Application of β-Glucosidase in a Biphasic System for the Efficient Conversion of Polydatin to Resveratrol
Abstract
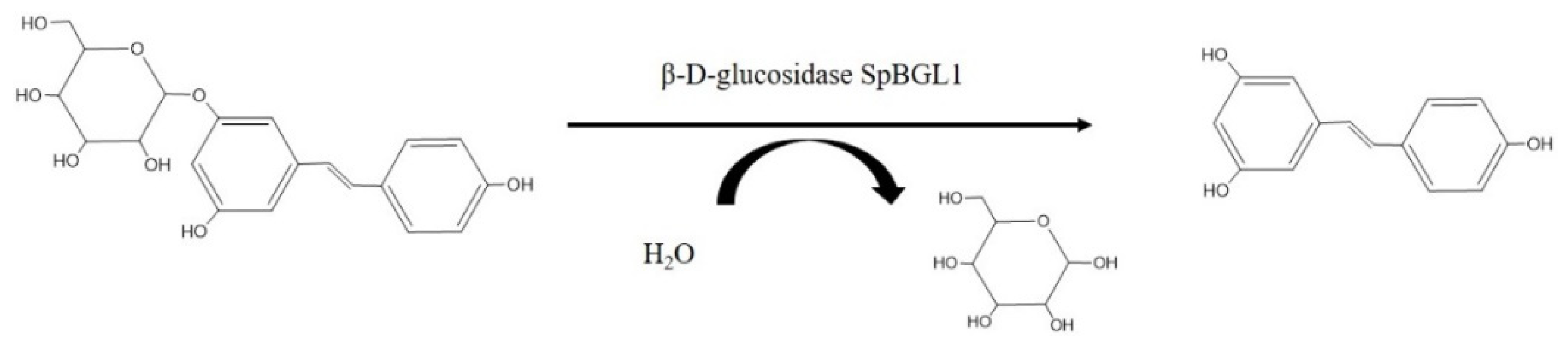
:1. Introduction
2. Results
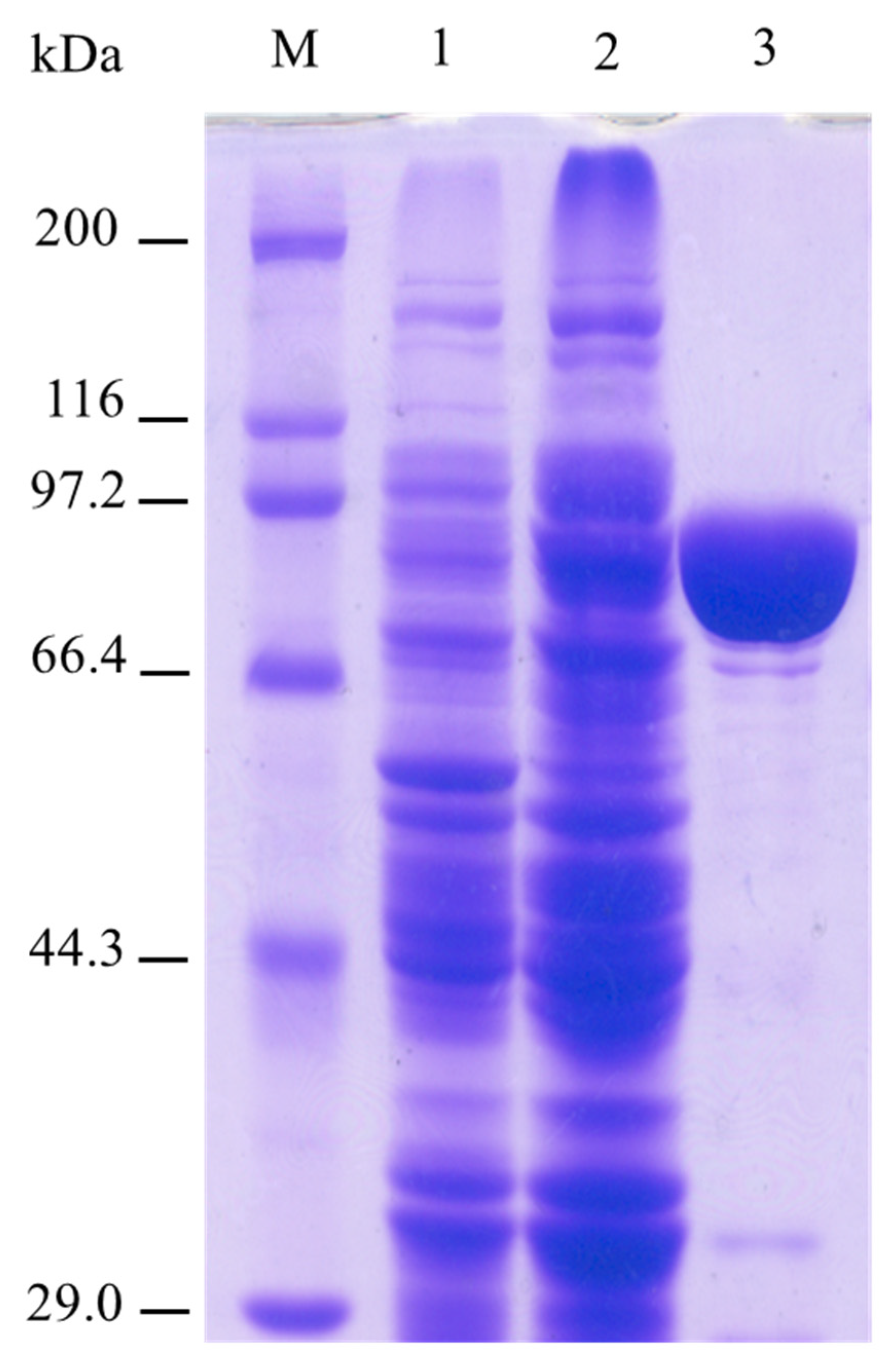
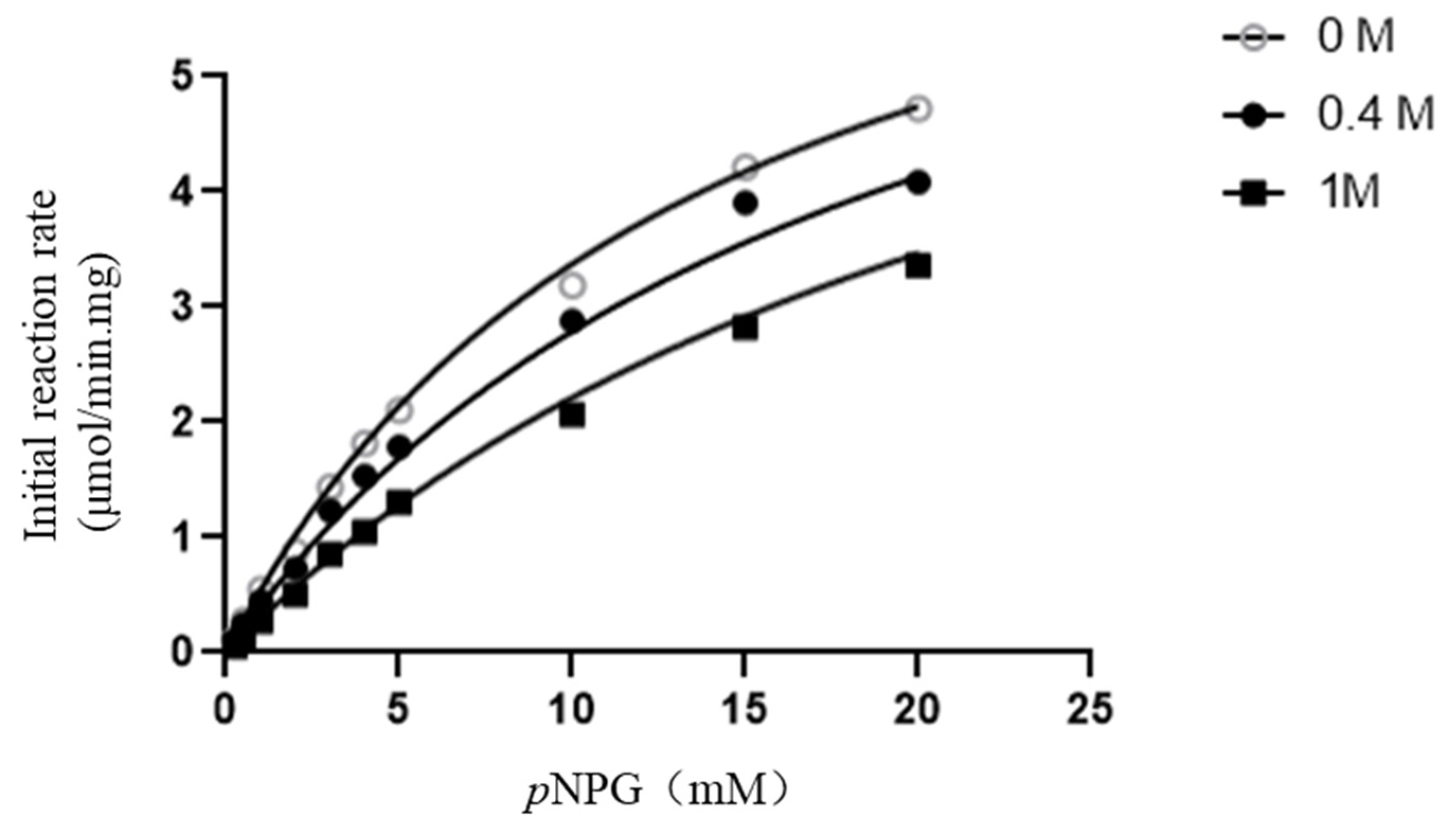
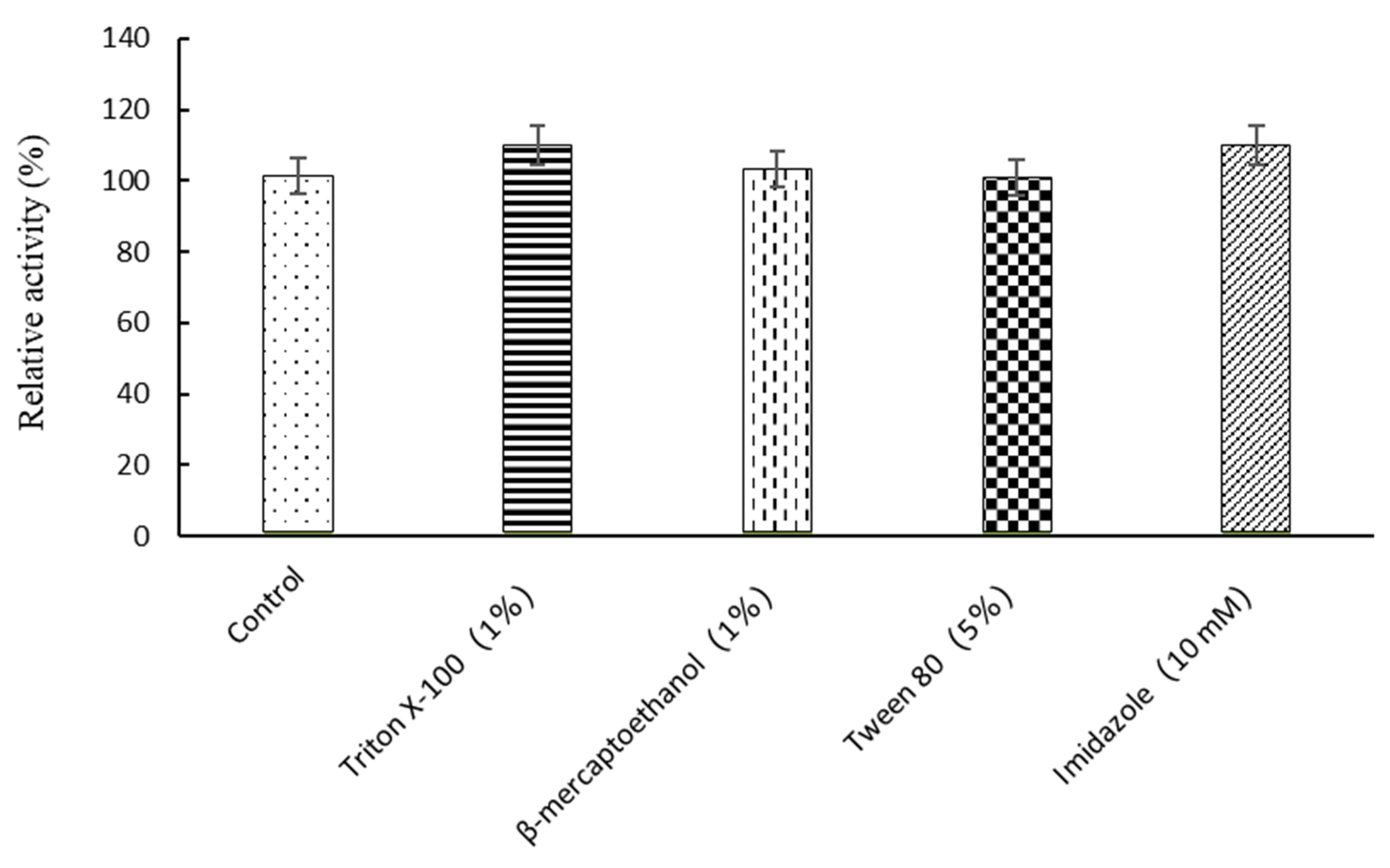
2.1. Enzymatic Properties of β-D-Glucosidase SpBGL1
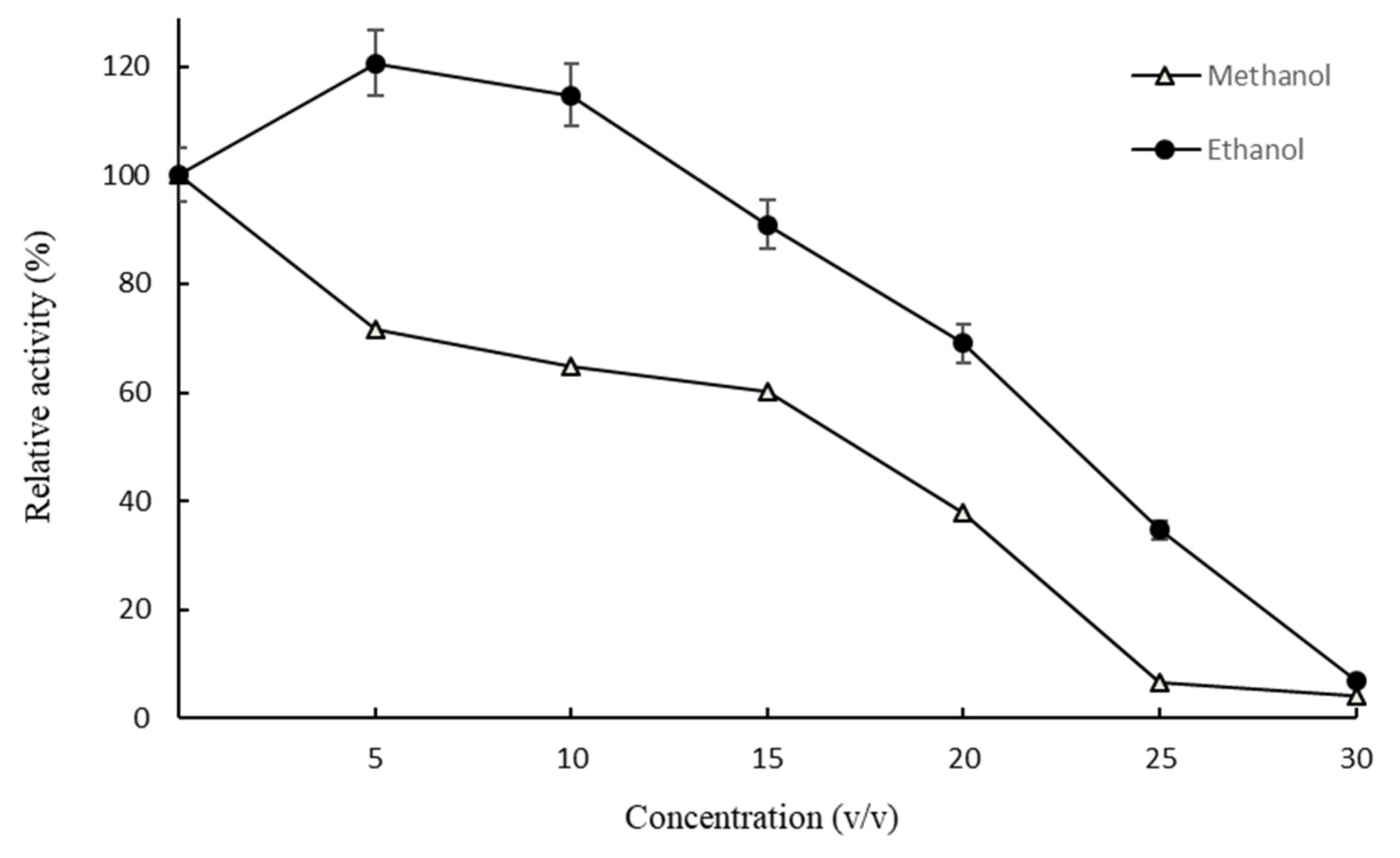
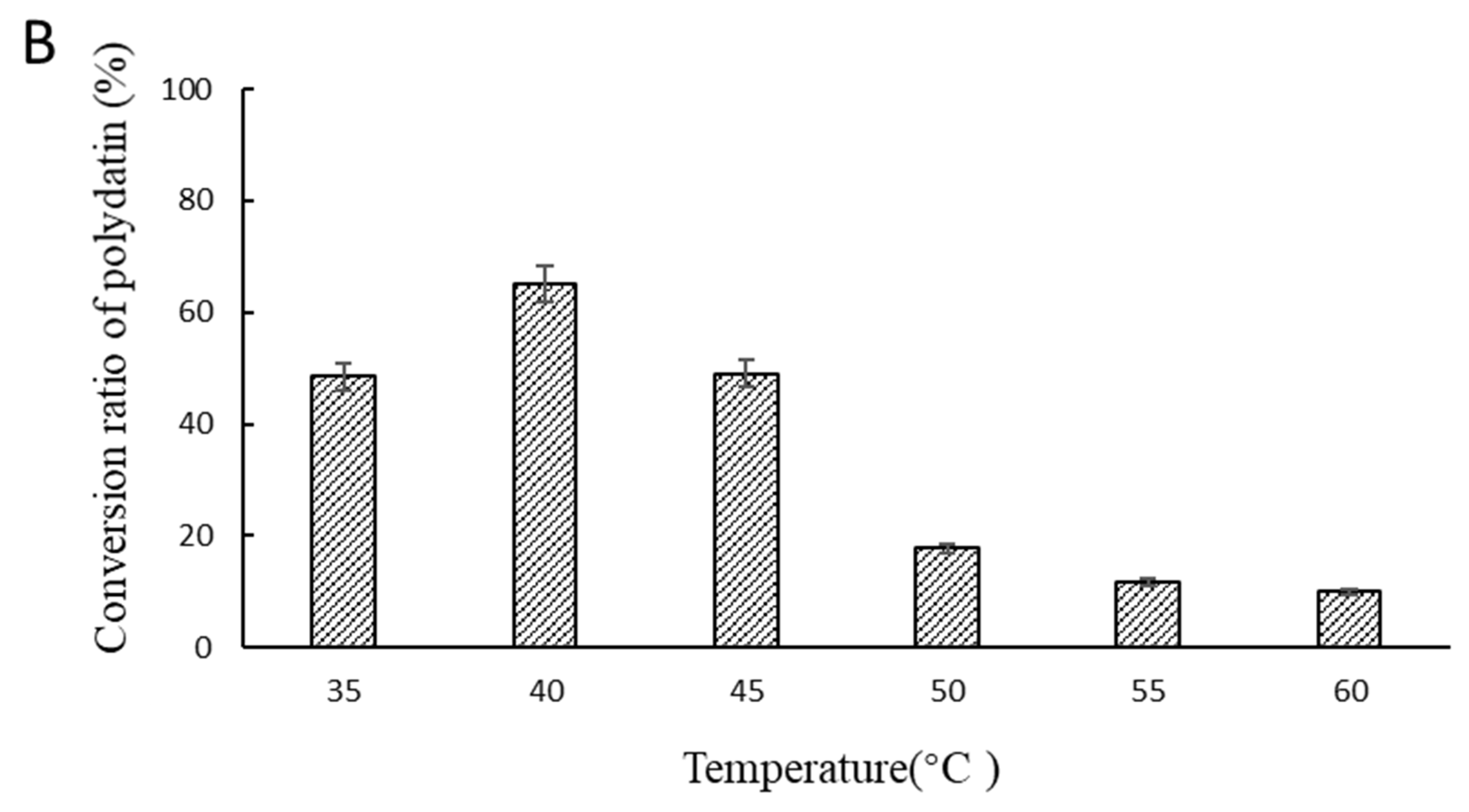
2.2. Optimal Reaction Conditions for Hydrolyzing Polydatin
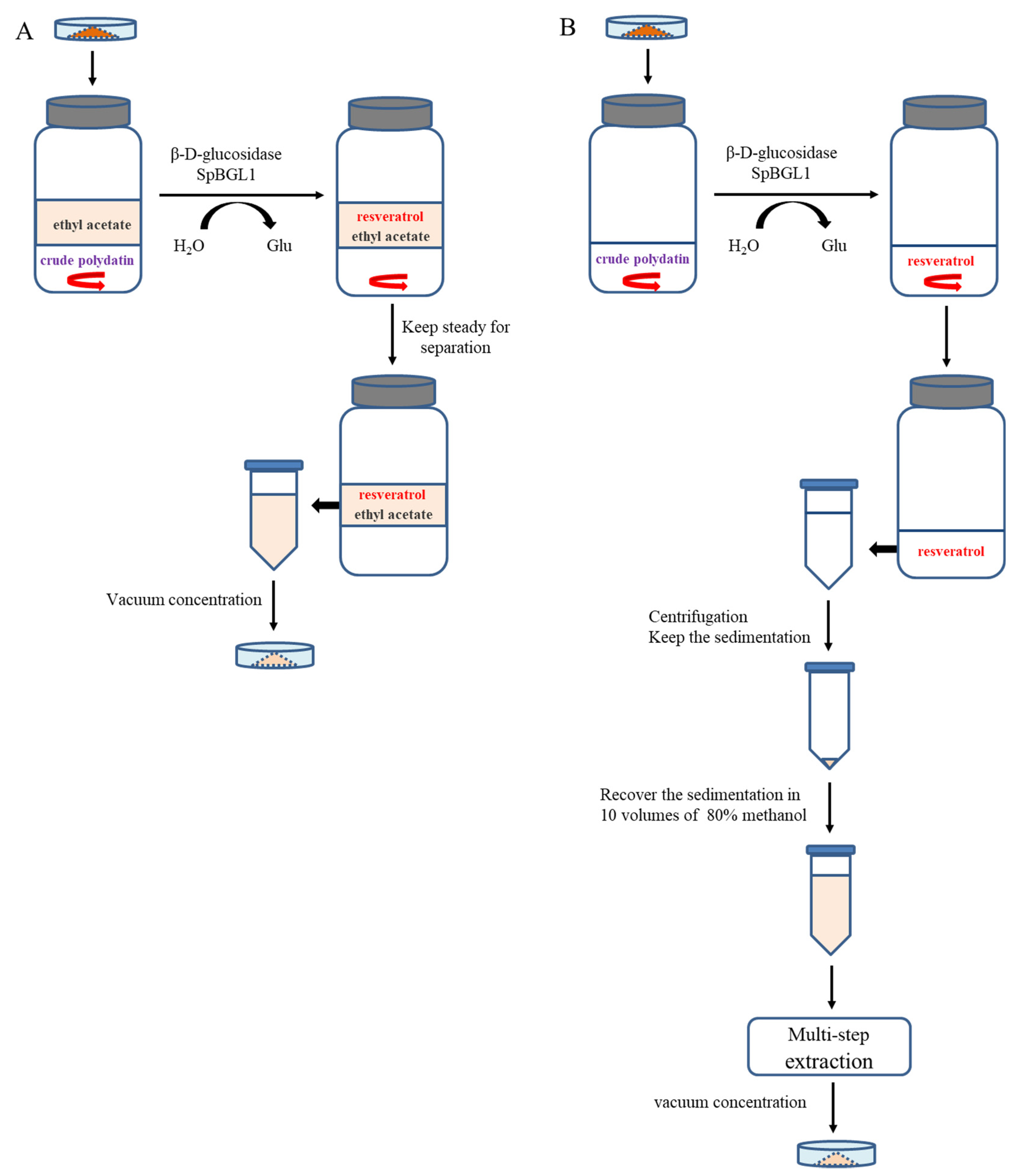
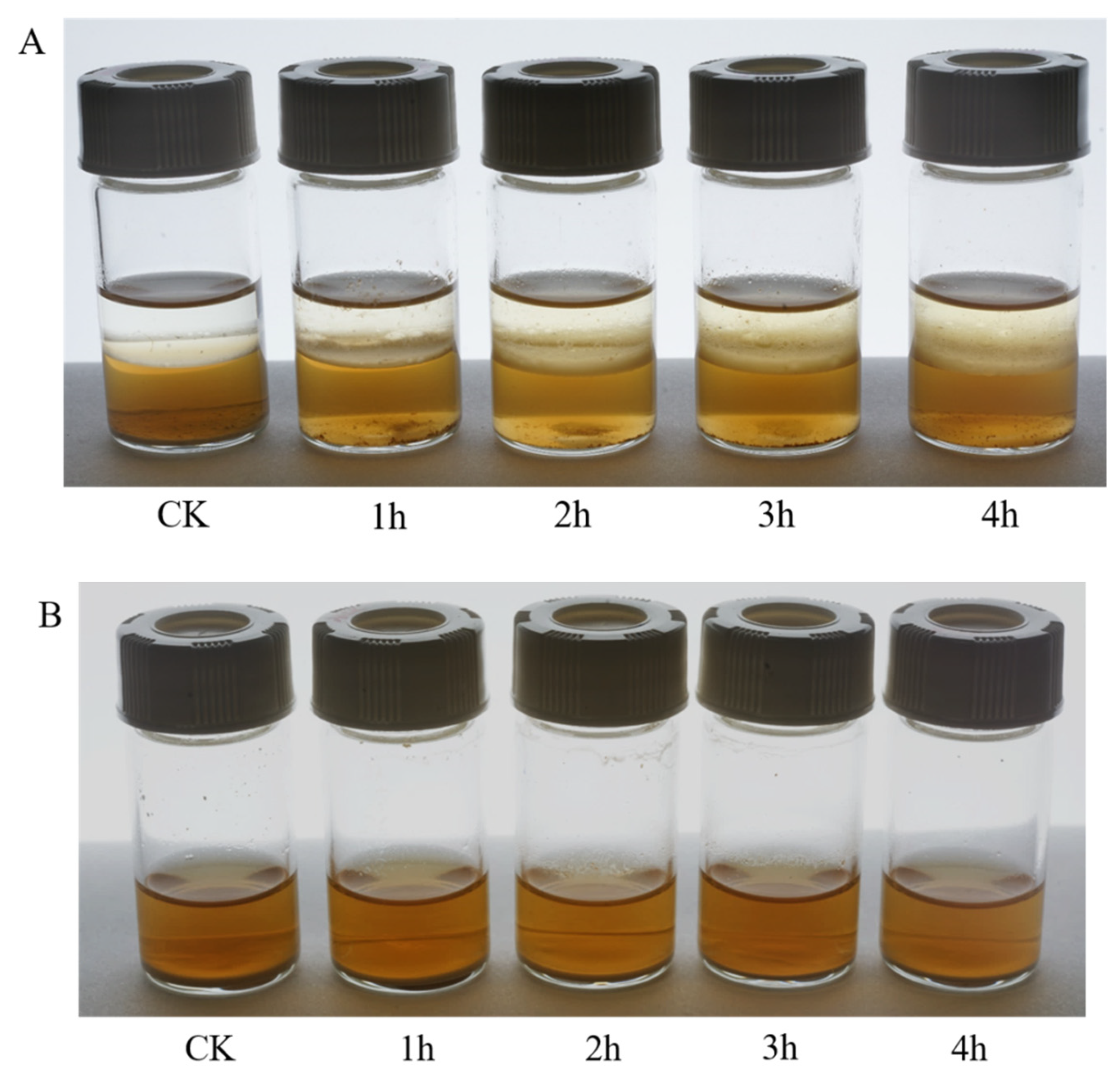
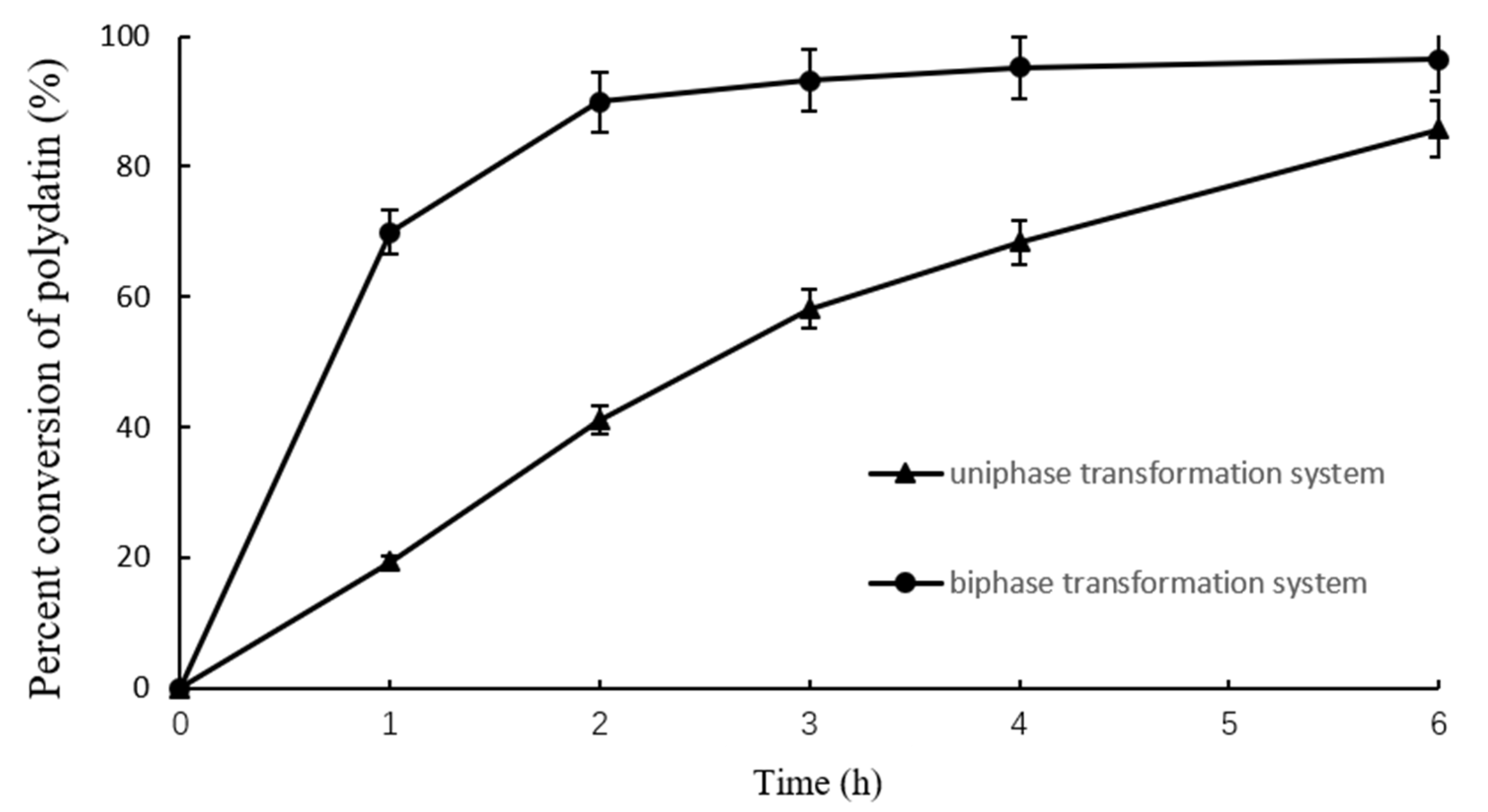
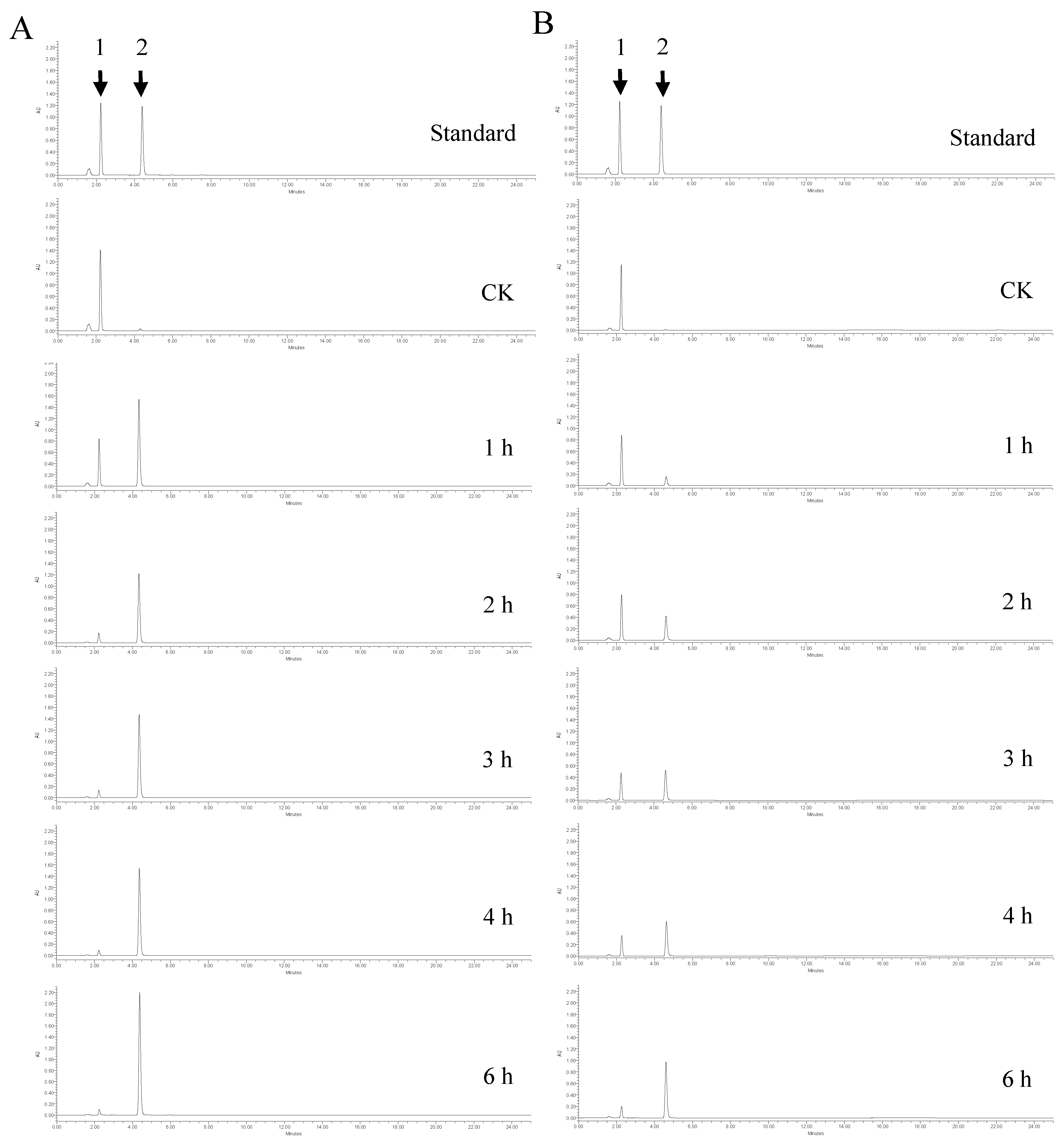
2.3. Comparison of Uniaphase and Biphase Enzymatic Conversion for Polydatin Hydrolysis
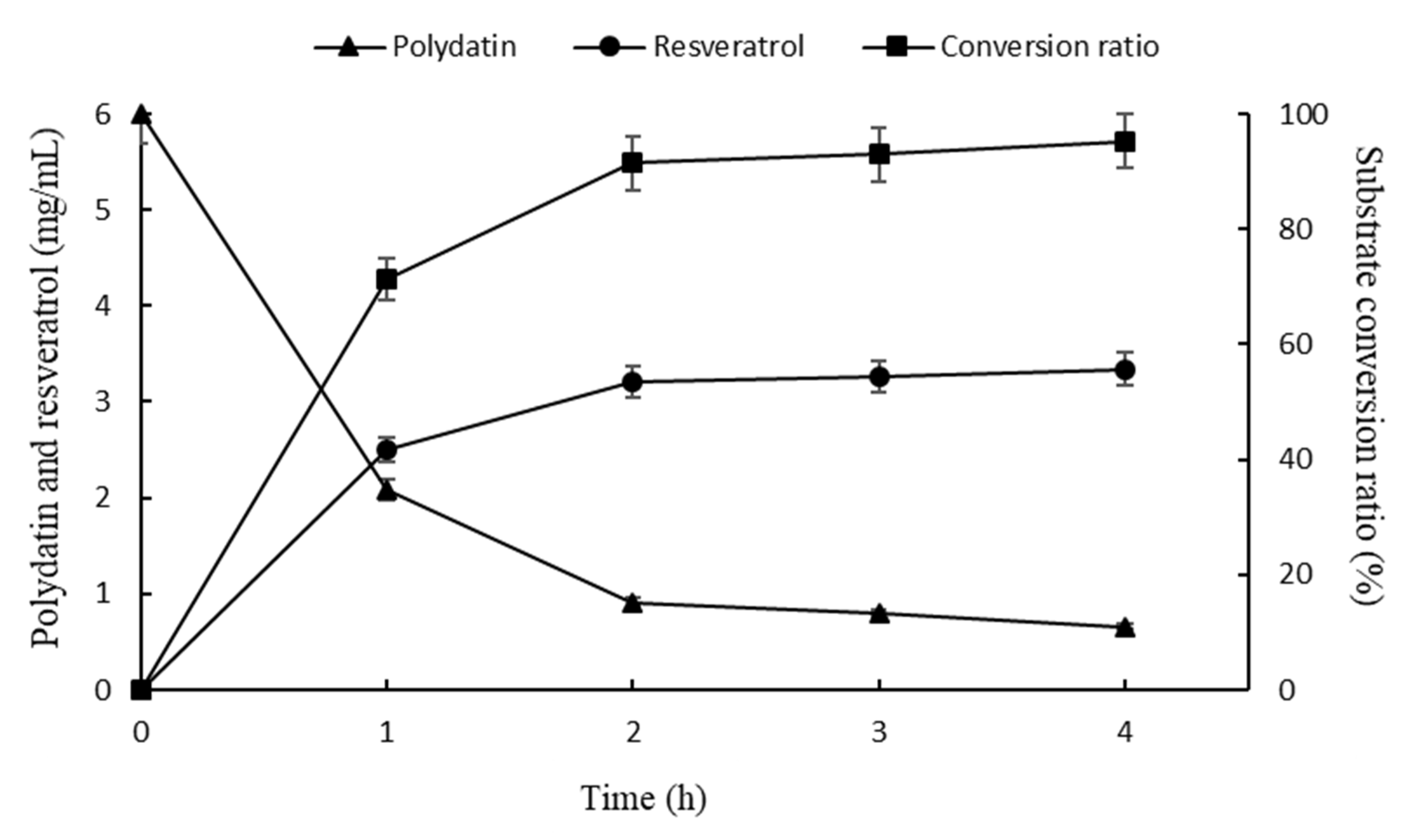
2.4. Optimization of Biphase Enzymatic Conversion System
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Growth Conditions
4.2. Cloning of SpBGL1
4.3. Heterologous Expression of SpBGL1
4.4. Enzyme Activity Assays
4.5. Enzymatic Characterization of SpBGL1
4.6. Optimal Enzymatic Conditions for Catalysis of Polydatin by SpBGL1
4.7. Enzymatic Hydrolysis Assay in The Uniphase and Biphasic Conversion Systems
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Jeandet, P.; Delaunois, B.; Conreux, A.; Donnez, D.; Nuzzo, V.; Cordelier, S.; Clement, C.; Courot, E. Biosynthesis, metabolism, molecular engineering and biological functions of stilbene phytoalexins in plants. Biofactors 2010, 36, 331–341. [Google Scholar] [CrossRef]
- Jeandet, P.; Douillt-Breuil, A.C.; Bessis, R.; Debord, S.; Sbaghi, M.; Adrian, M. Phytoalexins from the vitaceae: Biosynthesis, phytoalexin gene expression in transgenic plants, antifungal activity, and metabolism. J. Agric. Food Chem. 2002, 50, 2731–2741. [Google Scholar] [CrossRef]
- Jeandet, P.; Clément, C.; Courot, E. Use of plant cell suspensions as a basis for large-scale production of resveratrol in bioreactors. Eng. Life Sci. 2014, 14, 622–632. [Google Scholar] [CrossRef]
- Coutinho, D.D.; Pacheco, M.T.; Frozza, R.L.; Bernardi, A. Anti-Inflammatory Effects of Resveratrol: Mechanistic Insights. Int. J. Mol. Sci. 2018, 19, 1812. [Google Scholar] [CrossRef] [Green Version]
- Shen, C.Y.; Jiang, J.G.; Yang, L.; Wang, D.W.; Zhu, W. Anti-ageing active ingredients from herbs and nutraceuticals used in traditional Chinese medicine: Pharmacological mechanisms and implications for drug discovery. Br. J. Pharmacol. 2017, 174, 1395–1425. [Google Scholar] [CrossRef] [Green Version]
- Voloshyna, I.; Hussaini, S.M.; Reiss, A.B. Resveratrol in Cholesterol Metabolism and Atherosclerosis. J. Med. Food 2012, 15, 763–773. [Google Scholar] [CrossRef]
- Gambini, J.; Inglés, M.; Olaso, G.; Lopez-Grueso, R.; Bonet-Costa, V.; Gimeno-Mallench, L.; Mas-Bargues, C.; Abdelaziz, K.M.; Gomez-Cabrera, M.C.; Vina, J.; et al. Properties of Resveratrol: In Vitro and In Vivo Studies about Metabolism, Bioavailability, and Biological Effects in Animal Models and Humans. Oxid. Med. Cell. Longev. 2015, 2015, 837042. [Google Scholar] [CrossRef] [Green Version]
- Jeandet, P.; Bessis, R.; Gautheron, B. The production of resveratrol (3,5,4′-trihydroxystilbene) by grape berries in different developmental stages. Am. J. Enol. Vitic. 1991, 42, 41–46. [Google Scholar]
- Lyons, M.M.; Yu, C.W.; Toma, R.B.; Cho, S.Y.; Reiboldt, W.; Lee, J.; Van Breemen, R.B. Resveratrol in raw and baked blueberries and bilberries. J. Agric. Food Chem. 2003, 51, 5867–5870. [Google Scholar] [CrossRef]
- Mudnic, I.; Budimir, D.; Modun, D.; Gunjaca, G.; Generalic, I.; Skroza, D.; Katalinic, V.; Ljubenkov, I.; Boban, M. Antioxidant and Vasodilatory Effects of Blackberry and Grape Wines. J. Med. Food 2012, 15, 315–321. [Google Scholar] [CrossRef] [Green Version]
- Shrikanta, A.; Kumar, A.; Govindaswamy, V. Resveratrol content and antioxidant properties of underutilized fruits. J. Food Sci. Technol. 2015, 52, 383–390. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Catana, F.; Yang, Y.N.; Roderick, R.; van Breemen, R.B. An LC-MS method for analyzing total resveratrol in grape juice, cranberry juice, and in wine. J. Agric. Food Chem. 2002, 50, 431–435. [Google Scholar] [CrossRef]
- Nonomura, S.; Kanagawa, H.; Makimoto, A. Chemical constituents of polygonaceous plants. I. Studies on the components of Ko-jo-kon (Polygonum cuspidatum Sieb Et Zucc.). Yakugaku Zasshi 1963, 83, 988–990. [Google Scholar] [CrossRef] [Green Version]
- Henry, C.; Vitrac, X.; Decendit, A.; Ennamany, R.; Krisa, S.; Mérillon, J.-M. Cellular Uptake and Efflux of trans-Piceid and Its Aglycone trans-Resveratrol on the Apical Membrane of Human Intestinal Caco-2 Cells. J. Agric. Food Chem. 2005, 53, 798–803. [Google Scholar] [CrossRef]
- Signorelli, P.; Ghidoni, R. Resveratrol as an anticancer nutrient: Molecular basis, open questions and promises. J. Nutr. Biochem. 2005, 16, 449–466. [Google Scholar] [CrossRef]
- Wang, C.; Liu, X.; Zhang, M.; Shao, H.; Wang, X.; Wang, Q.; Bao, Z.; Fan, X.; Li, H. Efficient Enzyme-Assisted Extraction and Conversion of Polydatin to Resveratrol From Polygonum cuspidatum Using Thermostable Cellulase and Immobilized β-Glucosidase. Front. Microbiol. 2019, 10, 445. [Google Scholar] [CrossRef]
- Hu, X.; Liu, Y.; Li, D.; Feng, W.; Ni, H.; Cao, S.; Lu, F.; Li, Y. An innovative biotransformation to produce resveratrol by Bacillus safensis. RSC Adv. 2019, 9, 15448–15456. [Google Scholar] [CrossRef] [Green Version]
- Che, J.; Shi, J.; Gao, Z.; Zhang, Y. A New Approach to Produce Resveratrol by Enzymatic Bioconversion. J. Microbiol. Biotechnol. 2016, 26, 1348–1357. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.-G.; Liu, W.-Y.; Chen, G.-T. A simple method for the isolation and purification of resveratrol from Polygonum cuspidatum. J. Pharm. Anal. 2013, 3, 241–247. [Google Scholar] [CrossRef] [Green Version]
- Zada, N.S.; Belduz, A.O.; Güler, H.I.; Sahinkaya, M.; Khan, S.I.; Saba, M.; Bektas, K.I.; Kara, Y.; Kolaylı, S.; Badshah, M.; et al. Cloning, biochemical characterization and molecular docking of novel thermostable β-glucosidase BglA9 from Anoxybacillus ayderensis A9 and its application in de-glycosylation of Polydatin. Int. J. Biol. Macromol. 2021, 193, 1898–1909. [Google Scholar] [CrossRef]
- Wang, J.-D.; Fu, L.-N.; Wang, L.-T.; Cai, Z.-H.; Wang, Y.-Q.; Yang, Q.; Fu, Y.-J. Simultaneous transformation and extraction of resveratrol from Polygonum cuspidatum using acidic natural deep eutectic solvent. Ind. Crops Prod. 2021, 173, 114140. [Google Scholar] [CrossRef]
- Wang, H.; Liu, L.; Guo, Y.X.; Dong, Y.S.; Zhang, D.J.; Xiu, Z.L. Biotransformation of piceid in Polygonum cuspidatum to resveratrol by Aspergillus oryzae. Appl. Microbiol. Biotechnol. 2007, 75, 763–768. [Google Scholar] [CrossRef] [PubMed]
- Tian, T.L.; Sun, Q.L.; Shen, J.; Zhang, T.; Gao, P.; Sun, Q. Microbial transformation of polydatin and emodin-8-beta-D-glucoside of Polygonum cuspidatum Sieb. et Zucc into resveratrol and emodin respectively by Rhizopus microsporus. World J. Microbiol. Biotechnol. 2008, 24, 861–866. [Google Scholar] [CrossRef]
- Xu, N.E.; Luo, L.B.; Mei, J.F.; Jia, Y.Q.; Wang, Q.Z. Increasing the Content of Resveratrol in Chinese Herb Polygonum Cuspidatum by Bioconversion. Pharm. Biotechnol. 2012, 19, 3. [Google Scholar]
- Wan, H.-D.; Tao, G.-J.; Kim, D.; Xia, Y.-M. Enzymatic preparation of a natural sweetener rubusoside from specific hydrolysis of stevioside with β-galactosidase from Aspergillus sp. J. Mol. Catal. B Enzym. 2012, 82, 12–17. [Google Scholar] [CrossRef]
- Nguyen, T.T.; Jung, S.J.; Kang, H.K.; Kim, Y.M.; Moon, Y.H.; Kim, M.; Kim, D. Production of rubusoside from stevioside by using a thermostable lactase from Thermus thermophilus and solubility enhancement of liquiritin and teniposide. Enzym. Microb. Technol. 2014, 64–65, 38–43. [Google Scholar] [CrossRef]
- Chen, M.; Li, D.; Gao, Z.Q.; Zhang, C.Z. Enzymatic transformation of polydatin to resveratrol by piceid-beta-d-glucosidase from Aspergillus oryzae. Bioprocess Biosyst. Eng. 2014, 37, 1411–1416. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, X.; Yan, T.; Wang, F.; Li, J.; Jia, L.; Jia, J.; Hu, G. Screening of an Endophyte Transforming Polydatin to Resveratrol from Reynoutria Japonica Houtt and the Optimization of Its Transformation Parameters. Molecules 2020, 25, 4830. [Google Scholar] [CrossRef]
- Du, L.; Wang, Z.; Zhao, Y.; Huang, J.; Pang, H.; Wei, Y.; Lin, L.; Huang, R. A β-glucosidase from Novosphingobium sp. GX9 with high catalytic efficiency toward isoflavonoid glycoside hydrolysis and (+)-catechin transglycosylation. Appl. Microbiol. Biotechnol. 2014, 98, 7069–7079. [Google Scholar] [CrossRef]
- Huang, Z.F.; Yi, J.H.; Liu, Q.L.; Liu, Y.H.; Chen, Y.; Liu, Y.H. Rearch of Extracting and Purifying Process of Resveratrol from Polygonum caspidatum Extract by Enzymatic Hydrolysis. Nat. Prod. Res. Dev. 2009, 21, 1061–1064. [Google Scholar]
- Kakkar, T.; Boxenbaum, H.; Mayersohn, M. Estimation of Ki in a Competitive Enzyme-Inhibition Model: Comparisons Among Three Methods of Data Analysis. Drug Metab. Dispos. 1999, 27, 756. [Google Scholar] [PubMed]
- Shen, Y.; Wang, H.; Lu, Y.; Xu, L.; Yin, H.; Tam, J.P.; Yang, H.; Jia, X. Construction of a novel catalysis system for clean and efficient preparation of Baohuoside I from Icariin based on biphase enzymatic hydrolysis. J. Clean. Prod. 2018, 170, 727–734. [Google Scholar] [CrossRef]


| Substrate | Activity |
|---|---|
| Polydatin | + |
| Daidzin | + |
| Glycitin | + |
| Genistin | + |
| Icariin | + |
| Epimedin A | + |
| Epimedin B | + |
| Epimedin C | + |
| Rebaudioside A | − |
| Stevioside | − |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, J.; Liang, M.; Lin, Y.; Pang, H.; Wei, Y.; Huang, R.; Du, L. Application of β-Glucosidase in a Biphasic System for the Efficient Conversion of Polydatin to Resveratrol. Molecules 2022, 27, 1514. https://doi.org/10.3390/molecules27051514
Zhou J, Liang M, Lin Y, Pang H, Wei Y, Huang R, Du L. Application of β-Glucosidase in a Biphasic System for the Efficient Conversion of Polydatin to Resveratrol. Molecules. 2022; 27(5):1514. https://doi.org/10.3390/molecules27051514
Chicago/Turabian StyleZhou, Jie, Meng Liang, Yu Lin, Hao Pang, Yutuo Wei, Ribo Huang, and Liqin Du. 2022. "Application of β-Glucosidase in a Biphasic System for the Efficient Conversion of Polydatin to Resveratrol" Molecules 27, no. 5: 1514. https://doi.org/10.3390/molecules27051514





