Integrating Pharmacological and Computational Approaches for the Phytochemical Analysis of Syzygium cumini and Its Anti-Diabetic Potential
Abstract
:1. Introduction
2. Results
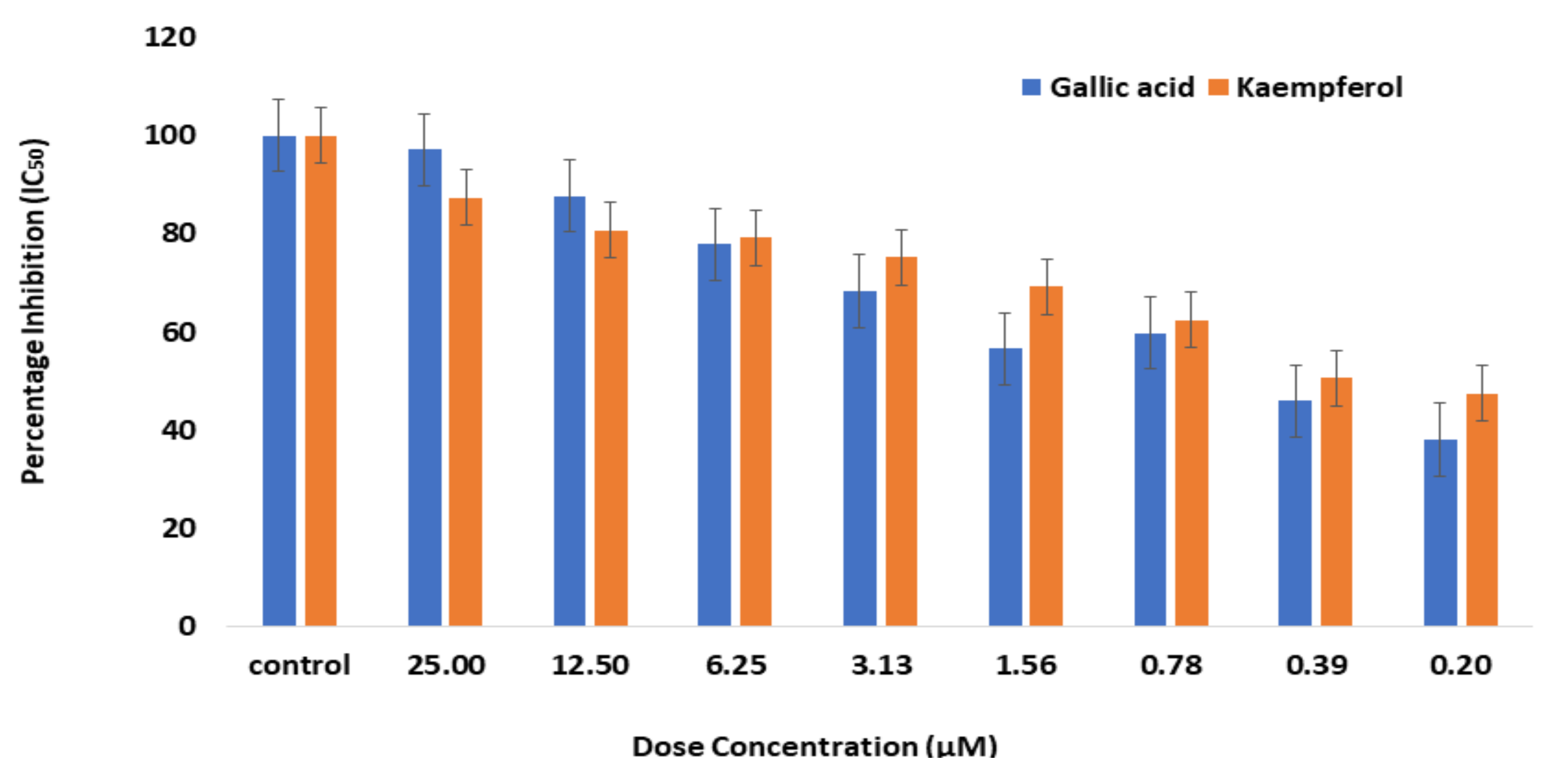
2.1. Enzyme Inhibition Assay
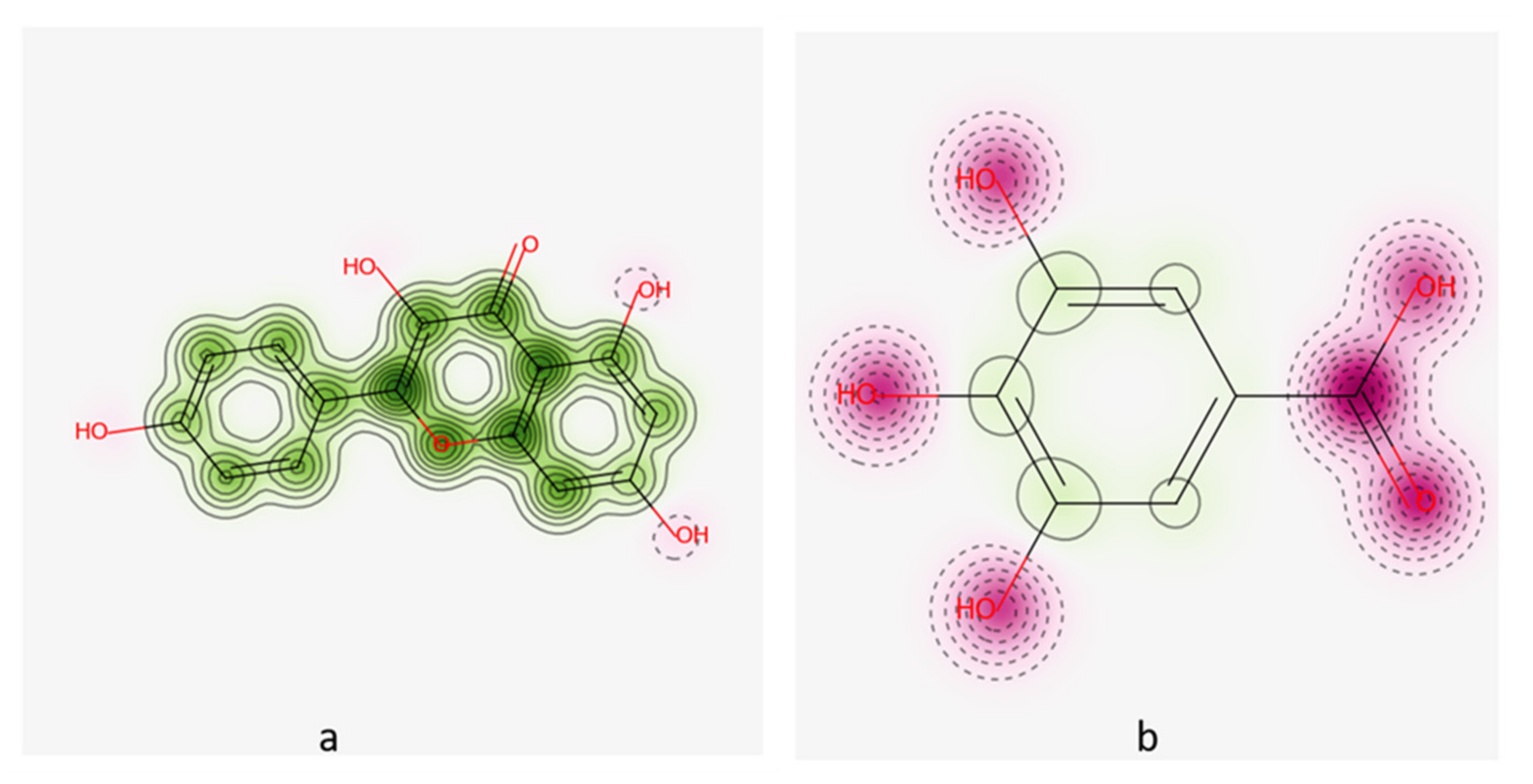
2.2. Molecular Docking
2.3. Drug Scan/ADMET Results
2.4. MD Simulation
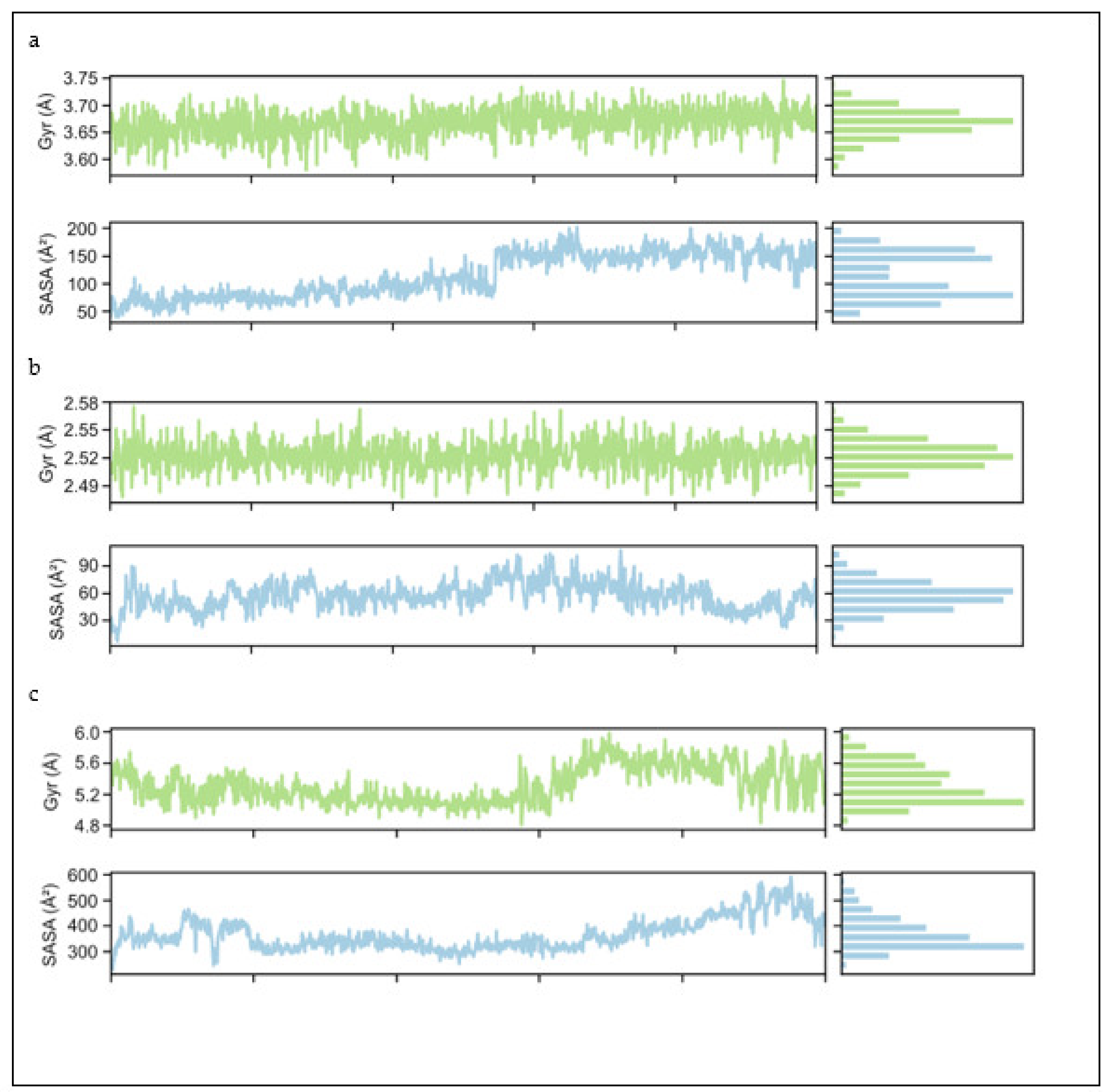
2.4.1. Root Mean Square Deviation and Root Mean Square Fluctuation
2.4.2. Radius of Gyration (Rg) and Solvent Accessible Surface Area (SASA)
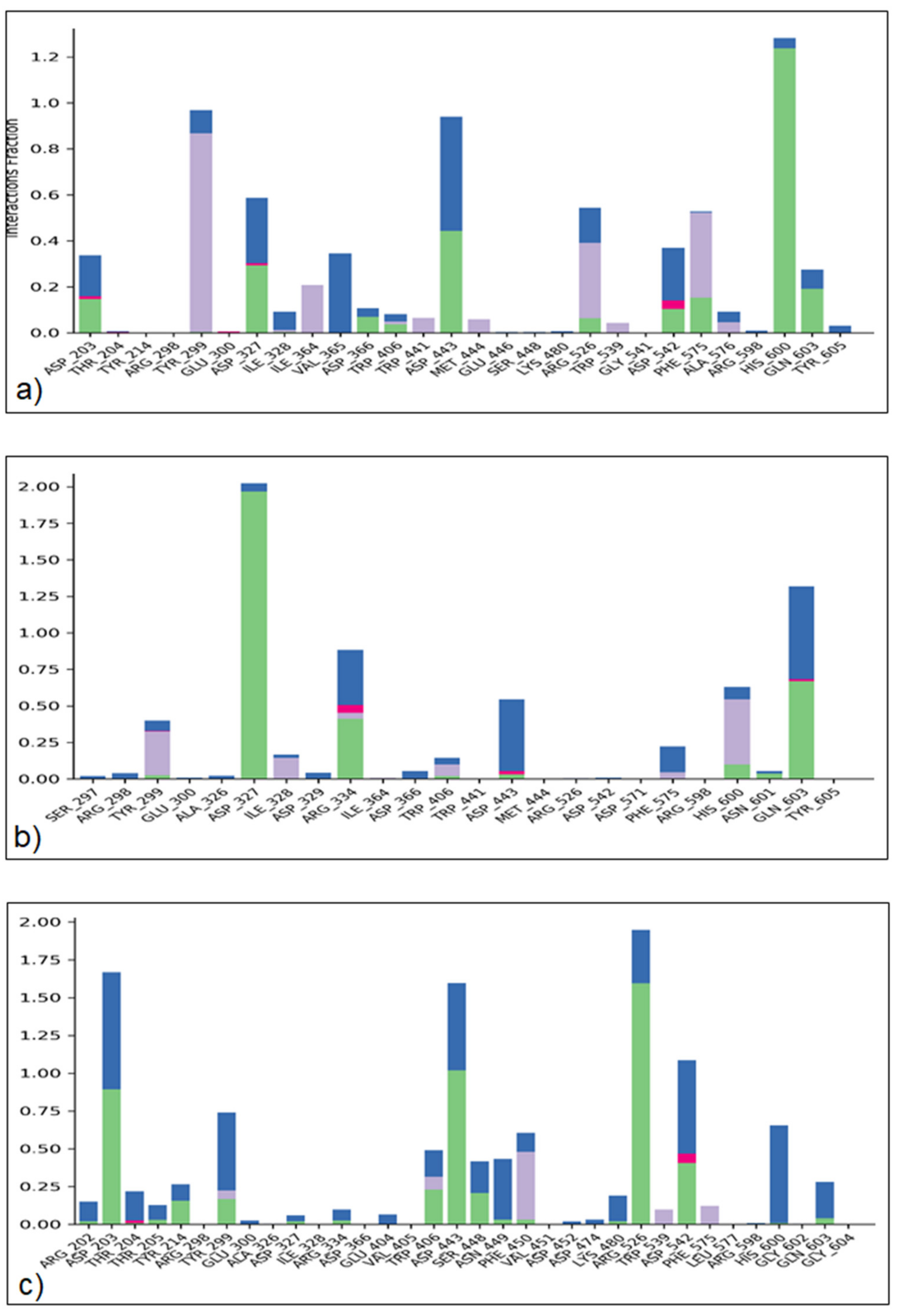
2.4.3. Protein–Ligand Interaction
2.5. Enzyme Inhibition Assay for Bioactive Phytochemicals
3. Discussion
4. Materials and Methods
4.1. Collection and Selection of Plant Sample
4.2. Plant Extract Preparation
4.3. α-. Glucosidase Inhibitory Activity
4.4. Molecular Docking
4.5. Drug-Likeness and ADMET Profiling
4.6. Molecular Dynamics (MD) Simulation
4.7. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Agesen, R.M.; Alibegovic, A.C.; Andersen, H.U.; Beck-Nielsen, H.; Gustenhoff, P.; Hansen, T.K.; Hedetoft, C.; Jensen, T.; Juhl, C.B.; Lerche, S.S.; et al. The effect of insulin degludec on risk of symptomatic nocturnal hypoglycaemia in adults with type 1 diabetes and high risk of nocturnal severe hypoglycaemia (the HypoDeg trial): Study rationale and design. BMC Endocr. Disord. 2019, 19, 78. [Google Scholar] [CrossRef] [PubMed]
- American Diabetes Association. Diagnosis and classification of diabetes mellitus. Diabetes Care 2006, 29, S43–S48. [Google Scholar]
- World Health Organization. Definition, Diagnosis and Classification of Diabetes Mellitus and Its Complications: Report of a WHO Consultation. Part 1, Diagnosis and Classification of Diabetes Mellitus; World Health Organization: Geneva, Switzerland, 1999. [Google Scholar]
- Khurshid, M.; Rashid, A.; Aslam, N.; Najam, S.; Siddique, T.; Islam, M.; Zain, M.; Khan, A.; Shah, A.; Awan, F. A Study of the Association between Diabetes Mellitus and Chronic Hepatitis B Virus Infection. West Indian Med. J. 2019, 68, 108. [Google Scholar]
- American Diabetes Association. Diagnosis and Classification of Diabetes Mellitus. Diabetes Care 2010, 33 (Suppl. S1), S62–S69. [Google Scholar]
- Babel, R.A.; Dandekar, M.P. A review on cellular and molecular mechanisms linked to the development of diabetes complications. Curr. Diabetes Rev. 2021, 17, 457–473. [Google Scholar]
- Hakeem, R.; Fawwad, A. Diabetes in Pakistan: Epidemiology, determinants and prevention. J. Diabetol. 2010, 1, 3. [Google Scholar]
- Schwartz, D.D.; Aikens, J.E.; Bussell, J.K.; Osborn, C.Y.; Safford, M.M. Seeing the person, not the illness: Promoting diabetes medication adherence through patient-centered collaboration. Clin. Diabetes 2017, 35, 35–42. [Google Scholar]
- Dirir, A.M.; Daou, M.; Yousef, A.F.; Yousef, L.F. A review of alpha-glucosidase inhibitors from plants as potential candidates for the treatment of type-2 diabetes. Phytochem. Rev. 2021, 21, 1049–1079. [Google Scholar]
- Jia, Y.; Lao, Y.; Leung, S.-W. Glycaemic control efficacy of oral anti-diabetic drugs in treating type 2 diabetes: A protocol for network meta-analysis. BMJ Open 2015, 5, e006139. [Google Scholar] [CrossRef]
- Wang, Y.; Curtis-Long, M.J.; Lee, B.W.; Yuk, H.J.; Kim, D.W.; Tan, X.F.; Park, K.H. Inhibition of tyrosinase activity by polyphenol compounds from Flemingia philippinensis roots. Bioorg. Med. Chem. 2014, 22, 1115–1120. [Google Scholar] [CrossRef]
- Kampkötter, A.; Gombitang Nkwonkam, C.; Zurawski, R.F.; Timpel, C.; Chovolou, Y.; Wätjen, W.; Kahl, R. Effects of the flavonoids kaempferol and fisetin on thermotolerance, oxidative stress and FoxO transcription factor DAF-16 in the model organism Caenorhabditis elegans. Arch. Toxicol. 2007, 81, 849–858. [Google Scholar] [PubMed]
- Park, M.J.; Lee, E.K.; Heo, H.-S.; Kim, M.-S.; Sung, B.; Kim, M.K.; Lee, J.; Kim, N.D.; Anton, S.; Choi, J.S.; et al. The anti-inflammatory effect of kaempferol in aged kidney tissues: The involvement of nuclear factor-κ B via nuclear factor-inducing kinase/I κ B kinase and mitogen-activated protein kinase pathways. J. Med. Food 2009, 12, 351–358. [Google Scholar]
- Nguyen, T.T.T.; Tran, E.; Ong, C.K.; Lee, S.K.; Do, P.T.; Huynh, T.T.; Nguyen, T.H.; Lee, J.J.; Tan, Y.; Ong, C.S.; et al. Kaempferol-induced growth inhibition and apoptosis in A549 lung cancer cells is mediated by activation of MEK-MAPK. J. Cell. Physiol. 2003, 197, 110–121. [Google Scholar]
- Peng, X.; Zhang, G.; Liao, Y.; Gong, D. Inhibitory kinetics and mechanism of kaempferol on α-glucosidase. Food Chem. 2016, 190, 207–215. [Google Scholar]
- Nayeem, N.; Smb, A. Gallic acid: A promising lead molecule for drug development. J. Appl. Pharm. 2016, 8, 1–4. [Google Scholar]
- Al-Numair, K.S.; Veeramani, C.; Alsaif, M.A.; Chandramohan, G. Influence of kaempferol, a flavonoid compound, on membrane-bound ATPases in streptozotocin-induced diabetic rats. Pharm. Biol. 2015, 53, 1372–1378. [Google Scholar]
- Abdel-Moneim, A.; Yousef, A.I.; El-Twab, S.M.A.; Reheim, E.S.A.; Ashour, M.B. Gallic acid and p-coumaric acid attenuate type 2 diabetes-induced neurodegeneration in rats. Metab. Brain Dis. 2017, 32, 1279–1286. [Google Scholar]
- Latha, R.C.R.; Daisy, P. Insulin-secretagogue, antihyperlipidemic and other protective effects of gallic acid isolated from Terminalia bellerica Roxb. in streptozotocin-induced diabetic rats. Chem. Biol. Interact. 2011, 189, 112–118. [Google Scholar]
- Rahimifard, M.; Baeeri, M.; Bahadar, H.; Moini-Nodeh, S.; Khalid, M.; Haghi-Aminjan, H.; Mohammadian, H.; Abdollahi, M. Therapeutic effects of gallic acid in regulating senescence and diabetes; an in vitro study. Molecules 2020, 25, 5875. [Google Scholar]
- Abdin, M.; Hamed, Y.S.; Akhtar, H.M.S.; Chen, D.; Mukhtar, S.; Wan, P.; Riaz, A.; Zeng, X. Extraction optimisation, antioxidant activity and inhibition on α-amylase and pancreatic lipase of polyphenols from the seeds of Syzygium cumini. Int. J. Food Sci. Technol. 2019, 54, 2084–2093. [Google Scholar]
- Zhang, X.; Shi, L.; Li, X.; Sheng, Q.; Yao, L.; Shen, D.; Lü, Z.-R.; Zhou, H.-M.; Park, Y.-D.; Lee, J.; et al. Effect of Ca2+ on the activity and structure of α-glucosidase: Inhibition kinetics and molecular dynamics simulations. J. Biosci. Bioeng. 2014, 117, 696–705. [Google Scholar] [PubMed]
- Chai, T.-T.; Kwek, M.-T.; Ong, H.-C.; Wong, F.-C. Water fraction of edible medicinal fern Stenochlaena palustris is a potent α-glucosidase inhibitor with concurrent antioxidant activity. Food Chem. 2015, 186, 26–31. [Google Scholar] [CrossRef] [PubMed]
- Salehi, B.; Ata, A.; Anil Kumar, N.V.; Sharopov, F.; Ramírez-Alarcón, K.; Ruiz-Ortega, A.; Ayatollahi, S.A.; Fokou, P.V.T.; Kobarfard, F.; Zakaria, Z.A.; et al. Antidiabetic Potential of Medicinal Plants and Their Active Components. Biomolecules 2019, 9, 551. [Google Scholar] [CrossRef] [PubMed]
- Bailey, C.; Day, C. Metformin: Its botanical background. Pract. Diabetes Int. 2004, 21, 115–117. [Google Scholar]
- Asano, N.; Oseki, K.; Kaneko, E.; Matsui, K. Enzymic synthesis of α-and β-D-glucosides of 1-deoxynojirimycin and their glycosidase inhibitory activities. Carbohydr. Res. 1994, 258, 255–266. [Google Scholar] [PubMed]
- Javaid, A.; Ashfaq, U.A.; Zafar, Z.; Akmal, A.; Taj, S.; Khalid, H. Phytochemical Analysis and Antidiabetic Potential of Armoracia rusticana: Pharmacological and Computational Approach. Comb. Chem. High Throughput Screen. 2021, 24, 465–471. [Google Scholar]
- Proma, N.M.; Naima, J.; Islam, M.R.; Papel, J.A.; Rahman, M.M.; Hossain, M.K. Phytochemical constituents and anti-diabetic properties of Syzygium cumini Linn. Seed. Int. J. Pharm. Sci. Res. 2018, 9, 1806–1814. [Google Scholar]
- Ramya, S.; Neethirajan, K.; Jayakumararaj, R. Profile of bioactive compounds in Syzygium cumini—A review. J. Pharm. Res. 2012, 5, 4548–4553. [Google Scholar]
- Cock, I.E.; Cheesman, M. The potential of plants of the genus Syzygium (Myrtaceae) for the prevention and treatment of arthritic and autoimmune diseases. In Bioactive Food as Dietary Interventions for Arthritis and Related Inflammatory Diseases; Elsevier: Amsterdam, The Netherlands, 2019; pp. 401–424. [Google Scholar]
- Nadkarni, K.; Nadkarni, A. Indian Materia Medica; Popular Prakashan Pvt. Ltd.: Bombay, India, 1976; Volume 1, p. 799. [Google Scholar]
- Caro-Ordieres, T.; Marín-Royo, G.; Opazo-Ríos, L.; Jiménez-Castilla, L.; Moreno, J.A.; Gómez-Guerrero, C.; Egido, J. The coming age of flavonoids in the treatment of diabetic complications. J. Clin. Med. 2020, 9, 346. [Google Scholar] [CrossRef]
- Yamaguchi, S.; Katahira, H.; Ozawa, S.; Nakamichi, Y.; Tanaka, T.; Shimoyama, T.; Takahashi, K.; Yoshimoto, K.; Imaizumi, M.O.; Nagamatsu, S.; et al. Activators of AMP-activated protein kinase enhance GLUT4 translocation and its glucose transport activity in 3T3-L1 adipocytes. Am. J. Physiol. Endocrinol. Metab. 2005, 289, E643–E649. [Google Scholar] [CrossRef] [Green Version]
- Thilagam, E.; Parimaladevi, B.; Kumarappan, C.; Mandal, S.C. α-Glucosidase and α-amylase inhibitory activity of Senna surattensis. J. Acupunct. Meridian Stud. 2013, 6, 24–30. [Google Scholar] [CrossRef] [PubMed]
- Bhandari, M.R.; Jong-Anurakkun, N.; Hong, G.; Kawabata, J. α-Glucosidase and α-amylase inhibitory activities of Nepalese medicinal herb Pakhanbhed (Bergenia ciliata, Haw.). Food Chem. 2008, 106, 247–252. [Google Scholar]
- Terstappen, G.C.; Reggiani, A. In silico research in drug discovery. Trends Pharmacol. Sci. 2001, 22, 23–26. [Google Scholar] [CrossRef]
- Lengauer, T.; Rarey, M. Computational methods for biomolecular docking. Curr. Opin. Struct. Biol. 1996, 6, 402–406. [Google Scholar] [CrossRef]
- Akmal, A.; Javaid, A.; Hussain, R.; Kanwal, A.; Zubair, M.; Ashfaq, U.A. Screening of phytochemicals against Keap1-NRF2 interaction to reactivate NRF2 Functioning: Pharmacoinformatics based approach. Pak. J. Pharm. Sci. 2019, 32, 2823–2828. [Google Scholar]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 1997, 23, 3–25. [Google Scholar] [CrossRef]
- Tsaioun, K.; Bottlaender, M.; Mabondzo, A. ADDME–Avoiding Drug Development Mistakes Early: Central nervous system drug discovery perspective. BMC Neurol. 2009, 9, S1. [Google Scholar] [CrossRef]
- Finch, A.; Pillans, P. P-glycoprotein and its role in drug-drug interactions. Aust. Prescr. 2014, 37, 137–139. [Google Scholar] [CrossRef]
- Hassan, S.S.U.; Abbas, S.Q.; Ali, F.; Ishaq, M.; Bano, I.; Hassan, M.; Jin, H.-Z.; Bungau, S.G. A Comprehensive In Silico Exploration of Pharmacological Properties, Bioactivities, Molecular Docking, and Anticancer Potential of Vieloplain F from Xylopia vielana Targeting B-Raf Kinase. Molecules 2022, 27, 917. [Google Scholar] [CrossRef]
- Limanto, A.; Simamora, A.; Santoso, A.W.; Timotius, K.H. Antioxidant, α-glucosidase inhibitory activity and molecular docking study of gallic acid, quercetin and rutin: A comparative study. Mol. Cell. Biomed. Sci. 2019, 3, 67–74. [Google Scholar] [CrossRef]
- Choudhary, D.K.; Chaturvedi, N.; Singh, A.; Mishra, A. Characterization, inhibitory activity and mechanism of polyphenols from faba bean (gallic-acid and catechin) on α-glucosidase: Insights from molecular docking and simulation study. Prep. Biochem. Biotechnol. 2020, 50, 123–132. [Google Scholar] [PubMed]
- Grewal, A.S.; Sharma, N.; Singh, S.; Arora, S. Molecular docking studies of phenolic compounds from Syzygium cumini with multiple targets of type 2 diabetes. J. Pharm. Technol. Res. Manag. 2018, 6, 125–133. [Google Scholar]
- Saddique, F.A.; Ahmad, M.; Ashfaq, U.A.; Ahmad, M.N.; Anjum, M.N.; Mohsin, N.A.; Aslam, S. Alpha-glucosidase inhibition and molecular docking studies of 1,2-benzothiazine 1,1-dioxide based carbohydrazides. Pak. J. Pharm. Sci. 2019, 32, 2829–2834. [Google Scholar] [PubMed]
- Tavani, C.; Bianchi, L.; De Palma, A.; Passeri, G.I.; Punzi, G.; Pierri, C.L.; Lovece, A.; Cavalluzzi, M.M.; Franchini, C.; Lentini, G.; et al. Nitro-substituted tetrahydroindolizines and homologs: Design, kinetics, and mechanism of α-glucosidase inhibition. Bioorg. Med. Chem. Lett. 2017, 27, 3980–3986. [Google Scholar]
- Ibraheem, F.; Ahmad, M.; Ashfaq, U.A.; Aslam, S.; Khan, Z.A.; Sultan, S. Synthesis, molecular docking and anti-diabetic studies of novel benzimidazole-pyrazoline hybrid molecules. Pak. J. Pharm. Sci. 2020, 33, 847–854. [Google Scholar]
- Prasad, C.V.; Anjana, T.; Banerji, A.; Gopalakrishnapillai, A. Gallic acid induces GLUT4 translocation and glucose uptake activity in 3T3-L1 cells. FEBS Lett. 2010, 584, 531–536. [Google Scholar]
- Ezzat, S.M.; Salama, M.M. A new α-glucosidase inhibitor from Achillea fragrantissima (Forssk.) Sch. Bip. growing in Egypt. Nat. Prod. Res. 2014, 28, 812–818. [Google Scholar]
- Taj, S.; Ashfaq, U.A.; Aslam, S.; Ahmad, M.; Bhatti, S.H. Alpha-glucosidase activity of novel pyrazolobenzothiazine 5,5-dioxide derivatives for the treatment of diabetes mellitus. Invitro combined with molecular docking approach. Biologia 2019, 74, 1523–1530. [Google Scholar]
- Taj, S.; Ahmad, M.; Ashfaq, U.A. Exploring of novel 4-hydroxy-2H-benzo [e][1,2] thiazine-3-carbohydrazide 1,1-dioxide derivative as a dual inhibitor of α-glucosidase and α-amylase: Molecular docking, biochemical, enzyme kinetic and in-vivo mouse model study. Int. J. Biol. Macromol. 2022, 207, 507–521. [Google Scholar]
- Vilar, S.; Cozza, G.; Moro, S. Medicinal chemistry and the molecular operating environment (MOE): Application of QSAR and molecular docking to drug discovery. Curr. Top. Med. Chem. 2008, 8, 1555–1572. [Google Scholar]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem 2019 update: Improved access to chemical data. Nucleic Acids Res. 2018, 47, D1102–D1109. [Google Scholar]
- Benet, L.Z.; Hosey, C.M.; Ursu, O.; Oprea, T.I. BDDCS, the rule of 5 and drugability. Adv. Drug Deliv. Rev. 2016, 101, 89–98. [Google Scholar] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [PubMed] [Green Version]
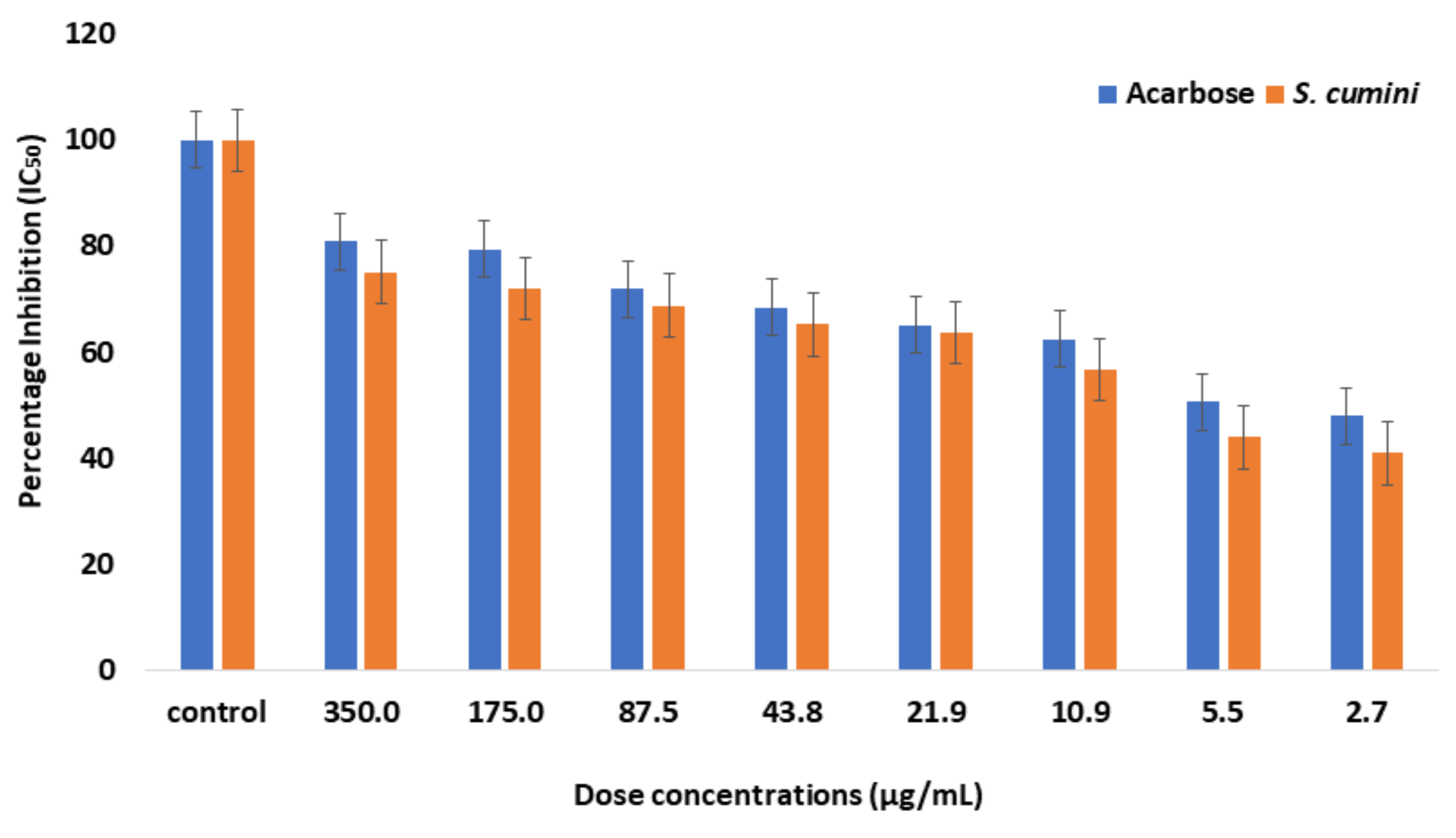


| Sr. No. | Plant Extract | IC50 |
|---|---|---|
| 1 | Syzygium cumini | 8.07 (µg/mL) |
| 2 | Acarbose | 5.26 (µM) |
| Sr. No. | PubChem ID | Chemical Name | Docking Score | RMSD Value | Receptor | Interaction |
|---|---|---|---|---|---|---|
| 1 | 5,280,863 | Kaempferol | −11.2 | 1.8 | Asp542 Arg526 His600 Asp327 | H-acceptor pi-H H-acceptor H-acceptor |
| 2 | 370 | Gallic acid | −10.3 | 1.0 | Asp327 Asp547 His600 | H-acceptor H-acceptor H-acceptor |
| 3 | 41,774 | Acarbose (Control) | −12.6 | 2.1 | Asp542 Asp327 His600 | H-acceptor H-acceptor H-acceptor |
| Compound | Molecular Weight (g/mol) | Number of HBA * (nON) | Number of HBD ** (nOHNH) | MlogP *** |
|---|---|---|---|---|
| Lipinski’s rule of five | <500 | <10 | <5 | <5 |
| Kaempferol | 286.24 | 6 | 4 | 2.17 |
| Gallic acid | 170.12 | 5 | 4 | 0.59 |
| Acarbose (Control) | 645.61 | 19 | 14 | −5.51 |
| Properties | Compounds | Acarbose | Gallic Acid | Kaempferol |
|---|---|---|---|---|
| Absorption | Water solubility | Soluble | Soluble | Soluble |
| P-glycoprotein substrate | × | × | √ | |
| Log Kp (skin permeation) cm/s | −16.29 | −6.84 | −8.73 | |
| Distribution | Blood-Brain Barrier | × | × | × |
| Gastro-Intestinal Absorption | low | ↑ | ↑ | |
| Metabolism CYP450 | CYP4501 A2 Inhibitor | × | × | × |
| CYP450 2C9 Inhibitor | × | × | × | |
| CYP450 2D6 Inhibitor | × | × | × | |
| CYP450 2C19 Inhibitor | × | × | × | |
| CYP450 3A4 Inhibitor | × | √ | √ | |
| Excretion | Renal OCT2 substrate | No | No | No |
| Toxicity | Oral Toxicity (LD50) (mg/kg) | 24,000 | 2851 | 475.5 |
| Oral Toxicity classification * | VI | V | IV |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rashid, F.; Javaid, A.; Mahmood-ur-Rahman; Ashfaq, U.A.; Sufyan, M.; Alshammari, A.; Alharbi, M.; Nisar, M.A.; Khurshid, M. Integrating Pharmacological and Computational Approaches for the Phytochemical Analysis of Syzygium cumini and Its Anti-Diabetic Potential. Molecules 2022, 27, 5734. https://doi.org/10.3390/molecules27175734
Rashid F, Javaid A, Mahmood-ur-Rahman, Ashfaq UA, Sufyan M, Alshammari A, Alharbi M, Nisar MA, Khurshid M. Integrating Pharmacological and Computational Approaches for the Phytochemical Analysis of Syzygium cumini and Its Anti-Diabetic Potential. Molecules. 2022; 27(17):5734. https://doi.org/10.3390/molecules27175734
Chicago/Turabian StyleRashid, Fatima, Anam Javaid, Mahmood-ur-Rahman, Usman Ali Ashfaq, Muhammad Sufyan, Abdulrahman Alshammari, Metab Alharbi, Muhammad Atif Nisar, and Mohsin Khurshid. 2022. "Integrating Pharmacological and Computational Approaches for the Phytochemical Analysis of Syzygium cumini and Its Anti-Diabetic Potential" Molecules 27, no. 17: 5734. https://doi.org/10.3390/molecules27175734







