Maize Flavonoid Biosynthesis, Regulation, and Human Health Relevance: A Review
Abstract
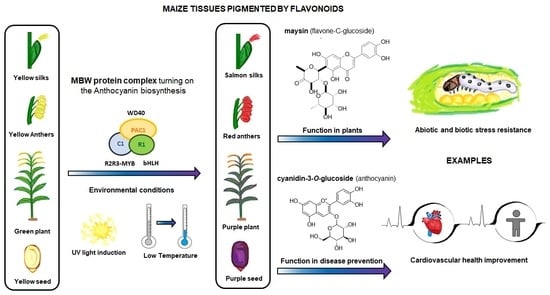
:1. Introduction
2. Structural Protein Genes of the Maize Flavonoid Pathway
2.1. Phenylpropanoid Pathway
2.2. Early Biosynthetic Genes of Flavonoids
2.2.1. Chalcone Synthase (ZmCHS, c2, EC 2.3.1.74)
2.2.2. Chalcone Isomerase (ZmCHI, chi1, EC 5.5.1.6)
2.2.3. Flavonoid 3-Dioxygenase (ZmF3H, fht1, EC 1.14.11.9)
2.2.4. Flavonoid 3′-Monooxygenase (ZmF3′H, pr1, EC 1.14.14.82)
2.3. Late Biosynthetic Genes of Maize Anthocyanins
2.3.1. Dihydroflavonol 4-Reductase (ZmDFR, a1, EC 1.1.1.219)
2.3.2. Anthocyanidin Synthase (ZmANS, a2, EC 1.14.20.4)
2.3.3. Anthocyanidin 3-O-Glucosyltransferase (ZmAGT, bz1, EC 2.4.1.115)
2.3.4. Malonyl-CoA: Anthocyanin 3-O-Glucoside-6′′-O-Malonyltransferase (Zm3MAT, aat1, EC 2.3.1.171)
2.3.5. Flavonoid 3′,5′-O-Methyltransferase, or Anthocyanin S-Adenosyl-l-Methionine-Dependent O-Methyltransferase (ZmFOMT or ZmAOMT, EC 2.1.1.267)
2.3.6. Glutathione-S-Transferase (ZmGST, bz2, EC 2.5.1.18)
2.3.7. Multidrug Resistance Protein (ZmABCC3 and -4, mrpa3, EC 7.6.2.2)
2.3.8. Flavanol-Anthocyanin Condensed Forms
2.4. Biosynthesis of Flavonols, Flavones C-Glycosides, and Phlobaphenes in Maize
2.4.1. Flavonol Synthase (ZmFLS1, fls1, EC 1.14.20.6)
2.4.2. Flavone Synthase I (ZmFNSI1-2, fnsi1, EC 1.14.20.5) and Flavone Synthase II (ZmFNSII-1, fnsii1, EC 1.14.19.76)
2.4.3. Flavanone 2-Hydroxylase (ZmF2H1, fns1, EC 1.14.14.162)

2.4.4. UDP-Glucose:2-Hydroxyflavanone C-Glucosyltransferase (ZmCGT, cgt1, EC 2.4.1.360)
2.4.5. UDP-Rhamnosyl Transferase (sm2, UGT91L1, EC 2.4.1.159)
2.4.6. Glucose-4,6 Dehydratase (ZmRHS1, sm1, EC 4.2.1.76)
3. Regulatory Factors of the Maize Flavonoid Pathway
3.1. The MBW Complex
3.2. Regulation of MBW Complex
4. Cyanidin-3-O-Glucoside, One of the Most Abundant Flavonoids in Maize, and Its Effects on Human Health
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Falcone Ferreyra, M.L.; Casas, M.I.; Questa, J.I.; Herrera, A.L.; DeBlasio, S.; Wang, J.; Jackson, D.; Grotewold, E.; Casati, P. Evolution and Expression of Tandem Duplicated Maize Flavonol Synthase Genes. Front. Plant Sci. 2012, 3, 101. [Google Scholar] [CrossRef] [PubMed]
- Righini, S.; Rodriguez, E.J.; Berosich, C.; Grotewold, E.; Casati, P.; Falcone Ferreyra, M.L. Apigenin Produced by Maize Flavone Synthase I and II Protects Plants against UV-B-Induced Damage. Plant Cell Environ. 2019, 42, 495–508. [Google Scholar] [CrossRef]
- Rius, S.P.; Emiliani, J.; Casati, P. P1 Epigenetic Regulation in Leaves of High Altitude Maize Landraces: Effect of UV-B Radiation. Front. Plant Sci. 2016, 7, 523. [Google Scholar] [CrossRef]
- Dewick, P.M. Medicinal Natural Products: A Biosynthetic Approach, 3rd ed.; John Wiley & Sons, Ltd.: Chichester, UK, 2009; ISBN 9780470742761. [Google Scholar]
- Rauter, A.P.; Ennis, M.; Hellwich, K.-H.; Herold, B.J.; Horton, D.; Moss, G.P.; Schomburg, I. Nomenclature of Flavonoids (IUPAC Recommendations 2017). Pure Appl. Chem. 2018, 90, 1429–1486. [Google Scholar] [CrossRef]
- Petroni, K.; Tonelli, C. Recent Advances on the Regulation of Anthocyanin Synthesis in Reproductive Organs. Plant Sci. 2011, 181, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Cone, K.C. Anthocyanin Synthesis in Maize Aleurone Tissue. In Endosperm. Plant Cell Monographs; Olsen, O.A., Ed.; Springer: Berlin/Heidelberg, Germany, 2007; pp. 305–320. ISBN 978-3-540-71235-0. [Google Scholar]
- Falcone Ferreyra, M.L.; Rius, S.P.; Casati, P. Flavonoids: Biosynthesis, Biological Functions, and Biotechnological Applications. Front. Plant Sci. 2012, 3, 1–15. [Google Scholar] [CrossRef]
- Faria, A.; Fernandes, I.; Norberto, S.; Mateus, N.; Calhau, C. Interplay between Anthocyanins and Gut Microbiota. J. Agric. Food Chem. 2014, 62, 6898–6902. [Google Scholar] [CrossRef]
- Hidalgo, M.; Oruna-Concha, M.J.; Kolida, S.; Walton, G.E.; Kallithraka, S.; Spencer, J.P.E.; Gibson, G.R.; De Pascual-Teresa, S. Metabolism of Anthocyanins by Human Gut Microflora and Their Influence on Gut Bacterial Growth. J. Agric. Food Chem. 2012, 60, 3882–3890. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Ling, W.; Du, Z.; Chen, Y.; Li, D.; Deng, S.; Liu, Z.; Yang, L. Effects of Anthocyanins on Cardiometabolic Health: A Systematic Review and Meta-Analysis of Randomized Controlled Trials. Adv. Nutr. Int. Rev. J. 2017, 8, 684–693. [Google Scholar] [CrossRef]
- Wallace, T.C.; Slavin, M.; Frankenfeld, C.L. Systematic Review of Anthocyanins and Markers of Cardiovascular Disease. Nutrients 2016, 8, 32. [Google Scholar] [CrossRef]
- Herrera-Sotero, M.Y.; Cruz-Hernández, C.D.; Trujillo-Carretero, C.; Rodríguez-Dorantes, M.; García-Galindo, H.S.; Chávez-Servia, J.L.; Oliart-Ros, R.M.; Guzmán-Gerónimo, R.I. Antioxidant and Antiproliferative Activity of Blue Corn and Tortilla from Native Maize. Chem. Cent. J. 2017, 11, 110. [Google Scholar] [CrossRef]
- Guzmán-Gerónimo, R.I.; Aparicio, E.A.; Barradas, O.G.; Chávez-Servia, J.; Alarcón-Zavaleta, T.M. Chemical, Antioxidant, and Cytotoxic Properties of Native Blue Corn Extract. In Natural Products and Cancer Drug Discovery; InTech: London, UK, 2017. [Google Scholar]
- Magaña-Cerino, J.M.; Tiessen, A.; Soto-Luna, I.C.; Peniche-Pavía, H.A.; Vargas-Guerrero, B.; Domínguez-Rosales, J.A.; García-López, P.M.; Gurrola-Díaz, C.M. Consumption of Nixtamal from a New Variety of Hybrid Blue Maize Ameliorates Liver Oxidative Stress and Inflammation in a High-Fat Diet Rat Model. J. Funct. Foods 2020, 72, 104075. [Google Scholar] [CrossRef]
- Yuan, W.; Jiang, T.; Du, K.; Chen, H.; Cao, Y.; Xie, J.; Li, M.; Carr, J.P.; Wu, B.; Fan, Z.; et al. Maize Phenylalanine Ammonia-Lyases Contribute to Resistance to Sugarcane Mosaic Virus Infection, Most Likely through Positive Regulation of Salicylic Acid Accumulation. Mol. Plant Pathol. 2019, 20, 1365–1378. [Google Scholar] [CrossRef] [PubMed]
- Ôba, K.; Conn, E.E. Induction of Cinnamic Acid 4-Hydroxylase in Developing Maize Seedlings. Phytochemistry 1988, 27, 2447–2450. [Google Scholar] [CrossRef]
- Yun, M.-S.; Chen, W.; Deng, F.; Yogo, Y. Differential Properties of 4-Coumarate: CoA Ligase Related to Growth Suppression by Chalcone in Maize and Rice. Plant Growth Regul. 2005, 46, 169–176. [Google Scholar] [CrossRef]
- Hu, C.; Li, Q.; Shen, X.; Quan, S.; Lin, H.; Duan, L.; Wang, Y.; Luo, Q.; Qu, G.; Han, Q.; et al. Characterization of Factors Underlying the Metabolic Shifts in Developing Kernels of Colored Maize. Sci. Rep. 2016, 6, 35479. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Zhang, W.; Yang, H.; Dong, Q.; Ren, J.; Fan, H.; Zhang, X.; Zhou, Y. Comparative Transcriptome Analysis Reveals Differentially Expressed Genes Related to the Tissue-Specific Accumulation of Anthocyanins in Pericarp and Aleurone Layer for Maize. Sci. Rep. 2019, 9, 2485. [Google Scholar] [CrossRef]
- Li, Y.; Kim, J.I.; Pysh, L.; Chapple, C. Four Isoforms of Arabidopsis Thaliana 4-Coumarate: CoA Ligase (4CL) Have Overlapping yet Distinct Roles in Phenylpropanoid Metabolism. Plant Physiol. 2015, 169, 838. [Google Scholar] [CrossRef] [PubMed]
- Xiong, W.; Wu, Z.; Liu, Y.; Li, Y.; Su, K.; Bai, Z.; Guo, S.; Hu, Z.; Zhang, Z.; Bao, Y.; et al. Mutation of 4-Coumarate: Coenzyme A Ligase 1 Gene Affects Lignin Biosynthesis and Increases the Cell Wall Digestibility in Maize Brown Midrib5 Mutants. Biotechnol. Biofuels 2019, 12, 82. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, S.; Brefort, T.; Neidig, N.; Djamei, A.; Kahnt, J.; Vermerris, W.; Koenig, S.; Feussner, K.; Feussner, I.; Kahmann, R. A Secreted Ustilago Maydis Effector Promotes Virulence by Targeting Anthocyanin Biosynthesis in Maize. eLife 2014, 3, e01355. [Google Scholar] [CrossRef]
- Franken, P.; Niesbach-Klösgen, U.; Weydemann, U.; Maréchal-Drouard, L.; Saedler, H.; Wienand, U. The Duplicated Chalcone Synthase Genes C2 and Whp (White Pollen) of Zea Mays Are Independently Regulated; Evidence for Translational Control of Whp Expression by the Anthocyanin Intensifying Gene In. EMBO J. 1991, 10, 2605–2612. [Google Scholar] [CrossRef]
- Szalma, S.J.; Snook, M.E.; Bushman, B.S.; Houchins, K.E.; McMullen, M.D. Duplicate Loci as QTL: The Role of Chalcone Synthase Loci in Flavone and Phenylpropanoid Biosynthesis in Maize. Crop Sci. 2002, 42, 1679–1687. [Google Scholar] [CrossRef]
- Grotewold, E.; Peterson, T. Isolation and Characterization of a Maize Gene Encoding Chalcone Flavonone Isomerase. MGG Mol. Gen. Genet. 1994, 242, 1–8. [Google Scholar] [CrossRef]
- Cheng, A.X.; Han, X.J.; Wu, Y.F.; Lou, H.X. The Function and Catalysis of 2-Oxoglutarate-Dependent Oxygenases Involved in Plant Flavonoid Biosynthesis. Int. J. Mol. Sci. 2014, 15, 1080–1095. [Google Scholar] [CrossRef] [PubMed]
- Sharma, M.; Cortes-Cruz, M.; Ahern, K.R.; McMullen, M.; Brutnell, T.P.; Chopra, S. Identification of the Pr1 Gene Product Completes the Anthocyanin Biosynthesis Pathway of Maize. Genetics 2011, 188, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Jeske, L.; Placzek, S.; Schomburg, I.; Chang, A.; Schomburg, D. BRENDA in 2019: A European ELIXIR Core Data Resource. Nucleic Acids Res. 2019, 47, D542–D549. [Google Scholar] [CrossRef]
- Portwood, J.L.; Woodhouse, M.R.; Cannon, E.K.; Gardiner, J.M.; Harper, L.C.; Schaeffer, M.L.; Walsh, J.R.; Sen, T.Z.; Cho, K.T.; Schott, D.A.; et al. Maizegdb 2018: The Maize Multi-Genome Genetics and Genomics Database. Nucleic Acids Res. 2019, 47, D1146–D1154. [Google Scholar] [CrossRef]
- Lim, Y.; Go, M.; Yew, W. Exploiting the Biosynthetic Potential of Type III Polyketide Synthases. Molecules 2016, 21, 806. [Google Scholar] [CrossRef] [PubMed]
- KEGG Flavonoid Biosynthesis. Available online: http://www.genome.jp/kegg-bin/show_pathway?ec00941 (accessed on 2 June 2020).
- Coe, E.H.; McCormick, S.M.; Modena, S.A. White Pollen in Maize. J. Hered. 1981, 72, 318–320. [Google Scholar] [CrossRef]
- Della Vedova, C.B.; Lorbiecke, R.; Kirsch, H.; Schulte, M.B.; Scheets, K.; Borchert, L.M.; Scheffler, B.E.; Wienand, U.; Cone, K.C.; Birchler, J.A. The Dominant Inhibitory Chalcone Synthase Allele C2-Idf (Inhibitor Diffuse) From Zea Mays (L.) Acts via an Endogenous RNA Silencing Mechanism. Genetics 2005, 170, 1989–2002. [Google Scholar] [CrossRef]
- Eloy, N.B.; Voorend, W.; Lan, W.; Saleme, M.d.L.S.; Cesarino, I.; Vanholme, R.; Smith, R.A.; Goeminne, G.; Pallidis, A.; Morreel, K.; et al. Silencing CHALCONE SYNTHASE in Maize Impedes the Incorporation of Tricin into Lignin and Increases Lignin Content. Plant Physiol. 2017, 173, 998–1016. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Ding, T.; Su, B.; Jiang, H. Genome-Wide Identification, Characterization and Expression Analysis of the Chalcone Synthase Family in Maize. Int. J. Mol. Sci. 2016, 17, 161. [Google Scholar] [CrossRef]
- Dowd, P.F.; Berhow, M.A.; Johnson, E.T. Enhanced Pest Resistance and Increased Phenolic Production in Maize Callus Transgenically Expressing a Maize Chalcone Isomerase -3 like Gene. Plant Gene 2018, 13, 50–55. [Google Scholar] [CrossRef]
- Deboo, G.B.; Albertsen, M.C.; Taylor, L.P. Flavanone 3-Hydroxylase Transcripts and Flavonol Accumulation Are Temporally Coordinate in Maize Anthers. Plant J. 1995, 7, 703–713. [Google Scholar] [CrossRef] [PubMed]
- Larson, R.L.; Bussard, J.B. Microsomal Flavonoid 3′-Monooxygenase from Maize Seedlings. Plant Physiol. 1986, 80, 483–486. [Google Scholar] [CrossRef]
- Larson, R.; Bussard, J.B.; Coe, E.H. Gene-Dependent Flavonoid 3′-Hydroxylation in Maize. Biochem. Genet. 1986, 24, 615–624. [Google Scholar] [CrossRef]
- Ford, R.H. Inheritance of Kernel Color in Corn: Explanations & Investigations. Source Am. Biol. Teach. Publ. By Natl. Assoc. Biol. Teach. 2000, 62, 181–188. [Google Scholar] [CrossRef]
- Sharma, M.; Chai, C.; Morohashi, K.; Grotewold, E.; Snook, M.E.; Chopra, S. Expression of Flavonoid 3′-Hydroxylase Is Controlled by P1, the Regulator of 3-Deoxyflavonoid Biosynthesis in Maize. BMC Plant Biol. 2012, 12, 1. [Google Scholar] [CrossRef]
- Casas, M.I.; Falcone Ferreyra, M.L.; Jiang, N.; Mejía Guerra, M.K.; Rodríguez, E.; Wilson, T.; Engelmeier, J.; Casati, P.; Grotewold, E. Identification and Characterization of Maize Salmon Silks Genes Involved in Insecticidal Maysin Biosynthesis. Plant Cell 2016, 28, 1297–1309. [Google Scholar] [CrossRef]
- Bernhardt, J.; Stich, K.; Schwarz-Sommer, Z.; Saedler, H.; Wienand, U. Molecular Analysis of a Second Functional A1 Gene (Dihydroflavonol 4-Reductase) in Zea Mays. Plant J. 1998, 14, 483–488. [Google Scholar] [CrossRef]
- Yang, F.; Li, W.; Jiang, N.; Yu, H.; Morohashi, K.; Ouma, W.Z.; Morales-Mantilla, D.E.; Gomez-Cano, F.A.; Mukundi, E.; Prada-Salcedo, L.D.; et al. A Maize Gene Regulatory Network for Phenolic Metabolism. Mol. Plant 2017, 10, 498–515. [Google Scholar] [CrossRef]
- Brown, J.; Sundaresan, V. A Recombination Hotspot in the Maize A1 Intragenic Region. Theor. Appl. Genet. 1991, 81, 185–188. [Google Scholar] [CrossRef]
- Yao, H.; Zhou, Q.; Li, J.; Smith, H.; Yandeau, M.; Nikolau, B.J.; Schnable, P.S. Molecular Characterization of Meiotic Recombination across the 140-Kb Multigenic A1-Sh2 Interval of Maize. Proc. Natl. Acad. Sci. USA 2002, 99, 6157–6162. [Google Scholar] [CrossRef]
- McMullen, M.D.; Snook, M.; Lee, E.A.; Byrne, P.F.; Kross, H.; Musket, T.A.; Houchins, K.; Coe, J. The Biological Basis of Epistasis between Quantitative Trait Loci for Flavone and 3-Deoxyanthocyanin Synthesis in Maize (Zea Mays L.). Genome 2001, 44, 667–676. [Google Scholar] [CrossRef]
- Guo, B.Z.; Zhang, Z.J.; Butrón, A.; Widstrom, N.W.; Snook, M.E.; Lynch, R.E.; Plaisted, D. Lost P1 Allele in Sh2 Sweet Corn: Quantitative Effects of P1 and A1 Genes on Concentrations of Maysin, Apimaysin, Methoxymaysin, and Chlorogenic Acid in Maize Silk. J. Econ. Entomol. 2004, 97, 2117–2126. [Google Scholar] [CrossRef]
- Lesnick, M.L.; Chandler, V.L. Activation of the Maize Anthocyanin Gene A2 Is Mediated by an Element Conserved in Many Anthocyanin Promoters. Plant Physiol. 1998, 117, 437–445. [Google Scholar] [CrossRef]
- Furtek, D.; Schiefelbein, J.W.; Johnston, F.; Nelson, O.E. Sequence Comparisons of Three Wild-Type Bronze-1 Alleles from Zea Mays. Plant Mol. Biol. 1988, 11, 473–481. [Google Scholar] [CrossRef]
- Paulsmeyer, M.N.; Brown, P.J.; Juvik, J.A. Discovery of Anthocyanin Acyltransferase1 (AAT1) in Maize Using Genotyping-by-Sequencing (GBS). G3 Genes Genomes Genet. 2018, 8, 3669–3678. [Google Scholar] [CrossRef]
- Hugueney, P.; Provenzano, S.; Verriès, C.; Ferrandino, A.; Meudec, E.; Batelli, G.; Merdinoglu, D.; Cheynier, V.; Schubert, A.; Ageorges, A. A Novel Cation-Dependent O- Methyltransferase Involved in Anthocyanin Methylation in Grapevine. Plant Physiol. 2009, 150, 2057–2070. [Google Scholar] [CrossRef]
- Chatham, L.A.; Juvik, J.A. Linking Anthocyanin Diversity, Hue, and Genetics in Purple Corn. G3 Genes Genomes Genet. 2021, 11, jkaa062. [Google Scholar] [CrossRef]
- Marrs, K.A.; Alfenito, M.R.; Lloyd, A.M.; Walbot, V. A Glutathione S-Transferase Involved in Vacuolar Transfer Encoded by the Maize Gene Bronze-2. Nature 1995, 375, 397–400. [Google Scholar] [CrossRef]
- Goodman, C.D.; Casati, P.; Walbot, V. A Multidrug Resistance–Associated Protein Involved in Anthocyanin Transport in Zea Mays. Plant Cell 2004, 16, 1812–1826. [Google Scholar] [CrossRef]
- Wilmouth, R.C.; Turnbull, J.J.; Welford, R.W.D.; Clifton, I.J.; Prescott, A.G.; Schofield, C.J. Structure and Mechanism of Anthocyanidin Synthase from Arabidopsis Thaliana. Structure 2002, 10, 93–103. [Google Scholar] [CrossRef]
- Ford, C.M.; Boss, P.K.; Hæj, P.B. Cloning and Characterization of Vitis Vinifera UDP-Glucose. Flavonoid 3- O-Glucosyltransferase, a Homologue of the Enzyme Encoded by the Maize Bronze-1 Locus That May Primarily Serve to Glucosylate Anthocyanidins in vivo. J. Biol. Chem. 1998, 273, 9224–9233. [Google Scholar] [CrossRef]
- Fukuchi-Mizutani, M.; Okuhara, H.; Fukui, Y.; Nakao, M.; Katsumoto, Y.; Yonekura-Sakakibara, K.; Kusumi, T.; Hase, T.; Tanaka, Y. Biochemical and Molecular Characterization of a Novel UDP-Glucose:Anthocyanin 3′-O-Glucosyltransferase, a Key Enzyme for Blue Anthocyanin Biosynthesis, from Gentian. Plant Physiol. 2003, 132, 1652–1663. [Google Scholar] [CrossRef]
- Roth, B.A.; Goff, S.A.; Klein, T.M.; Fromm, M.E. C1- and R-Dependent Expression of the Maize Bz1 Gene Requires Sequences with Homology to Mammalian Myb and Myc Binding Sites. Plant Cell 1991, 3, 317–325. [Google Scholar] [CrossRef]
- Rhoades, M.M. The Effect of the Bronze Locus on Anthocyanin Formation in Maize. Am. Nat. 1952, 86, 105–108. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, S.; Li, H.; Du, H.; Huang, H.; Li, Y.; Hu, Y.; Liu, H.; Liu, Y.; Yu, G.; et al. Identification of Transcription Factors ZmMYB111 and ZmMYB148 Involved in Phenylpropanoid Metabolism. Front. Plant Sci. 2016, 7, 148. [Google Scholar] [CrossRef]
- Wang, Q.; Dooner, H.K. Remarkable Variation in Maize Genome Structure Inferred from Haplotype Diversity at the Bz Locus. Proc. Natl. Acad. Sci. USA 2006, 103, 17644–17649. [Google Scholar] [CrossRef]
- Dooner, H.K.; He, L. Maize Genome Structure Variation: Interplay between Retrotransposon Polymorphisms and Genic Recombination. Plant Cell 2008, 20, 249–258. [Google Scholar] [CrossRef]
- Kass, L.B.; Chomet, P. Barbara McClintock. In Handbook of Maize; Springer: New York, NY, USA, 2009; pp. 17–52. ISBN 9780387778631. [Google Scholar]
- Jones, R.N. McClintock’s Controlling Elements: The Full Story. Cytogenet. Genome Res. 2005, 109, 90–103. [Google Scholar] [CrossRef] [PubMed]
- Paulsmeyer, M.; Juvik, J. Functional Characterization of an Anthocyanin Dimalonyltransferase in Maize. Molecules 2021, 26, 2020. [Google Scholar] [CrossRef] [PubMed]
- Paulsmeyer, M.; Chatham, L.; Becker, T.; West, M.; West, L.; Juvik, J. Survey of Anthocyanin Composition and Concentration in Diverse Maize Germplasms. J. Agric. Food Chem. 2017, 65, 4341–4350. [Google Scholar] [CrossRef] [PubMed]
- McGonigle, B.; Keeler, S.J.; Lau, S.M.C.; Koeppe, M.K.; O’Keefe, D.P. A Genomics Approach to the Comprehensive Analysis of the Glutathione S-Transferase Gene Family in Soybean and Maize. Plant Physiol. 2000, 124, 1105–1120. [Google Scholar] [CrossRef] [PubMed]
- Neuefeind, T.; Huber, R.; Reinemer, P.; Knäblein, J.; Prade, L.; Mann, K.; Bieseler, B. Cloning, Sequencing, Crystallization and X-ray Structure of Glutathione S-Transferase-III from Zea mays Var. Mutin: A Leading Enzyme in Detoxification of Maize Herbicides. J. Mol. Biol. 1997, 274, 577–587. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Gao, Q.; Xu, L.; Pang, S.; Liu, Z.; Wang, C.; Tan, W. Characterization of Glutathione S-Transferases in the Detoxification of Metolachlor in Two Maize Cultivars of Differing Herbicide Tolerance. Pestic. Biochem. Physiol. 2017, 143, 265–271. [Google Scholar] [CrossRef] [PubMed]
- Pang, K.; Li, Y.; Liu, M.; Meng, Z.; Yu, Y. Inventory and General Analysis of the ATP-Binding Cassette (ABC) Gene Superfamily in Maize (Zea mays L.). Gene 2013, 526, 411–428. [Google Scholar] [CrossRef]
- Alfenito, M.R.; Souer, E.; Goodman, C.D.; Buell, R.; Mol, J.; Koes, R.; Walbot, V. Functional Complementation of Anthocyanin Sequestration in the Vacuole by Widely Divergent Glutathione S-Transferases. Plant Cell 1998, 10, 1135–1149. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Li, H.; Huang, J.R. Arabidopsis TT19 Functions as a Carrier to Transport Anthocyanin from the Cytosol to Tonoplasts. Mol. Plant 2012, 5, 387–400. [Google Scholar] [CrossRef]
- Larsen, E.S.; Alfenito, M.R.; Briggs, W.R.; Walbot, V. A Carnation Anthocyanin Mutant Is Complemented by the Glutathione S-Transferases Encoded by Maize Bz2 and Petunia An9. Plant Cell Rep. 2003, 21, 900–904. [Google Scholar] [CrossRef]
- Schmitz, G.; Theres, K. Structural and Functional Analysis of the Bz2 Locus of Zea mays: Characterization of Overlapping Transcripts. MGG Mol. Gen. Genet. 1992, 233, 269–277. [Google Scholar] [CrossRef] [PubMed]
- Pang, S.; Duan, L.; Liu, Z.; Song, X.; Li, X.; Wang, C. Co-Induction of a Glutathione-S-Transferase, a Glutathione Transporter and an ABC Transporter in Maize by Xenobiotics. PLoS ONE 2012, 7, e40712. [Google Scholar] [CrossRef] [PubMed]
- Bruce, W.; Folkerts, O.; Garnaat, C.; Crasta, O.; Roth, B.; Bowen, B. Expression Profiling of the Maize Flavonoid Pathway Genes Controlled by Estradiol-Inducible Transcription Factors CRC and P. Curr. Opin. Plant Biol. 2000, 3, 173. [Google Scholar] [CrossRef]
- Francisco, R.M.; Regalado, A.; Ageorges, A.; Burla, B.J.; Bassin, B.; Eisenach, C.; Zarrouk, O.; Vialet, S.; Marlin, T.; Chaves, M.M.; et al. ABCC1, an ATP Binding Cassette Protein from Grape Berry, Transports Anthocyanidin 3-O-Glucosides. Plant Cell 2013, 25, 1840–1854. [Google Scholar] [CrossRef]
- Behrens, C.E.; Smith, K.E.; Iancu, C.V.; Choe, J.-y.; Dean, J.V. Transport of Anthocyanins and Other Flavonoids by the Arabidopsis ATP-Binding Cassette Transporter AtABCC2. Sci. Rep. 2019, 9, 437. [Google Scholar] [CrossRef]
- González-Manzano, S.; Pérez-Alonso, J.J.; Salinas-Moreno, Y.; Mateus, N.; Silva, A.M.S.; de Freitas, V.; Santos-Buelga, C. Flavanol-Anthocyanin Pigments in Corn: NMR Characterisation and Presence in Different Purple Corn Varieties. J. Food Compos. Anal. 2008, 21, 521–526. [Google Scholar] [CrossRef]
- Chatham, L.A.; West, L.; Berhow, M.A.; Vermillion, K.E.; Juvik, J.A. Unique Flavanol-Anthocyanin Condensed Forms in Apache Red Purple Corn. J. Agric. Food Chem. 2018, 66, 10844–10854. [Google Scholar] [CrossRef] [PubMed]
- Ferreyra, M.L.F.; Rius, S.; Emiliani, J.; Pourcel, L.; Feller, A.; Morohashi, K.; Casati, P.; Grotewold, E. Cloning and Characterization of a UV-B-Inducible Maize Flavonol Synthase. Plant J. 2010, 62, 77–91. [Google Scholar] [CrossRef]
- Falcone Ferreyra, M.L.; Emiliani, J.; Rodriguez, E.J.; Campos-Bermudez, V.A.; Grotewold, E.; Casati, P. The Identification of Maize and Arabidopsis Type I FLAVONE SYNTHASEs Links Flavones with Hormones and Biotic Interactions. Plant Physiol. 2015, 169, 1090–1107. [Google Scholar] [CrossRef] [PubMed]
- Falcone Ferreyra, M.L.; Rodriguez, E.; Casas, M.I.; Labadie, G.; Grotewold, E.; Casati, P. Identification of a Bifunctional Maize C-and O-Glucosyltransferase. J. Biol. Chem. 2013, 288, 31678–31688. [Google Scholar] [CrossRef]
- Vanegas, K.G.; Larsen, A.B.; Eichenberger, M.; Fischer, D.; Mortensen, U.H.; Naesby, M. Indirect and Direct Routes to C-Glycosylated Flavones in Saccharomyces Cerevisiae. Microb. Cell Fact. 2018, 17, 1–10. [Google Scholar] [CrossRef]
- McMullen, M.D. Salmon Silk Genes Contribute to the Elucidation of the Flavone Pathway in Maize (Zea mays L.). J. Hered. 2004, 95, 225–233. [Google Scholar] [CrossRef]
- Zhang, Z.; Liang, Z.; Yin, L.; Li, Q.X.; Wu, Z. Distribution of Four Bioactive Flavonoids in Maize Tissues of Five Varieties and Correlation with Expression of the Biosynthetic Genes. J. Agric. Food Chem. 2018, 66, 10431–10437. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Lv, W.; Zhang, H.; Ma, L.; Li, P.; Ge, L.; Li, G. Genome-Wide Analysis of the Basic Helix-Loop-Helix (BHLH) Transcription Factor Family in Maize. BMC Plant Biol. 2018, 18, 235. [Google Scholar] [CrossRef] [PubMed]
- Morohashi, K.; Casas, M.I.; Falcone Ferreyra, M.L.; Mejía Guerra, M.K.; Pourcel, L.; Yilmaz, A.; Feller, A.; Carvalho, B.; Emiliani, J.; Rodriguez, E.; et al. A Genome-Wide Regulatory Framework Identifies Maize Pericarp Color1 Controlled Genes. Plant Cell 2012, 24, 2745–2764. [Google Scholar] [CrossRef]
- Brazier-Hicks, M.; Evans, K.M.; Gershater, M.C.; Puschmann, H.; Steel, P.G.; Edwards, R. The C -Glycosylation of Flavonoids in Cereals. J. Biol. Chem. 2009, 284, 17926–17934. [Google Scholar] [CrossRef] [PubMed]
- McMullen, M.D.; Byrne, P.F.; Snook, M.E.; Wiseman, B.R.; Lee, E.A.; Widstrom, N.W.; Coe, E.H. Quantitative Trait Loci and Metabolic Pathways. Proc. Natl. Acad. Sci. USA 1998, 95, 1996–2000. [Google Scholar] [CrossRef] [PubMed]
- Riaz, B.; Chen, H.; Wang, J.; Du, L.; Wang, K.; Ye, X. Overexpression of Maize ZmC1 and ZmR Transcription Factors in Wheat Regulates Anthocyanin Biosynthesis in a Tissue-Specific Manner. Int. J. Mol. Sci. 2019, 20, 5806. [Google Scholar] [CrossRef] [PubMed]
- Das, P.K.; Shin, D.H.; Choi, S.-B.; Park, Y.-I. Sugar-Hormone Cross-Talk in Anthocyanin Biosynthesis. Mol. Cells 2012, 34, 501–507. [Google Scholar] [CrossRef]
- Yilmaz, A.; Nishiyama, M.Y.; Fuentes, B.G.; Souza, G.M.; Janies, D.; Gray, J.; Grotewold, E. GRASSIUS: A Platform for Comparative Regulatory Genomics across the Grasses. Plant Physiol. 2009, 149, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Peterson, T. Comparisons of Maize Pericarp Color1 Alleles Reveal Paralogous Gene Recombination and an Organ-Specific Enhancer Region. Plant Cell 2005, 17, 903–914. [Google Scholar] [CrossRef] [PubMed]
- Gray, J.; Bevan, M.; Brutnell, T.; Buell, C.R.; Cone, K.; Hake, S.; Jackson, D.; Kellogg, E.; Lawrence, C.; McCouch, S.; et al. A Recommendation for Naming Transcription Factor Proteins in the Grasses. Plant Physiol. 2009, 149, 4–6. [Google Scholar] [CrossRef] [PubMed]
- Schnable, J.C.; Springer, N.M.; Freeling, M. Differentiation of the Maize Subgenomes by Genome Dominance and Both Ancient and Ongoing Gene Loss. Proc. Natl. Acad. Sci. USA 2011, 108, 4069–4074. [Google Scholar] [CrossRef] [PubMed]
- Coe, E.H. Anthocyanin genetics. In The Maize Handbook; Springer: New York, NY, USA, 1994; pp. 279–281. [Google Scholar]
- Hollick, J.B.; Chandler, V.L. Genetic Factors Required to Maintain Repression of a Paramutagenic Maize Pl1 Allele. Genetics 2001, 157, 369–378. [Google Scholar] [CrossRef] [PubMed]
- Sidorenko, L.; Chandler, V. RNA-Dependent RNA Polymerase Is Required for Enhancer-Mediated Transcriptional Silencing Associated with Paramutation at the Maize P1 Gene. Genetics 2008, 180, 1983–1993. [Google Scholar] [CrossRef] [PubMed]
- Walker, E.L. Paramutation of the R1 Locus of Maize Is Associated with Increased Cytosine Methylation. Genetics 1998, 148, 1973–1981. [Google Scholar] [CrossRef] [PubMed]
- Alleman, M.; Sidorenko, L.; McGinnis, K.; Seshadri, V.; Dorweiler, J.E.; White, J.; Sikkink, K.; Chandler, V.L. An RNA-Dependent RNA Polymerase Is Required for Paramutation in Maize. Nature 2006, 442, 295–298. [Google Scholar] [CrossRef] [PubMed]
- Cone, K.C.; Cocciolone, S.M.; Burr, F.A.; Burr, B. Maize Anthocyanin Regulatory Gene Pl Is a Duplicate of C1 That Functions in the Plant. Plant Cell 1993, 5, 1795–1805. [Google Scholar] [CrossRef] [PubMed]
- Cone, K.C.; Cocciolone, S.M.; Moehlenkamp, C.A.; Weber, T.; Drummond, B.J.; Tagliani, L.A.; Bowen, B.A.; Perrot, G.H. Role of the Regulatory Gene Pl in the Photocontrol of Maize Anthocyanin Pigmentation. Plant Cell 1993, 5, 1807–1816. [Google Scholar] [CrossRef]
- Paz-Ares, J.; Ghosal, D.; Wienand, U.; Peterson, P.A.; Saedler, H. The Regulatory C1 Locus of Zea Mays Encodes a Protein with Homology to Myb Proto-Oncogene Products and with Structural Similarities to Transcriptional Activators. EMBO J. 1987, 6, 3553–3558. [Google Scholar] [CrossRef] [PubMed]
- Yonemaru, J.I.; Miki, K.; Choi, S.; Kiyosawa, A.; Goto, K. A Genomic Region Harboring the Pl1 Allele from the Peruvian Cultivar JC072A Confers Purple Cob on Japanese Flint Corn (Zea mays L.). Breed. Sci. 2018, 68, 582–586. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, J.M.; Feller, A.; Morohashi, K.; Frame, K.; Grotewold, E. The Basic Helix-Loop-Helix Domain of Maize R Links Transcriptional Regulation and Histone Modifications by Recruitment of an EMSY-Related Factor. Proc. Natl. Acad. Sci. USA 2007, 104, 17222–17227. [Google Scholar] [CrossRef] [PubMed]
- Sainz, M.B.; Grotewold, E.; Chandler, V.L. Evidence for Direct Activation of an Anthocyanin Promoter by the Maize C1 Protein and Comparison of DNA Binding by Related Myb Domain Proteins. Plant Cell 1997, 9, 611–625. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Chopra, S.; Peterson, T. A Segmental Gene Duplication Generated Differentially Expressed Myb -Homologous Genes in Maize. Plant Cell 2000, 12, 2311–2322. [Google Scholar] [CrossRef]
- Zhang, P.; Wang, Y.; Zhang, J.; Maddock, S.; Snook, M.; Peterson, T. A Maize QTL for Silk Maysin Levels Contains Duplicated Myb-Homologous Genes Which Jointly Regulate Flavone Biosynthesis. Plant Mol. Biol. 2003, 52, 1–15. [Google Scholar] [CrossRef]
- Landoni, M.; Puglisi, D.; Cassani, E.; Borlini, G.; Brunoldi, G.; Comaschi, C.; Pilu, R. Phlobaphenes Modify Pericarp Thickness in Maize and Accumulation of the Fumonisin Mycotoxins. Sci. Rep. 2020, 10, 1417. [Google Scholar] [CrossRef]
- Procissi, A.; Piazza, P.; Tonelli, C. A Maize R1 Gene Is Regulated Post-Transcriptionally by Differential Splicing of Its Leader. Plant Mol. Biol. 2002, 49, 239–248. [Google Scholar] [CrossRef]
- Lago, C.; Landoni, M.; Cassani, E.; Cantaluppi, E.; Doria, E.; Nielsen, E.; Giorgi, A.; Pilu, R. Study and Characterization of an Ancient European Flint White Maize Rich in Anthocyanins: Millo Corvo from Galicia. PLoS ONE 2015, 10, e0126521. [Google Scholar] [CrossRef]
- Chandler, V.L.; Radicella, J.P.; Robbins, T.P.; Chen, J.; Turks, D. Two Regulatory Genes of the Maize Anthocyanin Pathway Are Homologous: Isolation of B Utilizing R Genomic Sequences. Plant Cell 1989, 1, 1175–1183. [Google Scholar] [CrossRef]
- Goff, S.A.; Klein, T.M.; Roth, B.A.; Fromm, M.E.; Cone, K.C.; Radicella, J.P.; Chandler, V.L. Transactivation of Anthocyanin Biosynthetic Genes Following Transfer of B Regulatory Genes into Maize Tissues. EMBO J. 1990, 9, 2517–2522. [Google Scholar] [CrossRef]
- Burr, F.A.; Burr, B.; Scheffler, B.E.; Blewitt, M.; Wienand, U.; Matz, E.C. The Maize Repressor-like Gene Intensifier1 Shares Homology with the R1/B1 Multigene Family of Transcription Factors and Exhibits Missplicing. Plant Cell 1996, 8, 1249–1259. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.C.; Chezem, W.R.; Clay, N.K. Ternary WD40 Repeat-Containing Protein Complexes: Evolution, Composition and Roles in Plant Immunity. Front. Plant Sci. 2016, 6, 1108. [Google Scholar] [CrossRef] [PubMed]
- Lauter, N.; Gustus, C.; Westerbergh, A.; Doebley, J. The Inheritance and Evolution of Leaf Pigmentation and Pubescence in Teosinte. Genetics 2004, 167, 1949–1959. [Google Scholar] [CrossRef]
- Selinger, D.A.; Chandler, V.L. A Mutation in the Pale Aleurone Color1 Gene Identifies a Novel Regulator of the Maize Anthocyanin Pathway. Plant Cell 1999, 11, 5–14. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, A.; Brockman, A.; Aguirre, L.; Campbell, A.; Bean, A.; Cantero, A.; González, A. Advances in the MYB–BHLH–WD Repeat (MBW) Pigment Regulatory Model: Addition of a WRKY Factor and Co-Option of an Anthocyanin MYB for Betalain Regulation. Plant Cell Physiol. 2017, 58, 1431–1441. [Google Scholar] [CrossRef] [PubMed]
- Christie, P.J.; Alfenito, M.R.; Walbot, V. Impact of Low-Temperature Stress on General Phenylpropanoid and Anthocyanin Pathways: Enhancement of Transcript Abundance and Anthocyanin Pigmentation in Maize Seedlings. Planta 1994, 194, 541–549. [Google Scholar] [CrossRef]
- Petroni, K.; Cominelli, E.; Consonni, G.; Gusmaroli, G.; Gavazzi, G.; Tonelli, C. The Developmental Expression of the Maize Regulatory Gene Hopi Determines Germination-Dependent Anthocyanin Accumulation. Genetics 2000, 155, 323–336. [Google Scholar] [CrossRef]
- Piazza, P.; Procissi, A.; Jenkins, G.I.; Tonelli, C. Members of the C1/Pl1 Regulatory Gene Family Mediate the Response of Maize Aleurone and Mesocotyl to Different Light Qualities and Cytokinins. Plant Physiol. 2002, 128, 1077–1086. [Google Scholar] [CrossRef]
- Pietrini, F.; Iannelli, M.A.; Massacci, A. Anthocyanin Accumulation in the Illuminated Surface of Maize Leaves Enhances Protection from Photo-Inhibitory Risks at Low Temperature, without Further Limitation to Photosynthesis. Plant Cell Environ. 2002, 25, 1251–1259. [Google Scholar] [CrossRef]
- Hattori, T.; Vasil, V.; Rosenkrans, L.; Hannah, L.C.; McCarty, D.R.; Vasil, I.K. The Viviparous-1 Gene and Abscisic Acid Activate the C1 Regulatory Gene for Anthocyanin Biosynthesis during Seed Maturation in Maize. Genes Dev. 1992, 6, 609–618. [Google Scholar] [CrossRef]
- Kim, J.S.; Lee, B.H.; Kim, S.H.; Oh, K.H.; Kwang, Y.C. Responses to Environmental and Chemical Signals for Anthocyanin Biosynthesis in Non-Chlorophyllous Corn (Zea mays L.) Leaf. J. Plant Biol. 2006, 49, 16–25. [Google Scholar] [CrossRef]
- McCarty, D.R.; Carson, C.B.; Stinard, P.S.; Robertson, D.S. Molecular Analysis of Viviparous-1: An Abscisic Acid-Insensitive Mutant of Maize. Plant Cell 1989, 1, 523–532. [Google Scholar] [CrossRef]
- Wang, Y.; Yao, Q.; Zhang, Y.; Zhang, Y.; Xing, J.; Yang, B.; Mi, G.; Li, Z.; Zhang, M. The Role of Gibberellins in Regulation of Nitrogen Uptake and Physiological Traits in Maize Responding to Nitrogen Availability. Int. J. Mol. Sci. 2020, 21, 1824. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.S.; Bae, D.W.; Ryu, C.M. Aboveground Whitefly Infestation Modulates Transcriptional Levels of Anthocyanin Biosynthesis and Jasmonic Acid Signaling-Related Genes and Augments the Cope with Drought Stress of Maize. PLoS ONE 2015, 10, e0143879. [Google Scholar] [CrossRef]
- Oláh, C.; Ludmerszki, E.; Rácz, I.; Balassa, G.; Rudnóy, S. S-Methylmethionine-Salicylate Pretreatment Reduces Low Temperature Stress in Maize. Russ. J. Plant Physiol. 2018, 65, 63–68. [Google Scholar] [CrossRef]
- Rudnóy, S.; Majláth, I.; Pál, M.; Páldi, K.; Rácz, I.; Janda, T. Interactions of S-Methylmethionine and UV-B Can Modify the Defence Mechanisms Induced in Maize. Acta Physiol. Plant. 2015, 37, 148. [Google Scholar] [CrossRef]
- Hollick, J.B. Paramutation and Related Phenomena in Diverse Species. Nat. Rev. Genet. 2017, 18, 5–23. [Google Scholar] [CrossRef]
- Stam, M.; Belele, C.; Dorweiler, J.E.; Chandler, V.L. Differential Chromatin Structure within a Tandem Array 100 Kb Upstream of the Maize B1 Locus Is Associated with Paramutation. Genes Dev. 2002, 16, 1906–1918. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.H.; Wittmeyer, K.T.; Lee, T.F.; Meyers, B.C.; Chopra, S. Overlapping RdDM and Non-RdDM Mechanisms Work Together to Maintain Somatic Repression of a Paramutagenic Epiallele of Maize Pericarp Color1. PLoS ONE 2017, 12, e0187157. [Google Scholar] [CrossRef]
- Zhang, G.; Chen, D.; Zhang, T.; Duan, A.; Zhang, J.; He, C. Transcriptomic and Functional Analyses Unveil the Role of Long Non-Coding RNAs in Anthocyanin Biosynthesis during Sea Buckthorn Fruit Ripening. DNA Res. 2018, 25, 465–476. [Google Scholar] [CrossRef]
- Chialva, C.; Blein, T.; Crespi, M.; Lijavetzky, D. Insights into Long Non-Coding RNA Regulation of Anthocyanin Carrot Root Pigmentation. Sci. Rep. 2021, 11, 4093. [Google Scholar] [CrossRef] [PubMed]
- Waititu, J.K.; Zhang, C.; Liu, J.; Wang, H. Plant Non-Coding Rnas: Origin, Biogenesis, Mode of Action and Their Roles in Abiotic Stress. Int. J. Mol. Sci. 2020, 21, 8401. [Google Scholar] [CrossRef] [PubMed]
- Magaña Cerino, J.; Peniche Pavía, H.; Tiessen, A.; Gurrola Díaz, C. Pigmented Maize (Zea mays L.) Contains Anthocyanins with Potential Therapeutic Action Against Oxidative Stress-A Review. Pol. J. Food Nutr. Sci. 2019, 70, 85–99. [Google Scholar] [CrossRef]
- Ma, X.; Ning, S. Cyanidin-3-Glucoside Attenuates the Angiogenesis of Breast Cancer via Inhibiting STAT3/VEGF Pathway. Phyther. Res. 2019, 33, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.; Liu, X.; He, J.; Shao, Y.; Liu, J.; Wang, Z.; Xia, L.; Han, T.; Wu, P. Cyanidin-3-Glucoside Induces Mesenchymal to Epithelial Transition via Activating Sirt1 Expression in Triple Negative Breast Cancer Cells. Biochimie 2019, 162, 107–115. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Du, Y.; Li, H.; Wang, L.; Ponikwicka-Tyszko, D.; Lebiedzinska, W.; Pilaszewicz-Puza, A.; Liu, H.; Zhou, L.; Fan, H.; et al. Cyanidin-3-o-Glucoside Pharmacologically Inhibits Tumorigenesis via Estrogen Receptor β in Melanoma Mice. Front. Oncol. 2019, 9, 1110. [Google Scholar] [CrossRef]
- Baster, Z.; Li, L.; Kukkurainen, S.; Chen, J.; Pentikäinen, O.; Győrffy, B.; Hytönen, V.P.; Zhu, H.; Rajfur, Z.; Huang, C. Cyanidin-3-Glucoside Binds to Talin and Modulates Colon Cancer Cell Adhesions and 3D Growth. FASEB J. 2020, 34, 2227–2237. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Yuan, M.; Ye, Q.; Wang, X.; Xu, J.; Shi, G.; Hu, Z. Cyanidin-3-o-Glucoside Inhibits Epithelial-to-Mesenchymal Transition, and Migration and Invasion of Breast Cancer Cells by Upregulating Klf4. Food Nutr. Res. 2020, 64, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Wei, T.; Ji, X.; Xue, J.; Gao, Y.; Zhu, X.; Xiao, G. Cyanidin-3-: O-Glucoside Represses Tumor Growth and Invasion in Vivo by Suppressing Autophagy via Inhibition of the JNK Signaling Pathways. Food Funct. 2021, 12, 387–396. [Google Scholar] [CrossRef]
- Li, X.; Zhao, J.; Yan, T.; Mu, J.; Lin, Y.; Chen, J.; Deng, H.; Meng, X. Cyanidin-3-O-Glucoside and Cisplatin Inhibit Proliferation and Downregulate the PI3K/AKT/MTOR Pathway in Cervical Cancer Cells. J. Food Sci. 2021, 86, 2700–2712. [Google Scholar] [CrossRef] [PubMed]
- Matboli, M.; Hasanin, A.H.; Hussein, R.; El-Nakeep, S.; Habib, E.K.; Ellackany, R.; Saleh, L.A. Cyanidin 3-Glucoside Modulated Cell Cycle Progression in Liver Precancerous Lesion, in Vivo Study. World J. Gastroenterol. 2021, 27, 1435–1450. [Google Scholar] [CrossRef]
- Zhao, L.; Pan, F.; Zhou, N.; Zhang, H.; Wang, Y.; Hao, S.; Wang, C. Quantitative Proteomics and Bioinformatics Analyses Reveal the Protective Effects of Cyanidin-3-O-Glucoside and Its Metabolite Protocatechuic Acid against 2-Amino-3-Methylimidazo [4,5-f] Quinoline (IQ)-Induced Cytotoxicity in HepG2 Cells via Apoptosis-Relat. Food Chem. Toxicol. 2021, 153, 2256. [Google Scholar] [CrossRef]
- Li, X.; Mu, J.; Lin, Y.; Zhao, J.; Meng, X. Combination of Cyanidin-3-O-Glucoside and Cisplatin Induces Oxidative Stress and Apoptosis in HeLa Cells by Reducing Activity of Endogenous Antioxidants, Increasing Bax/Bcl-2 MRNA Expression Ratio, and Downregulating Nrf2 Expression. J. Food Biochem. 2021, 45, e13806. [Google Scholar] [CrossRef]
- Wang, Y.; Qi, W.; Huo, Y.; Song, G.; Sun, H.; Guo, X.; Wang, C. Cyanidin-3-Glucoside Attenuates 4-Hydroxynonenal-A Nd Visible Light-Induced Retinal Damage: In Vitro and in Vivo. Food Funct. 2019, 10, 2871–2880. [Google Scholar] [CrossRef] [PubMed]
- Takashina, Y.; Manabe, A.; Tabuchi, Y.; Ikari, A. Cyanidin Increases the Expression of Mg2+ Transport Carriers Mediated by the Activation of PPARα in Colonic Epithelial MCE301 Cells. Nutrients 2019, 11, 641. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.X.; Qi, S.S.; He, J.; Hu, C.Y.; Han, H.; Jiang, H.; Li, X.S. Cyanidin-3-Glucoside from Black Rice Ameliorates Diabetic Nephropathy via Reducing Blood Glucose, Suppressing Oxidative Stress and Inflammation, and Regulating Transforming Growth Factor Β1/Smad Expression. J. Agric. Food Chem. 2020, 68, 4399–4410. [Google Scholar] [CrossRef]
- Li, X.; Sun, M.; Long, Y. Cyanidin-3-O-Glucoside Attenuates Lipopolysaccharide-Induced Inflammation in Human Corneal Epithelial Cells by Inducing Let-7b-5p-Mediated HMGA2/PI3K/Akt Pathway. Inflammation 2020, 43, 1088–1096. [Google Scholar] [CrossRef]
- Tian, L.; Ning, H.; Shao, W.; Song, Z.; Badakhshi, Y.; Ling, W.; Yang, B.B.; Brubaker, P.L.; Jin, T. Dietary Cyanidin-3-Glucoside Attenuates High-Fat-Diet-Induced Body-Weight Gain and Impairment of Glucose Tolerance in Mice via Effects on the Hepatic Hormone FGF21. J. Nutr. 2020, 150, 2101–2111. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, M.; Wang, Z.; Guo, Z.; Wang, Z.; Chen, Q. Cyanidin-3-O-Glucoside Attenuates Endothelial Cell Dysfunction by Modulating MiR-204-5p/SIRT1-Mediated Inflammation and Apoptosis. BioFactors 2020, 46, 803–812. [Google Scholar] [CrossRef]
- Song, X.-L.; Li, M.-J.; Liu, Q.; Hu, Z.-X.; Xu, Z.-Y.; Li, J.-H.; Zheng, W.-L.; Huang, X.-M.; Xiao, F.; Cui, Y.-H.; et al. Cyanidin-3- O-Glucoside Protects Lens Epithelial Cells against High Glucose-Induced Apoptosis and Prevents Cataract Formation via Suppressing NF-ΚB Activation and Cox-2 Expression. J. Agric. Food Chem. 2020, 68, 8286–8294. [Google Scholar] [CrossRef]
- Molonia, M.S.; Occhiuto, C.; Muscarà, C.; Speciale, A.; Bashllari, R.; Villarroya, F.; Saija, A.; Cimino, F.; Cristani, M. Cyanidin-3-O-Glucoside Restores Insulin Signaling and Reduces Inflammation in Hypertrophic Adipocytes. Arch. Biochem. Biophys. 2020, 691, 8488. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Wu, C.; Kim, Y.-S.; Yang, S.O.; Kim, Y.; Kim, J.-S.; Jeong, M.-Y.; Lee, J.H.; Kim, B.; Lee, S.; et al. A Dietary Anthocyanin Cyanidin-3-O-Glucoside Binds to PPARs to Regulate Glucose Metabolism and Insulin Sensitivity in Mice. Commun. Biol. 2020, 3, 514. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Shi, Z.; Zhu, Y.; Shen, T.; Wang, H.; Shui, G.; Loor, J.J.; Fang, Z.; Chen, M.; Wang, X.; et al. Cyanidin-3-O-Glucoside Improves Non-Alcoholic Fatty Liver Disease by Promoting PINK1-Mediated Mitophagy in Mice. Br. J. Pharmacol. 2020, 177, 3591–3607. [Google Scholar] [CrossRef]
- Chen, L.; Li, K.; Liu, Q.; Quiles, J.L.; Filosa, R.; Kamal, M.A.; Wang, F.; Kai, G.; Zou, X.; Teng, H.; et al. Protective Effects of Raspberry on the Oxidative Damage in HepG2 Cells through Keap1/Nrf2-Dependent Signaling Pathway. Food Chem. Toxicol. 2019, 133, 110781. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.; Li, P.; Xue, H.; Li, Q. Cyanidin-3-Glucoside Prevents Hydrogen Peroxide (H2O2)-Induced Oxidative Damage in HepG2 Cells. Biotechnol. Lett. 2020, 42, 2453–2466. [Google Scholar] [CrossRef]
- Zhou, Y.; Wang, S.; Wan, T.; Huang, Y.; Pang, N.; Jiang, X.; Gu, Y.; Zhang, Z.; Luo, J.; Yang, L. Cyanidin-3-O-β-Glucoside Inactivates NLRP3 Inflammasome and Alleviates Alcoholic Steatohepatitis via SirT1/NF-ΚB Signaling Pathway. Free Radic. Biol. Med. 2020, 160, 334–341. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Tian, L.-M.; Liu, Y.; Guo, K.-S.; Lv, M.; Li, Q.-T.; Hao, S.-Y.; Ma, C.-H.; Chen, Y.-X.; Tanaka, M.; et al. Low Dose of Cyanidin-3-O-Glucoside Alleviated Dextran Sulfate Sodium-Induced Colitis, Mediated by CD169+ Macrophage Pathway. Inflamm. Bowel Dis. 2019, 25, 1510–1521. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Wang, G.; Zhu, C.; Jiang, X.; Sun, J.; Tian, L.; Bai, W. Effects of Cyanidin-3-O-Glucoside on 3-Chloro-1,2-Propanediol Induced Intestinal Microbiota Dysbiosis in Rats. Food Chem. Toxicol. 2019, 133, 110767. [Google Scholar] [CrossRef] [PubMed]
- Gan, Y.; Fu, Y.; Yang, L.; Chen, J.; Lei, H.; Liu, Q. Cyanidin-3-O-Glucoside and Cyanidin Protect against Intestinal Barrier Damage and 2,4,6-Trinitrobenzenesulfonic Acid-Induced Colitis. J. Med. Food 2020, 23, 90–99. [Google Scholar] [CrossRef]
- Bashllari, R.; Molonia, M.S.; Muscarà, C.; Speciale, A.; Wilde, P.J.; Saija, A.; Cimino, F. Cyanidin-3-O-Glucoside Protects Intestinal Epithelial Cells from Palmitate-Induced Lipotoxicity. Arch. Physiol. Biochem. 2020, 1–8. [Google Scholar] [CrossRef]
- Kaewmool, C.; Udomruk, S.; Phitak, T.; Pothacharoen, P.; Kongtawelert, P. Cyanidin-3-O-Glucoside Protects PC12 Cells Against Neuronal Apoptosis Mediated by LPS-Stimulated BV2 Microglial Activation. Neurotox. Res. 2020, 37, 111–125. [Google Scholar] [CrossRef]
- Sukprasansap, M.; Chanvorachote, P.; Tencomnao, T. Cyanidin-3-Glucoside Activates Nrf2-Antioxidant Response Element and Protects against Glutamate-Induced Oxidative and Endoplasmic Reticulum Stress in HT22 Hippocampal Neuronal Cells. BMC Complement. Med. Ther. 2020, 20, 46. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Zhao, F.; Wang, W.; Sang, J.; Jia, L.; Li, L.; Lu, F. Cyanidin-3-O-Glucoside Inhibits Aβ40 Fibrillogenesis, Disintegrates Preformed Fibrils, and Reduces Amyloid Cytotoxicity. Food Funct. 2020, 11, 2573–2587. [Google Scholar] [CrossRef]
- Jaiswal, V.; Park, M.; Lee, H.-J. Comparative Transcriptome Analysis of the Expression of Antioxidant and Immunity Genes in the Spleen of a Cyanidin 3-o-Glucoside-Treated Alzheimer’s Mouse Model. Antioxidants 2021, 10, 1435. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Lu, J.-L.; Sun, J.-X.; Jiang, X.-W.; Li, X.-S.; Li, Y.; Jiao, R.; Tian, L.-M.; Bai, W.-B. Cyanidin-3-O-Glucoside Promotes Progesterone Secretion by Improving Cells Viability and Mitochondrial Function in Cadmium-Sulfate-Damaged R2C Cells. Food Chem. Toxicol. 2019, 128, 97–105. [Google Scholar] [CrossRef]
- Li, X.; Guo, J.; Jiang, X.; Sun, J.; Tian, L.; Jiao, R.; Tang, Y.; Bai, W. Cyanidin-3-O-Glucoside Protects against Cadmium-Induced Dysfunction of Sex Hormone Secretion via the Regulation of Hypothalamus-Pituitary-Gonadal Axis in Male Pubertal Mice. Food Chem. Toxicol. 2019, 129, 13–21. [Google Scholar] [CrossRef]
- Ma, B.; Wu, Y.; Chen, B.; Yao, Y.; Wang, Y.; Bai, H.; Li, C.; Yang, Y.; Chen, Y. Cyanidin-3-O-β-Glucoside Attenuates Allergic Airway Inflammation by Modulating the IL-4Rα-STAT6 Signaling Pathway in a Murine Asthma Model. Int. Immunopharmacol. 2019, 69, 1–10. [Google Scholar] [CrossRef]
- Pitsillou, E.; Liang, J.; Ververis, K.; Lim, K.W.; Hung, A.; Karagiannis, T.C. Identification of Small Molecule Inhibitors of the Deubiquitinating Activity of the SARS-CoV-2 Papain-Like Protease: In Silico Molecular Docking Studies and in Vitro Enzymatic Activity Assay. Front. Chem. 2020, 8, 3971. [Google Scholar] [CrossRef]
- Pitsillou, E.; Liang, J.; Ververis, K.; Hung, A.; Karagiannis, T.C. Interaction of Small Molecules with the SARS-CoV-2 Papain-like Protease: In Silico Studies and in Vitro Validation of Protease Activity Inhibition Using an Enzymatic Inhibition Assay. J. Mol. Graph. Model. 2021, 104, 7851. [Google Scholar] [CrossRef]
- Ouyang, S.; Chen, W.; Gaofeng, Z.; Changcheng, L.; Guoping, T.; Minyan, Z.; Yang, L.; Min, Y.; Luo, J. Cyanidin-3-O-β-glucoside Protects against Pulmonary Artery Hypertension Induced by Monocrotaline via the TGF-β1/P38 MAPK/CREB Signaling Pathway. Mol. Med. Rep. 2021, 23, 1977. [Google Scholar] [CrossRef]
- Wang, H.; Li, S.; Zhang, G.; Wu, H.; Chang, X. Potential Therapeutic Effects of Cyanidin-3-O-Glucoside on Rheumatoid Arthritis by Relieving Inhibition of CD38+ NK Cells on Treg Cell Differentiation. Arthritis Res. Ther. 2019, 21, 220. [Google Scholar] [CrossRef]
- Aloud, B.M.; Petkau, J.C.; Yu, L.; McCallum, J.; Kirby, C.; Netticadan, T.; Blewett, H. Effects of Cyanidin 3-: O-Glucoside and Hydrochlorothiazide on T-Cell Phenotypes and Function in Spontaneously Hypertensive Rats. Food Funct. 2020, 11, 8560–8572. [Google Scholar] [CrossRef]
- Hiemori-Kondo, M.; Morikawa, E.; Fujikura, M.; Nagayasu, A.; Maekawa, Y. Inhibitory Effects of Cyanidin-3-O-Glucoside in Black Soybean Hull Extract on RBL-2H3 Cells Degranulation and Passive Cutaneous Anaphylaxis Reaction in Mice. Int. Immunopharmacol. 2021, 94, 7394. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Hu, Y.; Bai, W.; Zhao, J.; Huang, C.; Wen, C.; Deng, L.; Lu, D. Cyanidin-3-o-Glucoside Inhibits UVA-Induced Human Dermal Fibroblast Injury by Upregulating Autophagy. Photodermatol. Photoimmunol. Photomed. 2019, 35, 360–368. [Google Scholar] [CrossRef] [PubMed]
- Lian, W.; Chen, W. Cyanidin-3-o-Glucoside Improves Colonic Motility during Severe Acute Pancreatitis by Inhibiting the H2S-Regulated Ampk/Mtor Pathway. Drug Des. Dev. Ther. 2020, 14, 3385–3391. [Google Scholar] [CrossRef] [PubMed]
- Tri, A.; Safitri, A.; Fatchiyah, F. An in silico Approach Reveals the Potential Function of Cyanidin-3-o-Glucoside of Red Rice in Inhibiting the Advanced Glycation End Products (AGES)-Receptor (RAGE) Signaling Pathway. Acta Inform. Med. 2020, 28, 170–179. [Google Scholar] [CrossRef]
- Hu, B.; Chen, L.; Chen, Y.; Zhang, Z.; Wang, X.; Zhou, B. Cyanidin-3-Glucoside Regulates Osteoblast Differentiation via the ERK1/2 Signaling Pathway. ACS Omega 2021, 6, 4759–4766. [Google Scholar] [CrossRef]
- Chuntakaruk, H.; Kongtawelert, P.; Pothacharoen, P. Chondroprotective Effects of Purple Corn Anthocyanins on Advanced Glycation End Products Induction through Suppression of NF-ΚB and MAPK Signaling. Sci. Rep. 2021, 11, 1895. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, S.; Ferdousi, F.; Zheng, Y.-W.; Oda, T.; Isoda, H. Human Amniotic Epithelial Cells as a Tool to Investigate the Effects of Cyanidin 3-o-Glucoside on Cell Differentiation. Int. J. Mol. Sci. 2021, 22, 3768. [Google Scholar] [CrossRef] [PubMed]






| Gene Name | Locus | Enzyme/Protein Name | EC | Reference |
|---|---|---|---|---|
| (ZmPAL) | m* | Phenylalanine ammonium lyase | 4.3.1.24 | [16] |
| (ZmC4H) | 8L | Cinnamic acid 4-hydroxylase | 1.14.14.91 | [16,17] |
| bm5 (Zm4CL) | 5 | 4-Coumarate CoA ligase | 6.2.1.12 | [22] |
| c2 (ZmCHS) | 4L | Chalcone synthase | 2.3.1.74 | [24] |
| whp1 (ZmCHS) | 2L | Chalcone synthase | 2.3.1.74 | [25] |
| chi1 (ZmCHI) | 1L | Chalcone isomerase | 5.5.1.6 | [26] |
| fht1 (ZmF3H) | 2S | Flavonoid 3-dioxygenase | 1.14.11.9 | [27] |
| pr1 (ZmF3′H) | 5L | Flavonoid 3′-monooxygenase | 1.14.14.82 | [28] |
| Gene Name | Locus | Enzyme/Protein Name | EC | Reference |
|---|---|---|---|---|
| pr1 (ZmF3′H) | 5L | Flavonoid 3′-monooxygenase | 1.14.14.82 | [28] |
| a1 (ZmDFR) | 3L | Dihydroflavonol 4-reductase | 1.1.1.219 | [44] |
| -(ZmLAR) | - | Leucoanthocyanidin reductase | 1.17.1.3 | - * |
| a2 (ZmANS) | 5S | Anthocyanidin synthase | 1.14.20.4 | [50] |
| bz1 (ZmAGT) | 9S | Anthocyanidin 3-O-glucosyltransferase | 2.4.1.115 | [51] |
| aat1 (Zm3MAT) | 1L | Malonyl-CoA: anthocyanin 3-O-glucoside-6′′-O-malonyltransferase | 2.3.1.171 | [52] |
| omt1 and omt4- (ZmAOMT) | 4L | Anthocyanin S-adenosyl-l-methionine-dependent O- methyltransferase | 2.1.1.267 | [53,54] |
| bz2 (ZmGST) | 4L | Glutathione-S-transferase | 2.5.1.18 | [55] |
| mrpa3 (ZmABC3) mrpa4 (ZmABC4) | 9S 1S | Multidrug resistance-associated protein or ATP-binding cassette transporter | 7.6.2.2 | [56] |
| Gene Name | Locus | Enzyme/Protein Name | EC | Reference |
|---|---|---|---|---|
| fls1 (ZmFLS1) fls2 (ZmFLS2) | 5L 5L | Flavonol synthase | 1.14.20.6 | [1] |
| fnsi1 (ZmFNSI1) fnsi2 (ZmFNSI2) | 1S 1S | Flavone synthase I | 1.14.20.5 | [84] |
| fnsii1 (ZmFNSII1) | 10L | Flavone synthase II | 1.14.19.76 | [2] |
| fns1 (ZmF2H1) | 9L | Flavanone 2-hydroxylase | 1.14.14.162 | [85] |
| cgt1 (ZmCGT) | 6L | UDP-glucose:2-hydroxyflavanone C-glucosyltransferase | 2.4.1.360 | [86] |
| sm2 (UGT91L1) | 2L | flavonol-3-O-glucoside L-rhamnosyltransferase | 2.4.1.159 | [87] |
| sm1 (ZmRHS1) | 6L | Glucose-4,6 dehydratase | 4.2.1.76 | [43] |
| Gene Name | Family | Locus | Function | Regulates | Expression of Functional Allele | Paramutation |
|---|---|---|---|---|---|---|
| c1 (ZmMYB1) | R2R3-MYB | 9S | + | a1, a2, bz1, bz2, and c2 | Aleurone and scutellum | Not known |
| pl1 (ZmMYB2) | R2R3-MYB | 6L | + | Same as c1 | Sheaths, pericarp, husk, culms, cob, and anther glumes | Yes |
| p1 (ZmMYB3) | R2R3-MYB | 1S | + (works alone) | a1 and ch1 | Pericarp, silks, cob, and anther glumes | Yes |
| p2 (ZmMYB55) | R2R3-MYB | 1S | + (works alone) | Same as p1 | Silks and anther glumes | Not known |
| r1 (ZmbHLH1) | bHLH | 10L | + | a1, a2, bz1, bz2, and c2 | Anthers, brace roots, leaf blade tips, aleurone. and scutellum | Yes |
| b1 (ZmbHLH2) | bHLH | 2S | + | Same as r1 | Sheaths, pericarp, husk, culms, cob, and anther glumes | Yes |
| in1 | bHLH | 7S | - | r1 | Competition against r1 | Not known |
| pac1 (ZmWD40) | WD40 | 5L | + | a1, a2, bz1, bz2, and c2 | Any anthocyanin pigmented tissue | Not known |
| a3 | - | 3L | - | b1 | Dominant inhibitor | Not known |
| Biological Effects | Type of Study | Dose, Time, and Model | Main Biological Findings | Ref. |
|---|---|---|---|---|
| Antitumoral | In vitro | 10 and 20 µM; 24 h; human breast cancer MDA-MB-231 and Hs-578T cells | Attenuation of breast cancer-induced angiogenesis via inhibiting VEGF up-regulation of miR-124 reduces angiogenesis (inhibiting STAT3). | [140] |
| In vitro | 5, 10, 20, and 40 µM; 24 h; MDA-MB-231 and BT-549 cells | C3G induces reversion of EMT characterized by phenotype modulation with increased epithelial marker E-ca and ZO-1 and decreased mesenchymal marker vimentin, N-ca, and EMT-associated transcription factors Snail1 and Snail2. | [141] | |
| In vitro and in vivo | 5, 20, 50, 150, 300, and 500 µM; 12, 24, 48, and 72 h; human breast cancer cells, melanoma cells, human embryonic kidney 293 cells, mouse and human primary melanocytes, and human samples of melanoma | C3G treatment arrested the cell cycle at the G2/M phase by targeting cyclin B1 (CCNB1) and promoted apoptosis via ERβ in both mouse and human melanoma cell lines. | [142] | |
| In vivo | 10, 20, 40, and 80 µM; Chinese hamster ovary cells, human colon cancer cell lines, human breast cancer cell lines, and human melanoma cell line | C3G binds to talin (a key regulator of integrins and cell adhesion) and promotes the interaction of talin with β1A-integrin. | [143] | |
| In vitro | 10 and 40 μM; 24 h; MDA-MB-231 and MDA-MB-468 breast cancer cells | The EMT inhibition is related to the upregulation of KLF4, which has been reported to be an EMT suppressor in breast cancer cells. The upregulation of KLF4 expression by C3G involves transcriptional suppression of FBXO32. It was found that FBXO32 acted as a promoter of EMT and cell migration/invasion. | [144] | |
| In vivo | Drosophila malignant RafGOFscrib −/− model. Purified C3G was added to standard food. Doses: C3G: 0.1 mg/mL or 0.4 mg/mL. | Purified C3G inhibited tumor growth invasion, distant migration and prolongs the survival of tumor flies. C3G inhibited tumor invasion by reducing the MMP1 activity and through JNK pathway. | [145] | |
| In vitro | HeLa cells by evaluating cell proliferation assay (C3G doses: 0–800 μg/mL) during 24, 48, and 72 h; apoptosis (Cy3G dose: 400 μg/mL), cell cycle, cell migration, and invasion evaluation (C3G dose: 400 μg/mL). | C3G combined with DPP induced apoptosis associated with the suppressed PI3K/AKT/mTOR signaling. DDP plus C3G treatment of HeLa cells can inhibit cell proliferation through cyclin D1 downregulation. Finally, this combined treatment could inhibit the migration and invasion associated with decreasing the protein of TIMP-1. | [146] | |
| In vivo | Induction of hepatic precancerous lesion (PCL) with diethylnitrosamine/2-acetylaminofluorene (DEN/2-AAF) in a Wistar rat model. C3G (not described source) at 10, 15, and 20 mg/kg/day. The measured parameters were alpha fetoprotein (AFP) levels and liver function biochemical analytes, and RNA panel differential expression was evaluated via qPCR. Histopathological examination of liver sections stained with H&E was also conducted. | AFP levels were significantly decreased in the three C3G doses. Decreased ALT levels and increased serum albumin were found after C3G treatment. Moreover, C3G treatment decreases, in a dose-dependent manner, the mRNA expression of long non-coding RNA MALAT1 and tubulin gamma 1 and increases the miR-125b levels. Histologically, less discriminated dysplastic nodules were exhibited by liver sections of rats treated with C3G. | [147] | |
| In vitro | C3G or its metabolite protocatechuic acid (PCA) were tested at the following doses: 100, 200, and 400 μM in HepG2 cells. Cell viability after C3G and PCA treatments, LDH release, and apoptosis in HepG2 cells in which cytotoxicity was induced using 2-amino-3-methylimidazo [4,5-f]quinoline (IQ) were evaluated using CCK-8, LDH release, and flow cytometry assays, respectively. Tandem mass tag (TMT)-based proteomics was utilized to characterize the proteins and pathways associated with the improvement after C3G and PCA treatment. | Exposure to IQ increased cytotoxicity and apoptosis in HepG2 cells, which were alleviated by C3G and PCA. C3G was more effective than PCA in protecting HepG2 cells against IQ-induced cytotoxicity and regulating the related signaling pathways. Proteomics and bioinformatics analyses and Western blot validation revealed that apoptosis-related signaling pathways played pivotal roles in the protective effect of C3G against the cytotoxicity of IQ. Moreover, XIAP was identified as a key target. XIAP acts as a potent apoptotic inhibitor by hampering the activation of caspases 3, 7, and 9. Molecular docking provided evidence that C3G affected the bindings of IQ and its carcinogenic metabolites to XIAP-BIR3 and contributed to the inhibition of apoptosis. | [148] | |
| In vitro | Cisplatin (DDP) dose: 5 μg/mL. C3G (>98% purity) dose: 400 μg/mL. Cervical cancer HeLa cells. Measurements of oxidative stress (CAT, SOD, and GSH-Px) and quantitation of gene expression of bax, bcl-2, Nrf2, and Keap1 genes were performed. | C3G-DDP inhibited the activity of the antioxidant defense enzymes SOD, CAT, and GSH-Px. In parallel, C3G-DDP reduced GSH concentration while increasing the concentration of ROS and MDA. C3G-DDP reduces the expression of Nrf2 and Nrf2 target proteins: HO-1 and NQO1. Finally, C3G-DDP increased the mRNA expression ratio of bax/bcl-2 and activated the intrinsic apoptotic pathway of HeLa cells. | [149] | |
| Antidiabetic and protection against complications of diabetes | In vitro and in vivo | 1 and 5 µM; 4 h pre-treatment of ARPE-19 cells exposed to 30 µM 4-hydroxynonenal for 24 h. 50 (mg/kg)/day for 3 weeks (2 pre-illumination and 1 post-illumination) in rabbits in which retinal damage was induced by light exposure | Decreased apoptosis, lower senescence-associated beta-galactosidase, and lower VEGF release. Increased thickness of the neurosensory retina in rabbits exposed to light. | [150] |
| In vitro | 10 µM; 6 h; mouse colonic epithelial MCE301 cells | Higher gene expression of the Mg2+ transport carriers Trpm6 and Cnnm4. | [151] | |
| In vivo | 10 and 20 (mg/kg)/day for 8 weeks in Sprague Dawley rats in which diabetes was induced with a 45 mg/kg streptozotocin dose. | Reduced fasting glycemia and insulin levels, decreased serum creatinine and BUN, and lower urinary albumin. Improved antioxidant enzyme and reduced cytokine levels. Decreased fibrosis and glomerulosclerosis in renal tissue. | [152] | |
| In vitro | 100 µM; 24 h; human corneal epithelial cells (HCEC 6510) previously exposed to 10 µg/mL of LPS for 24 h | Reduced apoptosis and decreased production of cytokines. | [153] | |
| In vivo | 1.6 mg/mL in drinking water (∼6.4 mg/day), for 3 or 20 weeks, in C57BL/6J male mice fed a low- or high-fat diet | Decreased weight gain for high-fat diet, improved glucose tolerance, reduced hepatic and plasma triglycerides, and modulated hepatic FGF21 levels. | [154] | |
| In vitro and in vivo | 20 µM; 24 h; HUVEC cells exposed for 1 h to 100 ng/mL TNF-alpha before treatment with C3G. 50 or 100 mg/kg; 8 weeks; male New Zealand rabbits fed 8 weeks with a high-fat diet after balloon catheter injury was performed. | Reduced damage in the intima media; decreased levels of circulating cholesterol, low-density lipoprotein, and triglycerides; and increased high-density lipoprotein. Reduced levels of cytokines and lowered apoptosis rates. Higher expression of SIRT1. | [155] | |
| In vitro | 20 µM; 48 h (+1 h pre-treatment); lens epithelial SRA01/04 cells exposed to 100 mM glucose and Sprague Dawley rat lens tissue exposed to 50 mM glucose. | Reduced apoptosis rates, decreased NFkB levels, and lowered Cox-2 protein expression. Decreased opacity of rat’s lens tissue. | [156] | |
| In vitro | 5 and 10 µM pre-treatment; 24 h; 3T3-L1 cells and human SGBS cells exposed to 1 mM or 500 µM palmitate for 24 h | Reduced lipid content, lower PPARgamma and nuclear NFkB protein levels, improved levels of insulin signaling targets, and higher Adipoq gene expression. | [157] | |
| In vivo | 10 and 50 µM; 24 h; HepG2 pre-treated with 400 µM palmitic acid and 400 µM oleic acid for 24 h. 50 mg/day; 8 weeks; male C57BL 6J mice previously fed an HFD for 4 weeks and 8 additional weeks of HFD during C3G treatment. | Reduced plasma and liver triglycerides, reduced fatty acid synthesis, lower fasting plasma glucose and insulin, higher cell glucose uptake, activation of PPAR-alpha. | [158] | |
| Liver disease and hepato-protection | In vitro and in vivo | 100 µM; 12 h; HepG2 or AML-12 cells co-treated with 400 µM palmitic acid 0.2% (v/v) of C3G in the HFD, 4 weeks (after 12 weeks of HFD), male mice fed an HFD for 16 weeks | Reduced liver steatosis, lower fasting glucose and insulin levels, reduced NLRP3 inflammasome, higher antioxidative enzyme levels, lower ROS levels, increased mitophagy. | [159] |
| In vitro | 5 µg/mL; 12 h; HepG2 cells pre-exposed to 4 µM hydrogen peroxide for 6 h | Decreased ROS levels, increased glutathione content, and higher catalase activity. Increased Nrf2 and Keap1 protein levels. | [160] | |
| In vitro | 2.5–10 µM; 24 h; HepG2 cells previously treated with 400 µM hydrogen peroxide | Increased cell viability and antioxidative machinery. Decreased ROS, apoptosis rates, and apoptosis-related proteins. | [161] | |
| In vitro and in vivo | 200 (mg/kg)/day; 8 weeks; male C57BL 6J mice fed an HFC diet and 5% ethanol drinking solution during the C3G treatment. HepG2 and FL83B cells were treated with SIRT1 inhibitor EX527 at 10 µM, for 4 h, and 1 µM C3G for an additional 20 h. | Reduced liver lipid content, lower levels of proinflammatory cytokines and inflammasome proteins, reduced NFkB protein expression and acetylation, and increased SIRT1 protein levels. | [162] | |
| Colitis and gastrointestinal alterations | In vivo and in vitro | 1 ug i.p. on days 0, 3, and 6 of model induction; C57BL 6J mice in which colitis was induced with drinking water containing 3.5% of dextran sulfate sodium for 7 days. 1 µg/mL; 24 h; peritoneal macrophages activated with 1 ug mL−1 of LPS. | Reduced cytokine gene expression in the colon, induction of Treg cells, and reduction of peritoneal CD169+ macrophages. | [163] |
| In vivo | 500 and 1000 mg/kg of diet; 8 weeks; male Wistar rats in which dysbiosis and intestinal damage were parallelly induced with 20 mg/kg 3-chloro-1,2-propanediol for 8 weeks | Improved histological features, modulation of gut microbiota. | [164] | |
| In vivo and in vitro | 50, 100, or 200 µmol/kg; 3 days; female BALB c mice in which colitis was induced with 2.5 mg of 2,4,6-trinitrobenzen-osulfonic acid 12 h after the first dose of C3G. 50 and 100 µmol/L; 24 h pre-treatment; LPS-induced Caco-2 cells with 100 ng/mL for 24 h. | Prevention of histological damage, reduction of proinflammatory cytokines, and suppression of nitric oxide production. | [165] | |
| In vitro | 10 or 20 µmol/L; 24 h pre-treatment; Caco-2 cells induced with palmitic acid 100 µmol/L for 6 h | Decreased nuclear NFkB, reduction of cytokine IL6 and IL8 gene expression and COX2 protein, decrease in ROS, and increase in Nrf2 levels. | [166] | |
| Neuroprotective | In vitro | 2.5, 5, or 10 µmol/L; 4 h pre-treatment; microglial BV2 (macrophage) cells stimulated with 1 μg/mL LPS for 24–48 h | Decreased cytokine levels, reduced iNOS mRNA levels and lower NO production, suppression of NFkB activation and p38 signaling pathway, decreased neurotoxicity and apoptosis in PC12 cells exposed to conditioned media from LPS-activated BV2 cells. | [167] |
| In vitro | 0.05, 0.1, 0.25, 0.5, or 1 µmol/L; 24 h pre-treatment; HT22 neuronal cells exposed to 5 mM glutamate for 18 h | Reduction of apoptosis, decrease in ROS, increase in Nrf2 levels and antioxidative gene expression, reduction of ER stress biomarkers. | [168] | |
| In vitro | 1, 3, or 9 µmol/L; 24–48 h; PC12 neuronal cells exposed in parallel to amyloid beta fibrils | Increased cell viability, decreased necrosis, reduced ROS levels. | [169] | |
| In vitro | 30 mg kg-1 day-1; 38 weeks; APPswe/PS1dE9 mice modeling Alzheimer’s disease | Differential gene expression in the spleen of the treated animals, including upregulation of antioxidant and immune system-related molecular targets. | [170] | |
| Reproductive health | In vitro | 5, 20, 40, 80, or 160 µmg/L; 2 h pre-treatment; Leydig R2C cells exposed to 44.8 µmol/L cadmium sulfate for 24 h | Increased cell viability, reduced ROS levels, protection of mitochondrial potential, increased StAR protein and progesterone levels. | [171] |
| In vivo | 500 mg/kg of chow diet; 10, 20, or 30 days; Kunming male mice treated with 5 (mg/kg)/day of cadmium chloride | Decreased levels of circulating FSH and testosterone, increased LH circulating levels, differential modulation of gene expression in the hypothalamus, increased expression of proteins involved in testosterone biosynthesis. | [172] | |
| Respiratory system, antiviral, and anti-SARS-CoV2 | In vivo | Diet containing 0.4% C3G (~1.2 mg/day); 25 days; asthma model of BALB/c mice sensitized to ovalbumin intraperitoneally (20 μg on days 0, 7, and 14) and nasally (1% aerosols on days 21–25) | Decreased number of peripheral eosinophils; reduced inflammatory infiltration in the lungs; lower levels of IL-4, IL-5, and IL-13; inhibition of IL-4Ra-STAT6 pathway. | [173] |
| In silico and in vitro | 3–200 µmol/L; papain-like protease assay for determination of deubiquitinase activity | The molecular docking prediction showed a potential binding activity to the papain-like protease of SARS-CoV-2, concentration-dependent inhibition of papain-like protease deubiquitinase activity. | [174] | |
| In silico and in vitro | 3–200 µmol/L; papain-like protease assay for determination of total protease activity | Molecular docking prediction of binding to the papain-like protease of SARS-CoV2. Concentration-dependent inhibition of papain-like protease total protease activity. | [175] | |
| In vivo and in vitro | 200 or 400 mg/kg bw; oral administration from days 2–28; Sprague Dawley male rats injected intraperitoneally with monocrotaline 60 mg/kg bw on day 1 to induce a model of pulmonary artery hypertension. 10 or 20 µmol/L; 24 h pre-treatment; cells induced with TGF-beta1 8 ng/mL for additional 24 h. | Reduction of hemodynamic indicators of pulmonary artery hypertension, improved histological features and blood oxygenation, reduction of cytokines levels, reduced markers of proliferation in PASMC, inhibition of TGF-beta1-p38 MAPK-CREB signaling pathway. | [176] | |
| Anti-inflammatory and immune system modulation | In vivo and in vitro | 25 mg/kg; two tail-vein injections per week for a total of six injections starting ten days after the secondary immunization; Sprague-Dawley male rats in which arthritis was induced by three injections of bovine type II collagen. 25, 50, or 100 µmol/L; 24–48 h; rheumatoid arthritis synovial fibroblasts and mononuclear cells obtained from patients. | Increased Treg cells and decreased CD38+ NK cell proportion in blood and synovial fluid in murine model, increased apoptosis and decreased proliferation in human rheumatoid arthritis synovial fibroblasts, decrease in proinflammatory cytokines. | [177] |
| In vivo | 10 (mg/kg)/day; 15 weeks; spontaneously hypertensive male rats and Wistar-Kyoto rats. | No differences were observed either in the spleen weights or in the proportions of splenic T-cells and helper T-cells; modulation of CD62Lhi, CD62Llo, CD62L-, CD25+, and T-reg cells dependent on the genotype. | [178] | |
| In vitro and in vivo | 25, 50, 100, and 250 µmol/L, RBL-2H3 cells sensitized with anti-DNP IgE and exposed to DNP-BSA antigen 100 and 200 µmol/kg bw, orally administered 1 h before antigen exposure, and 40 mg/kg bw, intravenous administration 1 h before antigen challenge, male ICR mice sensitized with anti-DNP IgE (100 ng injection in the ear) 24 h before the experiment and then challenged with DNP-BSA antigen (140 µg/mouse). | Dose-dependent inhibition of histamine and beta-hexosaminidase release, decreased ear tissue response (measured as extravasation) after antigen challenge. | [179] | |
| Other studies | In vitro | 80 µmol/L; 2 h pre-treatment; primary human dermal fibroblast irradiated with 12 J/cm2 UVA light and treated with 3-methyladenine | Decreased apoptosis, increased expression of autophagy markers, reduced ROS levels. | [180] |
| In vivo | 100 mg/kg body weight, oral administration before induction; Wistar rats injected with 1 mL/kg of 5% taurocholate to induce a model of severe acute pancreatitis | Increased colonic motility, decreased serum levels of H2S and pro-inflammatory cytokines, activation of mTOR signaling, reduced protein levels of cystathionine-gamma-lyase. | [181] | |
| In silico | Molecular modeling to assess for potential interactions between C3G and the advanced glycation end product receptor and its ligands | The results suggest a potential interaction and subsequent inhibition of the receptor for advanced glycation end products. | [182] | |
| In vitro | 25–400 µM; 24–72 h; primary human osteoblasts and MC3T3-E1 osteoblast murine cell line | Increased cell proliferation, increased mineralization activity, activation of ERK1/2 signaling pathway, increased osteocalcin protein and mRNA levels. | [183] | |
| In vitro | 1.25, 2.5, and 5 µmol/L; 24 h co-treatment or 2 h pre-treatment; primary human articular chondrocytes exposed to advanced glycation end products 10 µg/mL for 24 h (parallel to C3G treatment) or 10 min (after 2 h pre-treatment). | Reduced protein and mRNA expression levels of matrix metalloproteinases, decreased NF-kB signaling, reduced ERK/MAPK signaling activation. | [184] | |
| In vitro | 20 µmol/L; six-day treatment renewed every 48 h; human amniotic epithelial cells | Differential modulation of genes including targets involved in adipocyte differentiation and muscle activity. | [185] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peniche-Pavía, H.A.; Guzmán, T.J.; Magaña-Cerino, J.M.; Gurrola-Díaz, C.M.; Tiessen, A. Maize Flavonoid Biosynthesis, Regulation, and Human Health Relevance: A Review. Molecules 2022, 27, 5166. https://doi.org/10.3390/molecules27165166
Peniche-Pavía HA, Guzmán TJ, Magaña-Cerino JM, Gurrola-Díaz CM, Tiessen A. Maize Flavonoid Biosynthesis, Regulation, and Human Health Relevance: A Review. Molecules. 2022; 27(16):5166. https://doi.org/10.3390/molecules27165166
Chicago/Turabian StylePeniche-Pavía, Héctor A., Tereso J. Guzmán, Jesús M. Magaña-Cerino, Carmen M. Gurrola-Díaz, and Axel Tiessen. 2022. "Maize Flavonoid Biosynthesis, Regulation, and Human Health Relevance: A Review" Molecules 27, no. 16: 5166. https://doi.org/10.3390/molecules27165166