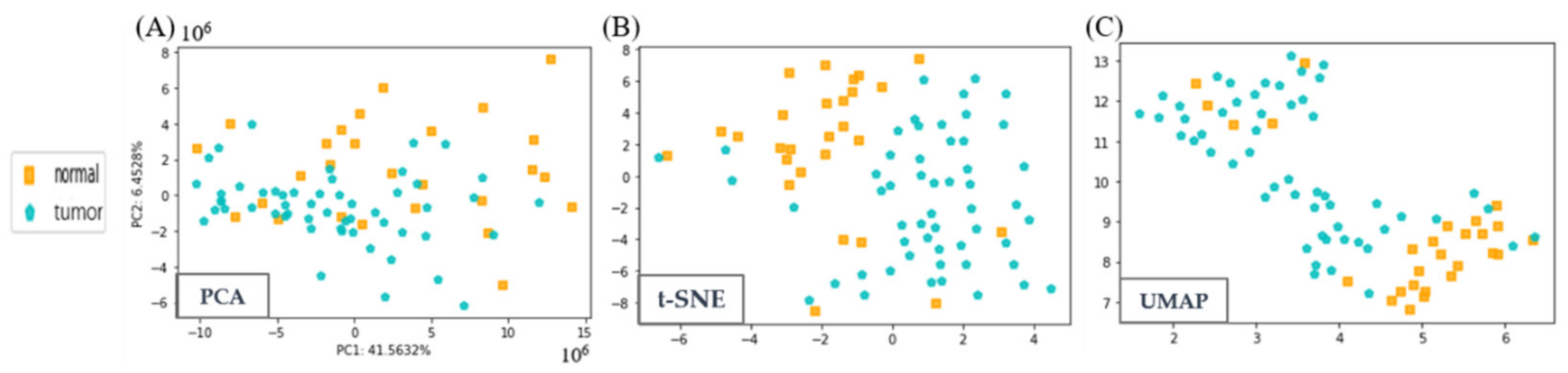
3.2. Visualization of Liquor and LCC Datasets
3.3. Comparison of Classification Models Based on KPIC2
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fiehn, O.; Kopka, J.; Dörmann, P.; Altmann, T.; Trethewey, R.N.; Willmitzer, L. Metabolite profiling for plant functional genomics. Nat. Biotechnol. 2000, 18, 1157–1161. [Google Scholar] [CrossRef] [PubMed]
- Musilová, J.; Glatz, Z. Metabolomics-Basic concepts, Strategies and Methodologies. Chemické Listy 2011, 105, 745–751. [Google Scholar]
- Nam, S.L.; Mata, A.; Dias, R.P.; Harynuk, J.J. Towards Standardization of Data Normalization Strategies to Improve Urinary Metabolomics Studies by GC× GC-TOFMS. Metabolites 2020, 10, 376. [Google Scholar] [CrossRef] [PubMed]
- De Vos, R.C.; Moco, S.; Lommen, A.; Keurentjes, J.J.; Bino, R.J.; Hall, R.D. Untargeted large-scale plant metabolomics using liquid chromatography coupled to mass spectrometry. Nat. Protoc. 2007, 2, 778–791. [Google Scholar] [CrossRef] [PubMed]
- James, J.T.; Tugizimana, F.; Steenkamp, P.A.; Dubery, I.A. Metabolomic analysis of methyl jasmonate-induced triterpenoid production in the medicinal herb Centella asiatica (L.) urban. Molecules 2013, 18, 4267–4281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zarate, E.; Boyle, V.; Rupprecht, U.; Green, S.; Villas-Boas, S.G.; Baker, P.; Pinu, F.R. Fully automated trimethylsilyl (TMS) derivatisation protocol for metabolite profiling by GC-MS. Metabolites 2017, 7, 1. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Jung, Y.; Shin, J.-H.; Kim, H.K.; Moon, B.C.; Ryu, D.H.; Hwang, G.-S. Secondary metabolite profiling of Curcuma species grown at different locations using GC/TOF and UPLC/Q-TOF MS. Molecules 2014, 19, 9535–9551. [Google Scholar] [CrossRef] [Green Version]
- Lebanov, L.; Ghiasvand, A.; Paull, B. Data handling and data analysis in metabolomic studies of essential oils using GC-MS. J. Chromatogr. A 2021, 1640, 461896. [Google Scholar] [CrossRef]
- von Roepenack-Lahaye, E.; Degenkolb, T.; Zerjeski, M.; Franz, M.; Roth, U.; Wessjohann, L.; Schmidt, J.; Scheel, D.; Clemens, S. Profiling of Arabidopsis secondary metabolites by capillary liquid chromatography coupled to electrospray ionization quadrupole time-of-flight mass spectrometry. Plant Physiol. 2004, 134, 548–559. [Google Scholar] [CrossRef] [Green Version]
- Tikunov, Y.; Lommen, A.; De Vos, C.R.; Verhoeven, H.A.; Bino, R.J.; Hall, R.D.; Bovy, A.G. A novel approach for nontargeted data analysis for metabolomics. Large-scale profiling of tomato fruit volatiles. Plant Physiol. 2005, 139, 1125–1137. [Google Scholar] [CrossRef] [Green Version]
- Roca, M.; Alcoriza, M.I.; Garcia-Canaveras, J.C.; Lahoz, A.n. Reviewing the metabolome coverage provided by LC-MS: Focus on sample preparation and chromatography-A tutorial. Anal. Chim. Acta 2020, 1147, 38–55. [Google Scholar] [CrossRef]
- Lommen, A. MetAlign: Interface-driven, versatile metabolomics tool for hyphenated full-scan mass spectrometry data preprocessing. Anal. Chem. 2009, 81, 3079–3086. [Google Scholar] [CrossRef]
- Lommen, A.; Kools, H.J. MetAlign 3.0: Performance enhancement by efficient use of advances in computer hardware. Metabolomics 2012, 8, 719–726. [Google Scholar] [CrossRef] [Green Version]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Orešič, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef] [Green Version]
- Katajamaa, M.; Miettinen, J.; Orešič, M. MZmine: Toolbox for processing and visualization of mass spectrometry based molecular profile data. Bioinformatics 2006, 22, 634–636. [Google Scholar] [CrossRef] [Green Version]
- Smith, C.A.; Want, E.J.; O’Maille, G.; Abagyan, R.; Siuzdak, G. XCMS: Processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 2006, 78, 779–787. [Google Scholar] [CrossRef]
- Wang, T.; Duedahl-Olesen, L.; Frandsen, H.L. Targeted and non-targeted unexpected food contaminants analysis by LC/HRMS: Feasibility study on rice. Food Chem. 2021, 338, 127957. [Google Scholar] [CrossRef]
- Röst, H.L.; Sachsenberg, T.; Aiche, S.; Bielow, C.; Weisser, H.; Aicheler, F.; Andreotti, S.; Ehrlich, H.-C.; Gutenbrunner, P.; Kenar, E. OpenMS: A flexible open-source software platform for mass spectrometry data analysis. Nat. Methods 2016, 13, 741–748. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, F.; Wu, P.; Bu, D.; Sun, S. OpenMS-Simulator: An open-source software for theoretical tandem mass spectrum prediction. BMC Bioinform. 2015, 16, 110. [Google Scholar] [CrossRef] [Green Version]
- Kutuzova, S.; Colaianni, P.; Rost, H.; Sachsenberg, T.; Alka, O.; Kohlbacher, O.; Burla, B.; Torta, F.; Schrubbers, L.; Kristensen, M. SmartPeak automates targeted and quantitative metabolomics data processing. Anal. Chem. 2020, 92, 15968–15974. [Google Scholar] [CrossRef]
- Tautenhahn, R.; Boettcher, C.; Neumann, S. Highly sensitive feature detection for high resolution LC/MS. BMC Bioinform. 2008, 9, 504. [Google Scholar] [CrossRef] [Green Version]
- Mihaleva, V.V.; Vorst, O.; Maliepaard, C.; Verhoeven, H.A.; de Vos, R.C.; Hall, R.D.; van Ham, R.C. Accurate mass error correction in liquid chromatography time-of-flight mass spectrometry based metabolomics. Metabolomics 2008, 4, 171–182. [Google Scholar] [CrossRef]
- Pedrioli, P.G.; Eng, J.K.; Hubley, R.; Vogelzang, M.; Deutsch, E.W.; Raught, B.; Pratt, B.; Nilsson, E.; Angeletti, R.H.; Apweiler, R. A common open representation of mass spectrometry data and its application to proteomics research. Nat. Biotechnol. 2004, 22, 1459–1466. [Google Scholar] [CrossRef]
- Wang, S.Y.; Kuo, C.H.; Tseng, Y.J. Ion trace detection algorithm to extract pure ion chromatograms to improve untargeted peak detection quality for liquid chromatography/time-of-flight mass spectrometry-based metabolomics data. Anal. Chem. 2015, 87, 3048–3055. [Google Scholar] [CrossRef] [PubMed]
- Åberg, K.M.; Torgrip, R.J.; Kolmert, J.; Schuppe-Koistinen, I.; Lindberg, J. Feature detection and alignment of hyphenated chromatographic–mass spectrometric data: Extraction of pure ion chromatograms using Kalman tracking. J. Chromatogr. A 2008, 1192, 139–146. [Google Scholar] [CrossRef]
- Conley, C.J.; Smith, R.; Torgrip, R.J.; Taylor, R.M.; Tautenhahn, R.; Prince, J.T. Massifquant: Open-source Kalman filter-based XC-MS isotope trace feature detection. Bioinformatics 2014, 30, 2636–2643. [Google Scholar] [CrossRef] [Green Version]
- Tengstrand, E.; Lindberg, J.; Åberg, K.M. TracMass 2: A Modular Suite of Tools for Processing Chromatography-Full Scan Mass Spectrometry Data. Anal. Chem. 2014, 86, 3435–3442. [Google Scholar] [CrossRef]
- Ji, H.; Lu, H.; Zhang, Z. Pure ion chromatogram extraction via optimal k-means clustering. Rsc Adv. 2016, 6, 56977–56985. [Google Scholar] [CrossRef]
- Ji, H.; Zeng, F.; Xu, Y.; Lu, H.; Zhang, Z. KPIC2: An effective framework for mass spectrometry-based metabolomics using pure ion chromatograms. Anal. Chem. 2017, 89, 7631–7640. [Google Scholar] [CrossRef] [PubMed]
- Jolliffe, I.T.; Cadima, J. Principal component analysis: A review and recent developments. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2016, 374, 20150202. [Google Scholar] [CrossRef]
- Koch, W.; Kukula-Koch, W.; Komsta, Å.u.; Marzec, Z.; Szwerc, W.; GÅ‚owniak, K. Green tea quality evaluation based on its catechins and metals composition in combination with chemometric analysis. Molecules 2018, 23, 1689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cebi, N.; Arici, M.; Sagdic, O. The famous Turkish rose essential oil: Characterization and authenticity monitoring by FTIR, Raman and GC-MS techniques combined with chemometrics. Food Chem. 2021, 354, 129495. [Google Scholar] [CrossRef] [PubMed]
- Maaten, L.v.d.; Hinton, G. Visualizing data using t-SNE. J. Mach. Learn. Res. 2008, 9, 2579–2605. [Google Scholar]
- Becht, E.; McInnes, L.; Healy, J.; Dutertre, C.A.; Kwok, I.W.; Ng, L.G.; Ginhoux, F.; Newell, E.W. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 2019, 37, 38–44. [Google Scholar] [CrossRef] [PubMed]
- McInnes, L.; Healy, J.; Melville, J. Umap: Uniform manifold approximation and projection for dimension reduction. arXiv Preprint 2018, arXiv:1802.03426 2018. [Google Scholar]
- Vermeulen, M.; Smith, K.; Eremin, K.; Rayner, G.; Walton, M. Application of Uniform Manifold Approximation and Projection (UMAP) in spectral imaging of artworks. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2021, 252, 1386–1425. [Google Scholar] [CrossRef]
- Thévenot, E.A.; Roux, A.; Xu, Y.; Ezan, E.; Junot, C. Analysis of the human adult urinary metabolome variations with age, body mass index, and gender by implementing a comprehensive workflow for univariate and OPLS statistical analyses. J. Proteome Res. 2015, 14, 3322–3335. [Google Scholar] [CrossRef]
- Bylesjö, M.; Eriksson, D.; Sjödin, A.; Jansson, S.; Moritz, T.; Trygg, J. Orthogonal projections to latent structures as a strategy for microarray data normalization. BMC Bioinform. 2007, 8, 207. [Google Scholar] [CrossRef] [Green Version]
- Belgiu, M.; Drăguţ, L. Random forest in remote sensing: A review of applications and future directions. ISPRS J. Photogramm. Remote Sens. 2016, 114, 24–31. [Google Scholar] [CrossRef]
- Barker, M.; Rayens, W. Partial least squares for discrimination. J. Chemom. A J. Chemom. Soc. 2003, 17, 166–173. [Google Scholar] [CrossRef]
- Trygg, J.; Wold, S. Orthogonal projections to latent structures (O-PLS). J. Chemom. A J. Chemom. Soc. 2002, 16, 119–128. [Google Scholar] [CrossRef]
- Madsen, R.; Lundstedt, T.; Trygg, J. Chemometrics in metabolomics—a review in human disease diagnosis. Anal. Chim. Acta 2010, 659, 23–33. [Google Scholar] [CrossRef]
- Verron, T.; Sabatier, R.; Joffre, R. Some theoretical properties of the O-PLS method. J. Chemom. A J. Chemom. Soc. 2004, 18, 62–68. [Google Scholar] [CrossRef]
- Lan, L.; Sun, W.; Chang, Q.; Sun, G. Comprehensive evaluation of Licorice Extract by five-dimensional quantitative profiling. J. Chromatogr. A 2021, 1644, 462105. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. Xgboost: A scalable tree boosting system. In Proceedings of the 22Nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining; ACM: New York, NY, USA, 2016; pp. 785–794. [Google Scholar]
- Feng, C.; Xu, Q.; Qiu, X.; Ji, J.; Lin, Y.; Le, S.; She, J.; Lu, D.; Wang, G. Evaluation and application of machine learning-based retention time prediction for suspect screening of pesticides and pesticide transformation products in LC-HRMS. Chemosphere 2021, 271, 129447. [Google Scholar] [CrossRef]
- Yu, Y.J.; Zheng, Q.X.; Zhang, Y.M.; Zhang, Q.; Zhang, Y.Y.; Liu, P.P.; Lu, P.; Fan, M.J.; Chen, Q.S.; Bai, C.C. Automatic data analysis workflow for ultra-high performance liquid chromatography-high resolution mass spectrometry-based metabolomics. J. Chromatogr. A 2019, 1585, 172–181. [Google Scholar] [CrossRef]
- Cai, Y.; Rattray, N.J.; Zhang, Q.; Mironova, V.; Santos-Neto, A.; Hsu, K.-S.; Rattray, Z.; Cross, J.R.; Zhang, Y.; Paty, P.B. Sex differences in colon cancer metabolism reveal a novel subphenotype. Sci. Rep. 2020, 10, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Kuhl, C.; Tautenhahn, R.; Bottcher, C.; Larson, T.R.; Neumann, S. CAMERA: An integrated strategy for compound spectra extraction and annotation of liquid chromatography/mass spectrometry data sets. Anal. Chem. 2012, 84, 283–289. [Google Scholar] [CrossRef] [Green Version]

| KPIC2 | XCMS/CAMERA | ||||
| Liquor Dataset | LCC Dataset | Liquor Dataset | LCC Dataset | ||
| Parameters | n_estimators | 11 | 20 | 11 | 22 |
| learning_rate | 0.01 | 0.01 | 0.1 | 0.01 | |
| silent | 1 | 1 | 1 | 1 | |
| subsample | 1 | 0.9 | 1 | 1 | |
| max_depth | 2 | 1 | 3 | 1 | |
| booster | gbtree | gbtree | gbtree | gbtree | |
| gamma | 0 | 0 | 0 | 0 | |
| min_child_weight | 1 | 3 | 1 | 1 | |
| colsample_bytree | 0.1 | 0.2 | 0.1 | 0.5 | |
| Model | Accuracy | 1.000 | 1.000 | 0.917 | 0.963 |
| Precision | 1.000 | 1.000 | 0.917 | 1.000 | |
| Recall/Sensitivity | 1.000 | 1.000 | 0.958 | 0.941 | |
| Specificity | 1.000 | 1.000 | 0.958 | 1.000 | |
| F1_score | 1.000 | 1.000 | 0.921 | 0.970 | |
| Actual | BJ-1 | BJ-2 | BJ-3 | BJ-4 | BJ-5 | BJ-6 | Class_Error | Error_Rate | |
|---|---|---|---|---|---|---|---|---|---|
| KPIC2 | BJ-1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0.00% |
| BJ-2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | ||
| BJ-3 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | ||
| BJ-4 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | ||
| BJ-5 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | ||
| BJ-6 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | ||
| XCMS/ CAMERA | BJ-1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 8.33% |
| BJ-2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | ||
| BJ-3 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | ||
| BJ-4 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | ||
| BJ-5 | 0 | 0 | 1 | 0 | 3 | 0 | 0.25 | ||
| BJ-6 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | ||
| Actual | Normal | Tumor | Class_Error | Error_Rate | |
|---|---|---|---|---|---|
| KPIC2 | Normal | 10 | 0 | 0 | 0.00% |
| Tumor | 0 | 17 | 0 | ||
| XCMS/CAMERA | Normal | 10 | 0 | 0 | 3.70% |
| Tumor | 1 | 16 | 0.06 | ||
| Dataset | Model | Accuracy | Precision | Recall | Specificity | F1_score |
|---|---|---|---|---|---|---|
| Liquor | PLS-DA | 0.667 | 0.611 | 0.833 | 0.833 | 0.689 |
| SVM | 0.833 | 0.889 | 0.917 | 0.917 | 0.861 | |
| RF | 0.750 | 0.875 | 0.875 | 0.875 | 0.780 | |
| XGBoost | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | |
| LCC | PLS-DA | 0.741 | 0.778 | 0.824 | 0.600 | 0.800 |
| SVM | 0.852 | 0.842 | 0.941 | 0.700 | 0.889 | |
| RF | 0.963 | 0.944 | 1.000 | 0.900 | 0.971 | |
| XGBoost | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tian, M.; Lin, Z.; Wang, X.; Yang, J.; Zhao, W.; Lu, H.; Zhang, Z.; Chen, Y. Pure Ion Chromatograms Combined with Advanced Machine Learning Methods Improve Accuracy of Discriminant Models in LC–MS-Based Untargeted Metabolomics. Molecules 2021, 26, 2715. https://doi.org/10.3390/molecules26092715
Tian M, Lin Z, Wang X, Yang J, Zhao W, Lu H, Zhang Z, Chen Y. Pure Ion Chromatograms Combined with Advanced Machine Learning Methods Improve Accuracy of Discriminant Models in LC–MS-Based Untargeted Metabolomics. Molecules. 2021; 26(9):2715. https://doi.org/10.3390/molecules26092715
Chicago/Turabian StyleTian, Miao, Zhonglong Lin, Xu Wang, Jing Yang, Wentao Zhao, Hongmei Lu, Zhimin Zhang, and Yi Chen. 2021. "Pure Ion Chromatograms Combined with Advanced Machine Learning Methods Improve Accuracy of Discriminant Models in LC–MS-Based Untargeted Metabolomics" Molecules 26, no. 9: 2715. https://doi.org/10.3390/molecules26092715






