Applications of Single-Molecule Vibrational Spectroscopic Techniques for the Structural Investigation of Amyloid Oligomers
Abstract
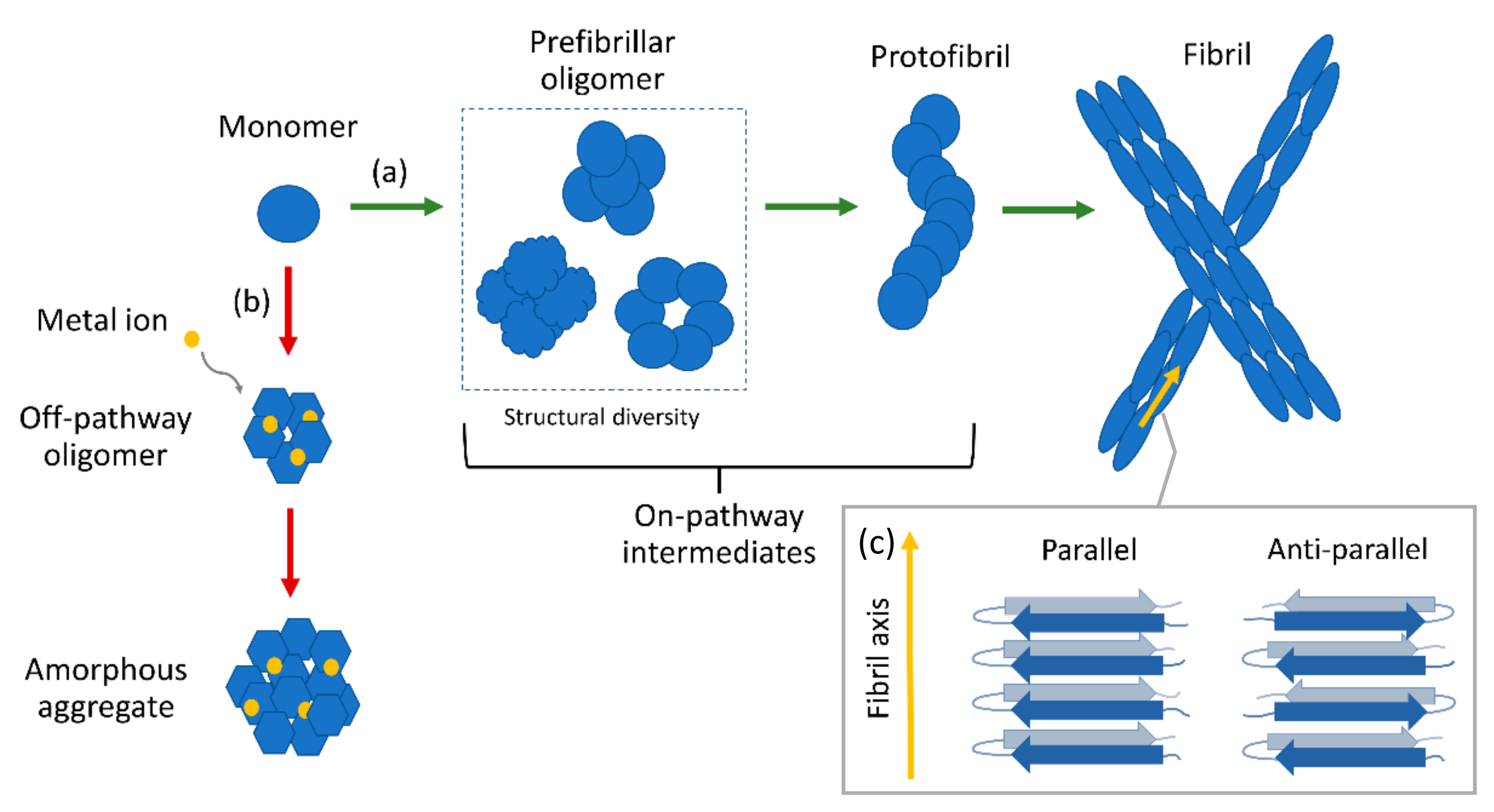
:1. Introduction
2. General Introduction and Comparison of Vibrational Spectroscopy and Other Common Spectroscopic Methods
3. Ensemble-Averaged Studies of Amyloid Oligomer Species
3.1. Infrared Spectroscopy
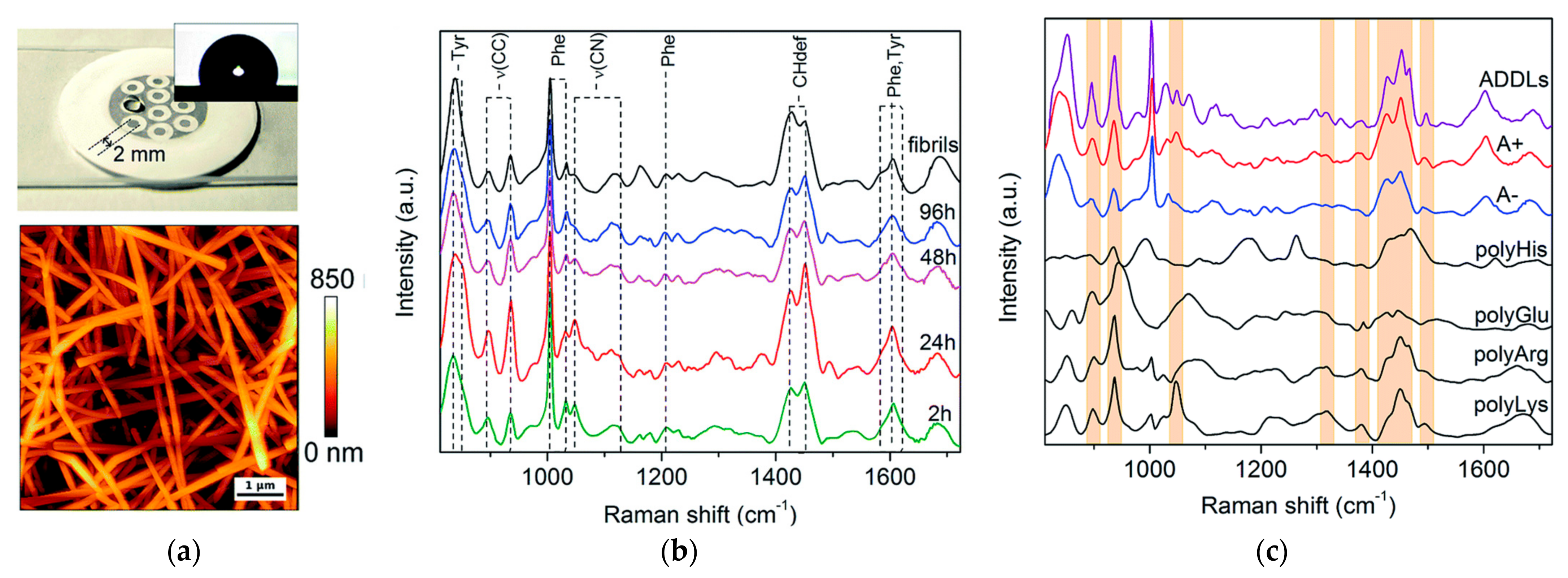
3.2. Raman Spectroscopy
4. Single-Molecule and Low-Copy Number Studies of Amyloid Oligomer Species
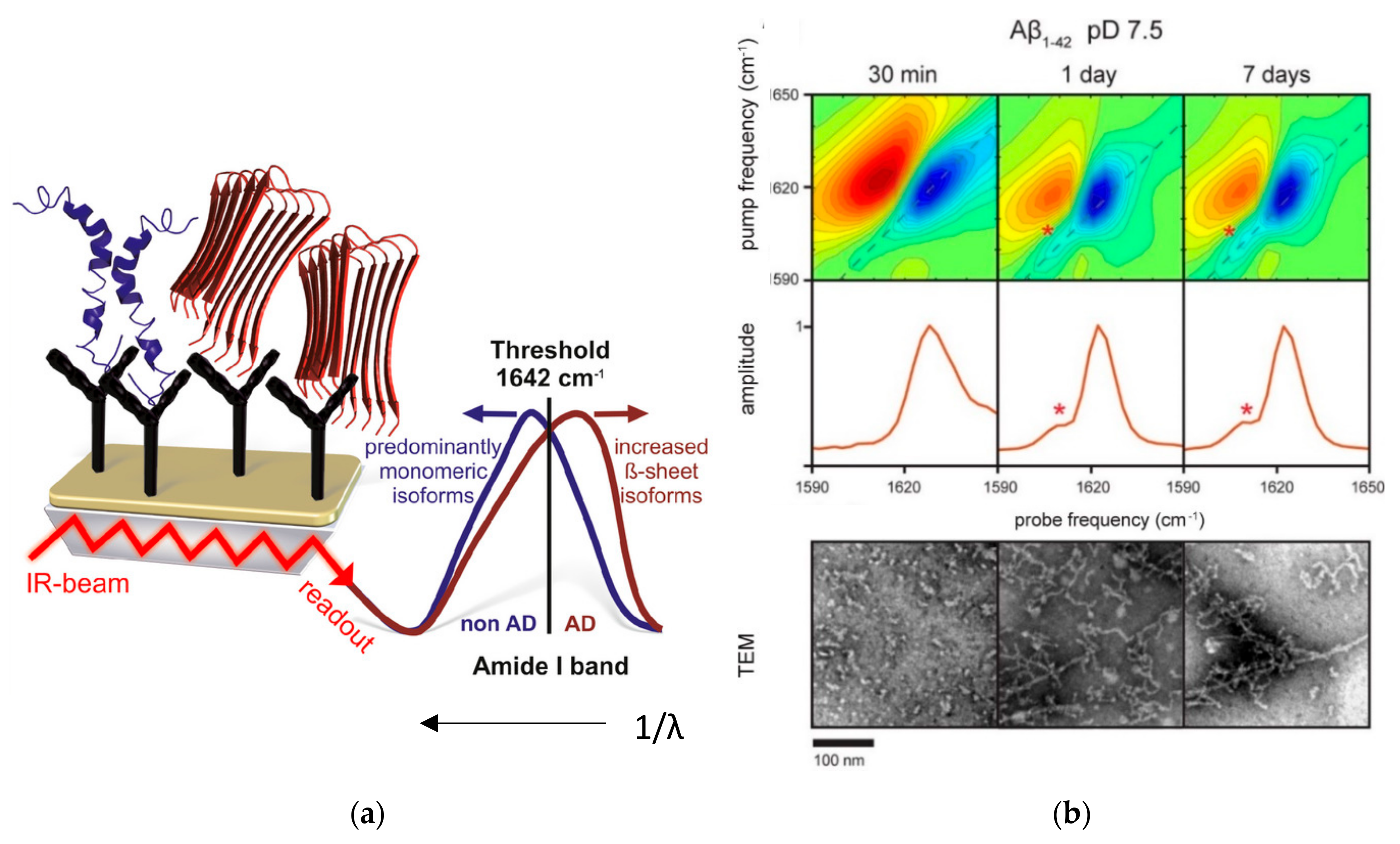
4.1. Infrared Spectroscopy
4.2. Raman Spectroscopy
5. Discussion
5.1. Comparing Various Single-Molecule Methods
5.2. Impact
6. Concluding Remarks and Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chiti, F.; Dobson, C.M. Protein Misfolding, Amyloid Formation, and Human Disease: A Summary of Progress Over the Last Decade. Annu. Rev. Biochem. 2017, 86, 27–68. [Google Scholar] [CrossRef]
- Sengupta, U.; Nilson, A.N.; Kayed, R. The Role of Amyloid-beta Oligomers in Toxicity, Propagation, and Immunotherapy. EBioMedicine 2016, 6, 42–49. [Google Scholar] [CrossRef]
- Bucciantini, M.; Giannoni, E.; Chiti, F.; Baroni, F.; Formigli, L.; Zurdo, J.; Taddei, N.; Ramponi, G.; Dobson, C.M.; Stefani, M. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature 2002, 416, 507–511. [Google Scholar] [CrossRef] [PubMed]
- Kayed, R.; Head, E.; Thompson, J.L.; McIntire, T.M.; Milton, S.C.; Cotman, C.W.; Glabe, C.G. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science 2003, 300, 486–489. [Google Scholar] [CrossRef]
- Selkoe, D.J. Soluble oligomers of the amyloid beta-protein impair synaptic plasticity and behavior. Behav. Brain. Res. 2008, 192, 106–113. [Google Scholar] [CrossRef] [PubMed]
- Shankar, G.M.; Li, S.; Mehta, T.H.; Garcia-Munoz, A.; Shepardson, N.E.; Smith, I.; Brett, F.M.; Farrell, M.A.; Rowan, M.J.; Lemere, C.A.; et al. Amyloid-beta protein dimers isolated directly from Alzheimer’s brains impair synaptic plasticity and memory. Nat. Med. 2008, 14, 837–842. [Google Scholar] [CrossRef] [PubMed]
- Welzel, A.T.; Maggio, J.E.; Shankar, G.M.; Walker, D.E.; Ostaszewski, B.L.; Li, S.; Klyubin, I.; Rowan, M.J.; Seubert, P.; Walsh, D.M.; et al. Secreted amyloid beta-proteins in a cell culture model include N-terminally extended peptides that impair synaptic plasticity. Biochemistry 2014, 53, 3908–3921. [Google Scholar] [CrossRef]
- Cizas, P.; Budvytyte, R.; Morkuniene, R.; Moldovan, R.; Broccio, M.; Losche, M.; Niaura, G.; Valincius, G.; Borutaite, V. Size-dependent neurotoxicity of beta-amyloid oligomers. Arch. Biochem. Biophys. 2010, 496, 84–92. [Google Scholar] [CrossRef]
- Guerrero-Munoz, M.J.; Gerson, J.; Castillo-Carranza, D.L. Tau Oligomers: The Toxic Player at Synapses in Alzheimer’s Disease. Front. Cell Neurosci. 2015, 9, 464. [Google Scholar] [CrossRef]
- Mroczko, B.; Groblewska, M.; Litman-Zawadzka, A. The Role of Protein Misfolding and Tau Oligomers (TauOs) in Alzheimer’s Disease (AD). Int. J. Mol. Sci. 2019, 20, 4661. [Google Scholar] [CrossRef] [Green Version]
- Julien, C.; Tomberlin, C.; Roberts, C.M.; Akram, A.; Stein, G.H.; Silverman, M.A.; Link, C.D. In vivo induction of membrane damage by beta-amyloid peptide oligomers. Acta Neuropathol. Commun. 2018, 6, 131. [Google Scholar] [CrossRef]
- Yasumoto, T.; Takamura, Y.; Tsuji, M.; Watanabe-Nakayama, T.; Imamura, K.; Inoue, H.; Nakamura, S.; Inoue, T.; Kimura, A.; Yano, S.; et al. High molecular weight amyloid beta1-42 oligomers induce neurotoxicity via plasma membrane damage. FASEB J. 2019, 33, 9220–9234. [Google Scholar] [CrossRef] [PubMed]
- Flach, K.; Hilbrich, I.; Schiffmann, A.; Gartner, U.; Kruger, M.; Leonhardt, M.; Waschipky, H.; Wick, L.; Arendt, T.; Holzer, M. Tau oligomers impair artificial membrane integrity and cellular viability. J. Biol. Chem. 2012, 287, 43223–43233. [Google Scholar] [CrossRef] [PubMed]
- Lasagna-Reeves, C.A.; Sengupta, U.; Castillo-Carranza, D.; Gerson, J.E.; Guerrero-Munoz, M.; Troncoso, J.C.; Jackson, G.R.; Kayed, R. The formation of tau pore-like structures is prevalent and cell specific: Possible implications for the disease phenotypes. Acta Neuropathol. Commun. 2014, 2, 56. [Google Scholar] [CrossRef] [PubMed]
- Winner, B.; Jappelli, R.; Maji, S.K.; Desplats, P.A.; Boyer, L.; Aigner, S.; Hetzer, C.; Loher, T.; Vilar, M.; Campioni, S.; et al. In vivo demonstration that alpha-synuclein oligomers are toxic. Proc. Natl. Acad. Sci. USA 2011, 108, 4194–4199. [Google Scholar] [CrossRef] [PubMed]
- Janson, J.; Ashley, R.H.; Harrison, D.; McIntyre, S.; Butler, P.C. The mechanism of islet amyloid polypeptide toxicity is membrane disruption by intermediate-sized toxic amyloid particles. Diabetes 1999, 48, 491–498. [Google Scholar] [CrossRef] [PubMed]
- Gurlo, T.; Ryazantsev, S.; Huang, C.J.; Yeh, M.W.; Reber, H.A.; Hines, O.J.; O’Brien, T.D.; Glabe, C.G.; Butler, P.C. Evidence for proteotoxicity in beta cells in type 2 diabetes: Toxic islet amyloid polypeptide oligomers form intracellularly in the secretory pathway. Am. J. Pathol. 2010, 176, 861–869. [Google Scholar] [CrossRef]
- Alies, B.; Hureau, C.; Faller, P. The role of metal ions in amyloid formation: General principles from model peptides. Metallomics 2013, 5, 183–192. [Google Scholar] [CrossRef]
- Viles, J.H. Metal ions and amyloid fiber formation in neurodegenerative diseases. Copper, zinc and iron in Alzheimer’s, Parkinson’s and prion diseases. Coord. Chem. Rev. 2012, 256, 2271–2284. [Google Scholar] [CrossRef]
- Chen, W.T.; Liao, Y.H.; Yu, H.M.; Cheng, I.H.; Chen, Y.R. Distinct effects of Zn2+, Cu2+, Fe3+, and Al3+ on amyloid-beta stability, oligomerization, and aggregation: Amyloid-beta destabilization promotes annular protofibril formation. J. Biol. Chem. 2011, 286, 9646–9656. [Google Scholar] [CrossRef] [Green Version]
- Deas, E.; Cremades, N.; Angelova, P.R.; Ludtmann, M.H.; Yao, Z.; Chen, S.; Horrocks, M.H.; Banushi, B.; Little, D.; Devine, M.J.; et al. Alpha-Synuclein Oligomers Interact with Metal Ions to Induce Oxidative Stress and Neuronal Death in Parkinson’s Disease. Antioxid. Redox Signal. 2016, 24, 376–391. [Google Scholar] [CrossRef]
- Lee, M.C.; Yu, W.C.; Shih, Y.H.; Chen, C.Y.; Guo, Z.H.; Huang, S.J.; Chan, J.C.; Chen, Y.R. Zinc ion rapidly induces toxic, off-pathway amyloid-beta oligomers distinct from amyloid-beta derived diffusible ligands in Alzheimer’s disease. Sci. Rep. 2018, 8, 4772. [Google Scholar] [CrossRef]
- Rezaei-Ghaleh, N.; Giller, K.; Becker, S.; Zweckstetter, M. Effect of zinc binding on beta-amyloid structure and dynamics: Implications for Abeta aggregation. Biophys. J. 2011, 101, 1202–1211. [Google Scholar] [CrossRef] [PubMed]
- Toyama, B.H.; Weissman, J.S. Amyloid structure: Conformational diversity and consequences. Annu. Rev. Biochem. 2011, 80, 557–585. [Google Scholar] [CrossRef] [PubMed]
- Larkin, P. Infrared and Raman Spectroscopy: Principles and Spectral Interpretation; Elsevier: Amsterdam, The Netherlands, 2011. [Google Scholar]
- Smith, E.; Dent, G. Modern Raman Spectroscopy: A Practical Approach; Wiley: Chichester, UK, 2019. [Google Scholar]
- Singh, M.K.; Singh, A. Chapter 14—Nuclear magnetic resonance spectroscopy. In Characterization of Polymers and Fibres; Singh, M.K., Singh, A., Eds.; Woodhead Publishing: Sawston, UK, 2022; pp. 321–339. [Google Scholar]
- Fleming, K.G. Fluorescence Theory. In Encyclopedia of Spectroscopy and Spectrometry, 3rd ed.; Lindon, J.C., Tranter, G.E., Koppenaal, D.W., Eds.; Academic Press: Oxford, UK, 2017; pp. 647–653. [Google Scholar]
- Price, N.C. Conformational issues in the characterization of proteins. Biotechnol. Appl. Biochem. 2000, 31, 29–40. [Google Scholar] [CrossRef] [PubMed]
- Biter, A.B.; Pollet, J.; Chen, W.H.; Strych, U.; Hotez, P.J.; Bottazzi, M.E. A method to probe protein structure from UV absorbance spectra. Anal. Biochem. 2019, 587, 113450. [Google Scholar] [CrossRef]
- Greenfield, N.J. Using circular dichroism spectra to estimate protein secondary structure. Nat. Protoc. 2006, 1, 2876–2890. [Google Scholar] [CrossRef]
- Kotler, S.A.; Brender, J.R.; Vivekanandan, S.; Suzuki, Y.; Yamamoto, K.; Monette, M.; Krishnamoorthy, J.; Walsh, P.; Cauble, M.; Holl, M.M.; et al. High-resolution NMR characterization of low abundance oligomers of amyloid-beta without purification. Sci. Rep. 2015, 5, 11811. [Google Scholar] [CrossRef]
- Staunton, D.; Owen, J.; Campbell, I.D. NMR and structural genomics. Acc. Chem. Res. 2003, 36, 207–214. [Google Scholar] [CrossRef]
- Karamanos, T.K.; Kalverda, A.P.; Thompson, G.S.; Radford, S.E. Mechanisms of amyloid formation revealed by solution NMR. Prog. Nucl. Magn. Reson. Spectrosc. 2015, 88–89, 86–104. [Google Scholar] [CrossRef] [Green Version]
- Puthenveetil, R.; Vinogradova, O. Solution NMR: A powerful tool for structural and functional studies of membrane proteins in reconstituted environments. J. Biol. Chem. 2019, 294, 15914–15931. [Google Scholar] [CrossRef] [PubMed]
- Pelton, J.T.; McLean, L.R. Spectroscopic methods for analysis of protein secondary structure. Anal. Biochem. 2000, 277, 167–176. [Google Scholar] [CrossRef] [PubMed]
- Tycko, R. Molecular structure of amyloid fibrils: Insights from solid-state NMR. Q. Rev. Biophys. 2006, 39, 1–55. [Google Scholar] [CrossRef] [PubMed]
- Tycko, R. Solid-state NMR studies of amyloid fibril structure. Annu. Rev. Phys. Chem. 2011, 62, 279–299. [Google Scholar] [CrossRef]
- Lee, Y.H.; Goto, Y. Kinetic intermediates of amyloid fibrillation studied by hydrogen exchange methods with nuclear magnetic resonance. Biochim. Biophys. Acta 2012, 1824, 1307–1323. [Google Scholar] [CrossRef]
- van der Wel, P.C.A. Insights into protein misfolding and aggregation enabled by solid-state NMR spectroscopy. Solid State Nucl. Magn. Reson. 2017, 88, 1–14. [Google Scholar] [CrossRef]
- Meier, B.H.; Riek, R.; Bockmann, A. Emerging Structural Understanding of Amyloid Fibrils by Solid-State NMR. Trends Biochem. Sci. 2017, 42, 777–787. [Google Scholar] [CrossRef] [PubMed]
- Kelly, S.M.; Price, N.C. Circular dichroism to study protein interactions. Curr. Protoc. Protein. Sci. 2006, 46. [Google Scholar] [CrossRef]
- Khrapunov, S. Circular dichroism spectroscopy has intrinsic limitations for protein secondary structure analysis. Anal. Biochem. 2009, 389, 174–176. [Google Scholar] [CrossRef]
- Calero, M.; Gasset, M. Fourier Transform Infrared and Circular Dichroism Spectroscopies for Amyloid Studies. In Amyloid Proteins: Methods and Protocols; Sigurdsson, E.M., Ed.; Humana Press: Totowa, NJ, USA, 2005; pp. 129–151. [Google Scholar]
- Barth, A. Infrared spectroscopy of proteins. Biochim. Biophys. Acta 2007, 1767, 1073–1101. [Google Scholar] [CrossRef] [Green Version]
- Shivu, B.; Seshadri, S.; Li, J.; Oberg, K.A.; Uversky, V.N.; Fink, A.L. Distinct beta-sheet structure in protein aggregates determined by ATR-FTIR spectroscopy. Biochemistry 2013, 52, 5176–5183. [Google Scholar] [CrossRef] [PubMed]
- Miura, T.; Suzuki, K.; Kohata, N.; Takeuchi, H. Metal binding modes of Alzheimer’s amyloid beta-peptide in insoluble aggregates and soluble complexes. Biochemistry 2000, 39, 7024–7031. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.-Y.; Yi, J.; Li, J.-F.; Ren, B.; Wu, D.-Y.; Panneerselvam, R.; Tian, Z.-Q. Nanostructure-based plasmon-enhanced Raman spectroscopy for surface analysis of materials. Nat. Rev. Mater. 2016, 1, 16021. [Google Scholar] [CrossRef]
- Jakubek, R.S.; Handen, J.; White, S.E.; Asher, S.A.; Lednev, I.K. Ultraviolet Resonance Raman Spectroscopic Markers for Protein Structure and Dynamics. Trends Analyt. Chem. 2018, 103, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Kneipp, K.; Kneipp, H.; Irving, I.; Ramachandra, R.D.; Michael, S.F. Surface-enhanced Raman scattering and biophysics. J. Phys. Condens. Matter 2002, 14, R597. [Google Scholar] [CrossRef]
- Langer, J.; Jimenez de Aberasturi, D.; Aizpurua, J.; Alvarez-Puebla, R.A.; Auguié, B.; Baumberg, J.J.; Bazan, G.C.; Bell, S.E.J.; Boisen, A.; Brolo, A.G.; et al. Present and Future of Surface-Enhanced Raman Scattering. ACS Nano 2019, 14, 28–117. [Google Scholar] [CrossRef]
- Khatib, O.; Wood, J.D.; McLeod, A.S.; Goldflam, M.D.; Wagner, M.; Damhorst, G.L.; Koepke, J.C.; Doidge, G.P.; Rangarajan, A.; Bashir, R.; et al. Graphene-Based Platform for Infrared Near-Field Nanospectroscopy of Water and Biological Materials in an Aqueous Environment. ACS Nano 2015, 9, 7968–7975. [Google Scholar] [CrossRef]
- Yong, Y.-C.; Wang, Y.-Z.; Zhong, J.-J. Nano-spectroscopic imaging of proteins with near-field scanning optical microscopy (NSOM). Curr. Opin. Biotechnol. 2018, 54, 106–113. [Google Scholar] [CrossRef]
- Schuler, B. Perspective: Chain dynamics of unfolded and intrinsically disordered proteins from nanosecond fluorescence correlation spectroscopy combined with single-molecule FRET. J. Chem. Phys. 2018, 149, 010901. [Google Scholar] [CrossRef]
- Ferreon, A.C.M.; Moran, C.R.; Gambin, Y.; Deniz, A.A. Single-Molecule Fluorescence Studies of Intrinsically Disordered Proteins. In Single Molecule Tools: Fluorescence Based Approaches, Part A; Academic Press: Cambridge, MA, USA, 2010; pp. 179–204. [Google Scholar]
- Wagele, J.; De Sio, S.; Voigt, B.; Balbach, J.; Ott, M. How Fluorescent Tags Modify Oligomer Size Distributions of the Alzheimer Peptide. Biophys. J. 2019, 116, 227–238. [Google Scholar] [CrossRef] [Green Version]
- Eftink, M.R. The use of fluorescence methods to monitor unfolding transitions in proteins. Biophys. J. 1994, 66, 482–501. [Google Scholar] [CrossRef]
- Siddhanta, S.; Narayana, C. Surface Enhanced Raman Spectroscopy of Proteins: Implications for Drug Designing. Nanomater. Nanotechnol. 2012, 2, 1. [Google Scholar] [CrossRef]
- Rygula, A.; Majzner, K.; Marzec, K.M.; Kaczor, A.; Pilarczyk, M.; Baranska, M. Raman spectroscopy of proteins: A review. J. Raman Spectrosc. 2013, 44, 1061–1076. [Google Scholar] [CrossRef]
- Milosevic, J.; Prodanovic, R.; Polovic, N. On the Protein Fibrillation Pathway: Oligomer Intermediates Detection Using ATR-FTIR Spectroscopy. Molecules 2021, 26, 970. [Google Scholar] [CrossRef]
- Nabers, A.; Ollesch, J.; Schartner, J.; Kotting, C.; Genius, J.; Haussmann, U.; Klafki, H.; Wiltfang, J.; Gerwert, K. An infrared sensor analysing label-free the secondary structure of the Abeta peptide in presence of complex fluids. J. Biophotonics 2016, 9, 224–234. [Google Scholar] [CrossRef]
- Garcia-Chame, M.A.; Gutierrez-Sanz, O.; Ercan-Herbst, E.; Haustein, N.; Filipiak, M.S.; Ehrnhofer, D.E.; Tarasov, A. A transistor-based label-free immunosensor for rapid detection of tau protein. Biosens. Bioelectron. 2020, 159, 112129. [Google Scholar] [CrossRef]
- Nabers, A.; Ollesch, J.; Schartner, J.; Kotting, C.; Genius, J.; Hafermann, H.; Klafki, H.; Gerwert, K.; Wiltfang, J. Amyloid-beta-Secondary Structure Distribution in Cerebrospinal Fluid and Blood Measured by an Immuno-Infrared-Sensor: A Biomarker Candidate for Alzheimer’s Disease. Anal. Chem. 2016, 88, 2755–2762. [Google Scholar] [CrossRef]
- Nabers, A.; Perna, L.; Lange, J.; Mons, U.; Schartner, J.; Guldenhaupt, J.; Saum, K.U.; Janelidze, S.; Holleczek, B.; Rujescu, D.; et al. Amyloid blood biomarker detects Alzheimer’s disease. EMBO Mol. Med. 2018, 10, e8763. [Google Scholar] [CrossRef]
- Lomont, J.P.; Rich, K.L.; Maj, M.; Ho, J.J.; Ostrander, J.S.; Zanni, M.T. Spectroscopic Signature for Stable beta-Amyloid Fibrils versus beta-Sheet-Rich Oligomers. J. Phys. Chem. B 2018, 122, 144–153. [Google Scholar] [CrossRef]
- Zhuang, W.; Sgourakis, N.G.; Li, Z.; Garcia, A.E.; Mukamel, S. Discriminating early stage A{beta}42 monomer structures using chirality-induced 2DIR spectroscopy in a simulation study. Proc. Natl. Acad. Sci. USA 2010, 107, 15687–15692. [Google Scholar] [CrossRef] [Green Version]
- Dicke, S.S.; Maj, M.; Fields, C.R.; Zanni, M.T. Metastable intermediate during hIAPP aggregation catalyzed by membranes as detected with 2D IR spectroscopy. RSC Chem. Biol. 2022, 3, 931–940. [Google Scholar] [CrossRef] [PubMed]
- Remorino, A.; Hochstrasser, R.M. Three-dimensional structures by two-dimensional vibrational spectroscopy. Acc. Chem. Res. 2012, 45, 1896–1905. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.; Ostrander, J.S.; Zanni, M.T. Watching Proteins Wiggle: Mapping Structures with Two-Dimensional Infrared Spectroscopy. Chem. Rev. 2017, 117, 10726–10759. [Google Scholar] [CrossRef] [PubMed]
- Grechko, M.; Zanni, M.T. Quantification of transition dipole strengths using 1D and 2D spectroscopy for the identification of molecular structures via exciton delocalization: Application to alpha-helices. J. Chem. Phys. 2012, 137, 184202. [Google Scholar] [CrossRef]
- Abedin, F.; Tatulian, S.A. Mutual structural effects of unmodified and pyroglutamylated amyloid beta peptides during aggregation. J. Pept. Sci. 2021, 27, e3312. [Google Scholar] [CrossRef]
- Baldassarre, M.; Baronio, C.M.; Morozova-Roche, L.A.; Barth, A. Amyloid beta-peptides 1-40 and 1-42 form oligomers with mixed beta-sheets. Chem. Sci 2017, 8, 8247–8254. [Google Scholar] [CrossRef]
- Baronio, C.M.; Baldassarre, M.; Barth, A. Insight into the internal structure of amyloid-beta oligomers by isotope-edited Fourier transform infrared spectroscopy. Phys. Chem. Chem. Phys. 2019, 21, 8587–8597. [Google Scholar] [CrossRef]
- Ostrander, J.S.; Lomont, J.P.; Rich, K.L.; Saraswat, V.; Feingold, B.R.; Petti, M.K.; Birdsall, E.R.; Arnold, M.S.; Zanni, M.T. Monolayer Sensitivity Enables a 2D IR Spectroscopic Immuno-biosensor for Studying Protein Structures: Application to Amyloid Polymorphs. J. Phys. Chem. Lett 2019, 10, 3836–3842. [Google Scholar] [CrossRef]
- Li, H.; Lantz, R.; Du, D. Vibrational Approach to the Dynamics and Structure of Protein Amyloids. Molecules 2019, 24, 186. [Google Scholar] [CrossRef] [PubMed]
- Sarroukh, R.; Goormaghtigh, E.; Ruysschaert, J.M.; Raussens, V. ATR-FTIR: A “rejuvenated” tool to investigate amyloid proteins. Biochim. Biophys. Acta 2013, 1828, 2328–2338. [Google Scholar] [CrossRef]
- Qi, Q.; Liu, C.; Liu, L.; Meng, Q.; Wei, S.; Ming, A.; Zhang, J.; Wang, Y.; Wu, L.; Zhu, X.; et al. Fabrication, Characterization, and Application of Large-Scale Uniformly Hybrid Nanoparticle-Enhanced Raman Spectroscopy Substrates. Micromachines 2019, 10, 282. [Google Scholar] [CrossRef] [PubMed]
- D’Urso, L.; Condorelli, M.; Puglisi, O.; Tempra, C.; Lolicato, F.; Compagnini, G.; La Rosa, C. Detection and characterization at nM concentration of oligomers formed by hIAPP, Abeta(1-40) and their equimolar mixture using SERS and MD simulations. Phys. Chem. Chem. Phys. 2018, 20, 20588–20596. [Google Scholar] [CrossRef] [PubMed]
- Bhowmik, D.; Mote, K.R.; MacLaughlin, C.M.; Biswas, N.; Chandra, B.; Basu, J.K.; Walker, G.C.; Madhu, P.K.; Maiti, S. Cell-Membrane-Mimicking Lipid-Coated Nanoparticles Confer Raman Enhancement to Membrane Proteins and Reveal Membrane-Attached Amyloid-beta Conformation. ACS Nano 2015, 9, 9070–9077. [Google Scholar] [CrossRef] [PubMed]
- White, S.H.; Wimley, W.C. Membrane protein folding and stability: Physical principles. Annu. Rev. Biophys. Biomol. Struct. 1999, 28, 319–365. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Bhatia, R.; Lal, R. Amyloid beta protein forms ion channels: Implications for Alzheimer’s disease pathophysiology. FASEB J. 2001, 15, 2433–2444. [Google Scholar] [CrossRef]
- Banchelli, M.; Cascella, R.; D’Andrea, C.; Cabaj, L.; Osticioli, I.; Ciofini, D.; Li, M.S.; Skupien, K.; de Angelis, M.; Siano, S.; et al. Nanoscopic insights into the surface conformation of neurotoxic amyloid beta oligomers. RSC Adv. 2020, 10, 21907–21913. [Google Scholar] [CrossRef]
- Devitt, G.; Howard, K.; Mudher, A.; Mahajan, S. Raman Spectroscopy: An Emerging Tool in Neurodegenerative Disease Research and Diagnosis. ACS Chem. Neurosci. 2018, 9, 404–420. [Google Scholar] [CrossRef]
- Hoffmann, W.; Folmert, K.; Moschner, J.; Huang, X.; von Berlepsch, H.; Koksch, B.; Bowers, M.T.; von Helden, G.; Pagel, K. NFGAIL Amyloid Oligomers: The Onset of Beta-Sheet Formation and the Mechanism for Fibril Formation. J. Am. Chem. Soc. 2018, 140, 244–249. [Google Scholar] [CrossRef]
- Ruggeri, F.S.; Charmet, J.; Kartanas, T.; Peter, Q.; Chia, S.; Habchi, J.; Dobson, C.M.; Vendruscolo, M.; Knowles, T.P. Microfluidic deposition for resolving single-molecule protein architecture and heterogeneity. Nat. Commun. 2018, 9, 3890. [Google Scholar] [CrossRef]
- Ruggeri, F.S.; Sneideris, T.; Vendruscolo, M.; Knowles, T.P.J. Atomic force microscopy for single molecule characterisation of protein aggregation. Arch. Biochem Biophys 2019, 664, 134–148. [Google Scholar] [CrossRef]
- Ruggeri, F.S.; Mannini, B.; Schmid, R.; Vendruscolo, M.; Knowles, T.P. Single molecule secondary structure determination of proteins through infrared absorption nanospectroscopy. Nat. Commun. 2020, 11, 2945. [Google Scholar] [CrossRef] [PubMed]
- Ruggeri, F.S.; Habchi, J.; Chia, S.; Horne, R.I.; Vendruscolo, M.; Knowles, T.P.J. Infrared nanospectroscopy reveals the molecular interaction fingerprint of an aggregation inhibitor with single Abeta42 oligomers. Nat. Commun. 2021, 12, 688. [Google Scholar] [CrossRef] [PubMed]
- Feuillie, C.; Lambert, E.; Ewald, M.; Azouz, M.; Henry, S.; Marsaudon, S.; Cullin, C.; Lecomte, S.; Molinari, M. High Speed AFM and NanoInfrared Spectroscopy Investigation of Abeta1-42 Peptide Variants and Their Interaction with POPC/SM/Chol/GM1 Model Membranes. Front. Mol. Biosci. 2020, 7, 571696. [Google Scholar] [CrossRef] [PubMed]
- Dou, T.; Zhou, L.; Kurouski, D. Unravelling the Structural Organization of Individual alpha-Synuclein Oligomers Grown in the Presence of Phospholipids. J. Phys. Chem. Lett. 2021, 12, 4407–4414. [Google Scholar] [CrossRef] [PubMed]
- Waeytens, J.; Van Hemelryck, V.; Deniset-Besseau, A.; Ruysschaert, J.M.; Dazzi, A.; Raussens, V. Characterization by Nano-Infrared Spectroscopy of Individual Aggregated Species of Amyloid Proteins. Molecules 2020, 25, 2899. [Google Scholar] [CrossRef]
- Banerjee, S.; Ghosh, A. Structurally Distinct Polymorphs of Tau Aggregates Revealed by Nanoscale Infrared Spectroscopy. J. Phys. Chem. Lett. 2021, 12, 11035–11041. [Google Scholar] [CrossRef]
- Banerjee, S.; Holcombe, B.; Ringold, S.; Foes, A.; Naik, T.; Baghel, D.; Ghosh, A. Nanoscale Infrared Spectroscopy Identifies Structural Heterogeneity in Individual Amyloid Fibrils and Prefibrillar Aggregates. J. Phys. Chem. B 2022, 126, 5832–5841. [Google Scholar] [CrossRef]
- Nie, S.; Emory, S.R. Probing Single Molecules and Single Nanoparticles by Surface-Enhanced Raman Scattering. Science 1997, 275, 1102–1106. [Google Scholar] [CrossRef]
- Kneipp, K.; Wang, Y.; Kneipp, H.; Perelman, L.T.; Itzkan, I.; Dasari, R.R.; Feld, M.S. Single Molecule Detection Using Surface-Enhanced Raman Scattering (SERS). Phys. Rev. Lett. 1997, 78, 1667–1670. [Google Scholar] [CrossRef]
- Lesser-Rojas, L.; Ebbinghaus, P.; Vasan, G.; Chu, M.-L.; Erbe, A.; Chou, C.-F. Low-Copy Number Protein Detection by Electrode Nanogap-Enabled Dielectrophoretic Trapping for Surface-Enhanced Raman Spectroscopy and Electronic Measurements. Nano Lett. 2014, 14, 2242–2250. [Google Scholar] [CrossRef]
- Blum, C.; Opilik, L.; Atkin, J.M.; Braun, K.; Kämmer, S.B.; Kravtsov, V.; Kumar, N.; Lemeshko, S.; Li, J.-F.; Luszcz, K.; et al. Tip-enhanced Raman spectroscopy—An interlaboratory reproducibility and comparison study. J. Raman Spectrosc. 2014, 45, 22–31. [Google Scholar] [CrossRef]
- Wang, X.; Huang, S.-C.; Hu, S.; Yan, S.; Ren, B. Fundamental understanding and applications of plasmon-enhanced Raman spectroscopy. Nat. Rev. Phys. 2020, 2, 253–271. [Google Scholar] [CrossRef]
- Khater, I.M.; Nabi, I.R.; Hamarneh, G. A Review of Super-Resolution Single-Molecule Localization Microscopy Cluster Analysis and Quantification Methods. Patterns 2020, 1, 100038. [Google Scholar] [CrossRef] [PubMed]
- Tahir, M.A.; Dina, N.E.; Cheng, H.; Valev, V.K.; Zhang, L. Surface-enhanced Raman spectroscopy for bioanalysis and diagnosis. Nanoscale 2021, 13, 11593–11634. [Google Scholar] [CrossRef] [PubMed]
- Kurouski, D.; Deckert-Gaudig, T.; Deckert, V.; Lednev, I.K. Surface characterization of insulin protofilaments and fibril polymorphs using tip-enhanced Raman spectroscopy (TERS). Biophys. J. 2014, 106, 263–271. [Google Scholar] [CrossRef]
- vandenAkker, C.C.; Deckert-Gaudig, T.; Schleeger, M.; Velikov, K.P.; Deckert, V.; Bonn, M.; Koenderink, G.H. Nanoscale Heterogeneity of the Molecular Structure of Individual hIAPP Amyloid Fibrils Revealed with Tip-Enhanced Raman Spectroscopy. Small 2015, 11, 4131–4139. [Google Scholar] [CrossRef]
- Darussalam, E.Y.; Peterfi, O.; Deckert-Gaudig, T.; Roussille, L.; Deckert, V. pH-dependent disintegration of insulin amyloid fibrils monitored with atomic force microscopy and surface-enhanced Raman spectroscopy. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2021, 256, 119672. [Google Scholar] [CrossRef]
- Louros, N.N.; Tsiolaki, P.L.; Baltoumas, F.A.; Chryssikos, G.D.; Gionis, V.; Hamodrakas, S.J.; Iconomidou, V.A. Tracking the amyloidogenic core of IAPP amyloid fibrils: Insights from micro-Raman spectroscopy. J. Struct. Biol. 2017, 199, 140–152. [Google Scholar] [CrossRef]
- Flynn, J.D.; McGlinchey, R.P.; Walker, R.L., 3rd; Lee, J.C. Structural features of alpha-synuclein amyloid fibrils revealed by Raman spectroscopy. J. Biol. Chem. 2018, 293, 767–776. [Google Scholar] [CrossRef]
- Flynn, J.D.; Lee, J.C. Raman fingerprints of amyloid structures. Chem. Commun. 2018, 54, 6983–6986. [Google Scholar] [CrossRef]
- Zikic, B.; Bremner, A.; Talaga, D.; Lecomte, S.; Bonhommeau, S. Tip-enhanced Raman spectroscopy of Aβ(1-42) fibrils. Chem. Phys. Lett. 2021, 768, 138400. [Google Scholar] [CrossRef]
- Deckert-Gaudig, T.; Kurouski, D.; Hedegaard, M.A.; Singh, P.; Lednev, I.K.; Deckert, V. Spatially resolved spectroscopic differentiation of hydrophilic and hydrophobic domains on individual insulin amyloid fibrils. Sci. Rep. 2016, 6, 33575. [Google Scholar] [CrossRef]
- Bonhommeau, S.; Talaga, D.; Hunel, J.; Cullin, C.; Lecomte, S. Tip-Enhanced Raman Spectroscopy to Distinguish Toxic Oligomers from Abeta1-42 Fibrils at the Nanometer Scale. Angew. Chem. Int. Ed. Engl. 2017, 56, 1771–1774. [Google Scholar] [CrossRef] [PubMed]
- Devitt, G.; Rice, W.; Crisford, A.; Nandhakumar, I.; Mudher, A.; Mahajan, S. Conformational Evolution of Molecular Signatures during Amyloidogenic Protein Aggregation. ACS Chem. Neurosci. 2019, 10, 4593–4611. [Google Scholar] [CrossRef]
- D’Andrea, C.; Foti, A.; Cottat, M.; Banchelli, M.; Capitini, C.; Barreca, F.; Canale, C.; de Angelis, M.; Relini, A.; Marago, O.M.; et al. Nanoscale Discrimination between Toxic and Nontoxic Protein Misfolded Oligomers with Tip-Enhanced Raman Spectroscopy. Small 2018, 14, e1800890. [Google Scholar] [CrossRef] [PubMed]
- Vu, K.H.P.; Lee, M.C.; Blankenburg, G.H.; Chang, Y.J.; Chu, M.L.; Erbe, A.; Lesser-Rojas, L.; Chen, Y.R.; Chou, C.F. Time-Evolved SERS Signatures of DEP-Trapped Abeta and Zn(2+)Abeta Peptides Revealed by a Sub-10 nm Electrode Nanogap. Anal. Chem. 2021, 93, 16320–16329. [Google Scholar] [CrossRef] [PubMed]
- Holzel, R.; Calander, N.; Chiragwandi, Z.; Willander, M.; Bier, F.F. Trapping single molecules by dielectrophoresis. Phys. Rev. Lett. 2005, 95, 128102. [Google Scholar] [CrossRef] [PubMed]
- Tong, J.; Suo, F.; Ma, J.; Tobing, L.Y.; Qian, L.; Zhang, D.H. Surface plasmon enhanced infrared photodetection. Opto-Electron. Adv. 2019, 2, 18002601–18002610. [Google Scholar] [CrossRef]
- Yang, X.; Sun, Z.; Low, T.; Hu, H.; Guo, X.; Garcia de Abajo, F.J.; Avouris, P.; Dai, Q. Nanomaterial-Based Plasmon-Enhanced Infrared Spectroscopy. Adv. Mater. 2018, 30, e1704896. [Google Scholar] [CrossRef]
- Dazzi, A.; Prater, C.B. AFM-IR: Technology and Applications in Nanoscale Infrared Spectroscopy and Chemical Imaging. Chem. Rev. 2017, 117, 5146–5173. [Google Scholar] [CrossRef]
- Lu, Y.H.; Larson, J.M.; Baskin, A.; Zhao, X.; Ashby, P.D.; Prendergast, D.; Bechtel, H.A.; Kostecki, R.; Salmeron, M. Infrared Nanospectroscopy at the Graphene-Electrolyte Interface. Nano Lett. 2019, 19, 5388–5393. [Google Scholar] [CrossRef]
- Paulite, M.; Fakhraai, Z.; Li, I.T.; Gunari, N.; Tanur, A.E.; Walker, G.C. Imaging secondary structure of individual amyloid fibrils of a beta2-microglobulin fragment using near-field infrared spectroscopy. J. Am. Chem. Soc. 2011, 133, 7376–7383. [Google Scholar] [CrossRef] [PubMed]
- Balandin, A.A.; Ghosh, S.; Bao, W.; Calizo, I.; Teweldebrhan, D.; Miao, F.; Lau, C.N. Superior Thermal Conductivity of Single-Layer Graphene. Nano Lett. 2008, 8, 902–907. [Google Scholar] [CrossRef] [PubMed]
- Clement, J.E.; Leray, A.; Bouhelier, A.; Finot, E. Spectral pointillism of enhanced Raman scattering for accessing structural and conformational information on single protein. Phys. Chem. Chem. Phys. 2016, 19, 458–466. [Google Scholar] [CrossRef] [PubMed]
- Leray, A.; Clement, J.E.; Bouhelier, A.; Finot, E. Conformational Changes and Charge Transfer in Biomolecules Resolved Using Dynamic Enhanced Raman Correlation Spectroscopy. J. Phys. Chem. B 2019, 123, 1931–1938. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.A.; Mousavi, M.Z.; Giovannini, G.; Zhao, Y.; Hubarevich, A.; Soler, M.A.; Rocchia, W.; Garoli, D.; De Angelis, F. Multiplexed Discrimination of Single Amino Acid Residues in Polypeptides in a Single SERS Hot Spot. Angew. Chem. Int. Ed. Engl. 2020, 59, 11423–11431. [Google Scholar] [CrossRef]
- Holzel, R.; Pethig, R. Protein Dielectrophoresis: I. Status of Experiments and an Empirical Theory. Micromachines 2020, 11, 533. [Google Scholar] [CrossRef]


| Fluorescence | NMR | CD | UV–Vis | FTIR | Raman | |
|---|---|---|---|---|---|---|
| Basic principle | Light emission by residual aromatic amino acids | Nuclear spin relaxation | Differential absorption of circular polarized light | Electronic transitions | Vibrations of molecular bonds (changes in dipole moments) | Vibrations of molecular bonds (changes in polarizability) |
| Resolution | Medium (tertiary structure on a local level) | High (secondary and tertiary structure on a global and local level) | Low to medium (secondary and tertiary structure on a global level) | Low to medium (tertiary structure on a global level) | Low to medium (secondary structure on a global level; tertiary structure on a local level with isotope-labeling) | Medium to high (secondary and tertiary structure on a global level) |
| Sensitivity | Single molecule (extrinsic FS)–μM (intrinsic FS) | 0.1–1 mM | μM–mM | μM | 0.1–1 mM (proteins), 1–100 mM (small molecules) | Single molecule (PERS)–mM (bulk Raman) |
| Limitations | Photostability issues, limited fluorophore lifespan, auto-fluorescence; fluorescent labeling might affect protein aggregation and structure (extrinsic FS) | High sample purity, sample size limit ≤100 kDa (solution NMR); high amount of sample, lyophilized and isotopically labeled samples (ssNMR) | Less accurate predictions for β-structure than for α-helices | Stray light and light scattering interferences, overlapping of spectral peaks | Water interference, overlapping of spectral peaks | Fluorescence interference, photodecomposition and low signal (bulk Raman); requires appropriate substrate/plasmonic structures (PERS) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vu, K.H.P.; Blankenburg, G.H.; Lesser-Rojas, L.; Chou, C.-F. Applications of Single-Molecule Vibrational Spectroscopic Techniques for the Structural Investigation of Amyloid Oligomers. Molecules 2022, 27, 6448. https://doi.org/10.3390/molecules27196448
Vu KHP, Blankenburg GH, Lesser-Rojas L, Chou C-F. Applications of Single-Molecule Vibrational Spectroscopic Techniques for the Structural Investigation of Amyloid Oligomers. Molecules. 2022; 27(19):6448. https://doi.org/10.3390/molecules27196448
Chicago/Turabian StyleVu, Katrin Ha Phuong, Gerhard Heinrich Blankenburg, Leonardo Lesser-Rojas, and Chia-Fu Chou. 2022. "Applications of Single-Molecule Vibrational Spectroscopic Techniques for the Structural Investigation of Amyloid Oligomers" Molecules 27, no. 19: 6448. https://doi.org/10.3390/molecules27196448







