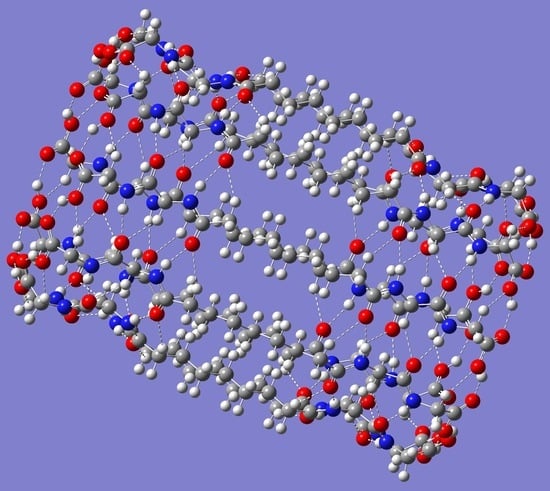
2.1. (P-CH2-P)n Bolaamphiphile Nanotubes
A bolaamphiphile molecule contains two functional head groups connected by a hydrophobic chain. This study focuses on two peptide-based amphiphilic carboxy groups symmetrically placed at the two ends of the molecule. The smaller bolaamphiphile molecule consists of two N-α amido glycylglycine head groups linked by a single CH2 group, HOOC-[CH2-NHCO]2-CH2-[CONH-CH2]2-COOH, or P-CH2-P for short. The larger one has seven CH2 groups separating the two head groups, P-(CH2)7-P. For convenience, the former is discussed in this subsection, while the latter is discussed in the next subsection.
Geometry optimizations showed that formation of tube-like self-assembly (P-CH
2-P)
n structures requires a minimum of four monomers (
n = 4), or a tetramer. Tubular geometries containing more than four building molecules were also optimized, namely, hexamer, octamer, and decamer. The optimized geometries had C
nh symmetry, with
n being the number of self-assembled molecules. Frequency calculations confirmed that all geometries are indeed minimum energy structures with no imaginary frequencies. Geometry optimizations of even larger sizes,
n = 12 and
n = 16 were also carried out. Top views for both the smallest and the largest optimized (P-CH
2-P)
n nanotubes are shown in
Figure 1. Inspection of
Figure 1 reveals the inner cavity of the tubes, with some hydrogen atoms from the CH
2 groups pointing inward while others point outward.
Corresponding side views of the nanotubes are shown in
Figure 2. Both
Figure 1 and
Figure 2 readily reveal that intratubular H-bonds drive the tubular formation, specifically, H-bonds between adjacent molecules involving the amide motifs, N-H· · ·O=C, and those involving the carboxy groups, O-H· · ·O=C. Weaker C-H· · ·O=C bonds with the C-H group as H-bond donor appear to be present as well.
Binding energies and average binding energies for the different (P-CH
2-P)
n nanotubes are listed in
Table 1, in kcal/mol. Binding energies, ΔE, are the negative values of the interaction energies. The interaction energies are calculated as the difference between the total energy of the nanotube and the sum of the energies of the individual monomers with the same geometry as they have in the optimized nanotubes. Dividing the binding energy by the number of molecules in the nanotube structure yields the corresponding average binding energy, ΔE/
n.
Table 1 shows that both DFT methods predict large binding energies that increase linearly with the size of the molecules making up the nanotube. The B3LYP method tends to underestimate the calculated binding energies relative to those obtained with the wB97XD method. The gap in the binding energies between the two methods increases with the size of the systems. For example, for
n = 4, the gap is 59.20 kcal/mol, while, for
n = 16, the gap is more than three times that amount, 186.27 kcal/mol.
Table 1 also shows that the average binding energies significantly increase as the system size grows from
n = 4 to
n = 6, regardless of the DFT method used. A small additional increase is seen at
n = 8, which is followed by a steady but small decline in the following larger systems. The difference in average binding energies between the two DFT methods becomes somewhat smaller with
n. Accordingly, the difference decreases from 14.80 to 11.64 kcal/mol as the systems grows from
n = 4 to
n = 16.
It is important to notice that the binding energies in
Table 1 do not account for the energy cost or deformation energy associated with the change in the geometry from the isolated optimized monomer to the geometry adopted upon nanotube formation. To gauge the magnitude of this deformation energy, the geometry of the free monomer was first optimized at the B3LYP/6-31G(d) level. Corresponding frequency calculations resulted in no imaginary frequencies, confirming the minimum energy nature of the optimized monomer. Single-point energy calculations on the optimized monomer were performed with both the B3LYP/6-31G(d) and the wB97XD/DGDZVP methods. Comparing the energies of the monomer in each nanotube with that of the optimized monomer resulted in a relatively large average deformation energy of 17.1 ± 0.6 kcal/mol with the B3LYP/method. Likewise, the average deformation energy with the wB97XD method is 19.4 ± 0.7 kcal/mol. Thus, the binding energies based on the optimized monomer as a reference point can be obtained by subtracting the average deformation energies from the average binding energies listed in
Table 1. Using this approach, for example, the average binding energy for the largest nanotube,
n = 16, is 23.6 kcal/mol (B3LYP) or 32.9 kcal/mol (wB97XD).
Part of the stability of the nanosystems presented in
Table 1 can be attributed to the H-bonding interactions, O-H· · ·O=C, between the carboxy groups of adjacent molecules. The contribution from these interactions may be estimated by calculating the binding energies after removal of the carboxy groups, replacing them with hydrogen atoms and keeping the geometry of each remaining structure as that in the original optimized system. The results shown in
Table 2 demonstrate that removal of the carboxy groups greatly reduces the wB97XD binding energies of the nanotubes to about 62% of its original value for the smallest system, and to about 66–70% for all the other system sizes. That is, the O-H· · ·O=C hydrogen bonds account for approximately a third or more of the wB97XD binding energies of the (P-CH
2-P)
n nanotubes. This is also true for the related average binding energies. The B3LYP binding energies of the nanotubes are similarly reduced upon removal of the carboxy groups, namely, to 50% for the tetramer and to about 60–68% for the others.
Geometrical parameters (distances and angles) for the various H-bonds driving the formation of the (P-CH
2-P)
n nanotubes are displayed in
Table 3. Inspection of
Figure 2, from one end to the center of the nanotube, makes it apparent that any monomer in a nanotube participates in the following six interactions as an H-bond donor or acceptor with adjacent molecules: O-H· · ·O=C, between the carboxy groups; C-H· · ·O-H, between a methylene group and the carboxylic acid OH group; two N-H· · ·O=C, between amide groups; C-H· · ·O=C, between a methylene group and an amide O=C group; and, lastly, a bifurcated C-H· · ·O=C, between the central methylene and amide O=C groups. Note that these interactions repeat, passing from the center all the way to the other end of the nanotube, due to the symmetry of the molecule. Not listed in
Table 3 are the dimensions of the nanotubes. Although the length remains constant at about 18 Å, the diameter of the nanotubes increases from about 5 Å in the tetramer to about 22 Å in the hexadecamer.
As seen in
Table 3, the lengthening of the interaction distances concomitant with the narrowing of the respective angles for the O-H· · ·O=C hydrogen bonds indicates a weakening of these interactions with the increasing size of the nanotubes. For example, from the smallest to the largest system, this H-bond distance is increased by 0.216 Å, while the angle is reduced by 18.0 degrees. Although its angles change little, the changes in H-bond distances suggest that the bifurcated C-H· · ·O=C interaction weakens with
n. In contrast, all other H-bonds are strengthened as the size of the nanotube grows, although the rate of change becomes smaller with
n.
The observations made previously on geometrical grounds can be further supported by examining the calculated electron density at the critical point of each of the H-bonds, ρ
cp, driving the formation of the (P-CH
2-P)
n nanotubes. The H-bond critical electron densities calculated with the wB97XD/DGDZVP method are shown in
Table 4. Inspection of
Table 4 reveals, for example, that the weakening of the O-H· · ·O=C interaction is accompanied by a sizeable reduction in the corresponding ρ
cp with increasing size of the nanotubes. Moreover, the critical bond density for the bifurcated C-H· · ·O=C interaction exhibits an important decrease as the system grows from
n = 4 to
n = 6. This decrease is followed by a steady yet small increase in ρ
cp as the system size increases gradually from
n = 6 to
n = 16. The results for the bifurcated H-bond are consistent with the geometrical changes mentioned before, namely, a large elongation of the H-bond followed by a comparatively small but consistent reduction in the H-bond distance. The bond critical densities for the other H-bonds tend to increase with the size of the nanotube, with some already nearing plateau. Lastly, the H-bond between a methylene group and the carboxylic acid OH group appears so weak when
n = 4 that it actually lacks the corresponding H-bond critical point. Nonetheless, this C-H· · ·O-H interaction does gain considerable strength with increasing size of the nanotube, as demonstrated by the presence of the H-bond critical point for
n > 4. Geometrically, the very weak C-H· · ·O-H interaction when
n = 4 is consistent with a relatively large H-bond distance and narrow angle (
Table 3). For
n > 4, this H-bond distance consistently shortens, while the H-bond angle widens greatly, indicating that the H-bond is indeed becoming stronger.
2.2. (P-(CH2)7-P)n Bolaamphiphile Nanotubes
The self-assembly into nanotubes of the larger bolaamphiphile molecule is discussed in this section. The building molecules consist of two N-α amido glycylglycine head groups linked by seven methylene groups, HOOC-[CH
2-NHCO]
2-(CH
2)
7-[CONH-CH
2]
2-COOH, or P-(CH
2)
7-P for short. Matsui and Gologan reported that, in two weeks and at pH 4, this heptane bolaamphiphile undergoes self-assembly into a crystalline tubule [
97]. These authors suggested an assembly of the building motifs into a planar multilayer of intermolecular H-bonds between adjacent amide groups, accompanied by H-bonds between carboxy groups that bring two monomers together, extending the system in the direction perpendicular to that of the amide–amide H-bonds. At low pH, the planar multilayer curls tightly into tubular shapes.
A sheet-like arrangement of up to 12 P-(CH
2)
7-P monomers, positioned relative to one another as shown in
Figure 3 (top panel), was built as the initial guess for geometry optimization. It is important to note that, in this approach, the H-bonds between carboxy groups run parallel with the H-bonds between amide groups. Consequently, the idea is first to examine the possible folding of the initial sheet-like geometry into nanotubes with the length of the given monomer or building block, along with a diameter that increases with the number of monomers in the system. The tubular lengthening via carboxylic acid H-bonds is discussed in the next subsection. Geometry optimizations for the smaller (P-(CH
2)
7-P)
n systems of size
n = 4 and
n = 6 resulted in minimum energy structures with partial folding, as shown in the middle and bottom panels, respectively, in
Figure 3.
For the larger-size monomers, geometry optimizations resulted in complete tubular folding. It was, nonetheless, possible to obtain the tubular shapes for the
n = 4 and
n = 6 systems by using a guess geometry built by bringing the corresponding end monomers in the optimized structures shown in
Figure 3 closer. The open structure for
n = 4 is just 2.18 kcal/mol lower in energy than the completely folded one. In sharp contrast, the open structure for
n = 6 is much higher in energy (by 39.48 kcal/mol) than the completely folded one. In general, the optimized self-assembled (P-(CH
2)
7-P)
n nanotubes for
n = 4, 6, 8, and 10 resulted in structures of symmetry C
nh, with
n being the number of self-assembled molecules. Frequency calculations confirm that all these optimized geometries, including the partially folded ones, are indeed minimum energy structures with no imaginary frequencies. Likewise, geometry optimization for the larger dodecamer was also successfully carried out. Side views of the optimized geometries are shown in
Figure 4 for the nanotubes of sizes
n = 4 and
n = 12.
Figure 4 shows the H-bonds driving the tubular formation. Not surprisingly, these are the same type of interactions seen in the smaller nanotubes (P-CH
2-P)
n discussed earlier. One difference that is worthy of notice is the potential for the dispersion interactions in adjacent molecules emerging from the longer hydrocarbon chain in each P-(CH
2)
7-P monomer.
Binding energies and average binding energies for the different (P-(CH
2)
7-P)
n nanotubes of size
n are listed in
Table 5. The results parallel those of the smaller (P-CH
2-P)
n nanotubes (
Table 1). For example,
Table 5 shows that both DFT methods predict large binding energies that increase linearly with the size of the molecules making up the nanotube, and that the B3LYP method underestimates the calculated binding energies relative to those obtained with the wB97XD method. Moreover, the increase in the average binding energies seems to level off rather quickly, and the difference in average binding energies between the two DFT methods becomes smaller with
n. To obtain the average binding energies using the geometry of the optimized monomer as a reference point, the deformation energies were calculated for the monomer with the geometry in each of the nanotubes. For the smallest nanotube, the deformation energy is 10.53 kcal/mol (B3LYP) or 11.22 kcal/mol (wB97XD). The corresponding values for the other nanotubes,
n > 4, are somewhat larger, but much closer to one another, with an average deformation energy of 13.0 ± 0.1 kcal/mol (B3LYP) or 13.6 ± 0.1 kcal/mol (wB97XD). Despite the relatively large deformation energies, the average binding energies remain significant. For example, the average binding energy for the largest nanotube,
n = 12, is 26.6 kcal/mol (B3LYP) or 40.6 kcal/mol (wB97XD) after accounting for the deformation energy.
Table 6 displays the difference between the binding energies (and average binding energies) of the (P-(CH
2)
7-P)
n nanotubes and those of the (P-CH
2-P)
n nanotubes. The results are obtained by subtracting the values listed in
Table 1 from the related ones in
Table 5. These changes in binding energies are important because they help provide insight into the role played by the hydrocarbon chain in stabilizing the nanotubes. The negative changes in binding energies seen in
Table 6 for the B3LYP method suggest that the hydrocarbon chain plays a destabilizing role; the opposite, however, is true for the wB97XD method, which shows corresponding positive changes in the binding energies. The inability of the B3LYP hybrid functional to account for dispersion interactions helps explain the negative changes in binding energies. On the other hand, the wB97XD method is better equipped to capture the important dispersion interactions taking place among the hydrocarbon chains of adjacent monomers. Interestingly, the B3LYP change in the average binding energy remains basically constant for the larger nanotubes. In contrast, the corresponding changes with the wB97XD method show a steady decrease with increasing system size, although this change seems to stabilize at about 0.6 kcal/mol for the largest
n.
Given that the various types of H-bond interactions driving the formation of both (P-CH
2-P)
n and (P-(CH
2)
7-P)
n nanotubes are basically the same, it was of interest to examine how the presence of the longer hydrocarbon chain modifies the geometrical parameters (distances and angles) for these interactions.
Table 7 shows changes in the geometrical parameters listed in
Table 3 upon addition of the longer hydrocarbon chain for nanotubes of size up to
n = 12. Examination of
Table 7 reveals that changes are minimal, within the second and third decimal place, for most H-bond distances. Larger changes, within the first decimal place, occur for the C-H· · ·O=C hydrogen bonds which were originally involved in bifurcated H-bonds. Changes in corresponding H-bond angles are also small, within the tenths, for half of the H-bonds, and within the ones for the other half. Again, larger angle changes are seen in the originally bifurcated C-H· · ·O=C hydrogen bond and the N-H· · ·O=C closer to it. Lastly, the lengths of the (P-(CH
2)
7-P)
n nanotubes lie between 26 and 25 Å, that is, about 8 Å longer than in their smaller (P-CH
2-P)
n counterparts.
The H-bond critical electron densities calculated with the wB97XD/DGDZVP method for the (P-(CH
2)
7-P)
n nanotubes are shown in
Table 8. Cross-examination of
Table 8 and
Table 4 shows that the calculated H-bond critical bond densities are quite similar (within the third or fourth decimal place) for both the larger and smaller bolaamphiphiles considered here, in agreement with the similar geometrical characteristics discussed above.
2.3. Intertubular Elongation through O-H· · ·O=C Hydrogen Bonds
Each nanotubular assembly of the (P-(CH
2)
7-P)
n bolaamphiphile peptide could in principle be extended via intertubular O-H· · ·O=C hydrogen bond interactions, as proposed by Matsui and Gologan [
97]. That is, the H-bonds between adjacent carboxy groups in each parent nanotube are disrupted and replaced with the same type of H-bonds but now between adjacent parent nanotubes, which would result in a tubular elongation. To test this hypothesis, a geometry optimization of two (P-(CH
2)
7-P)
n nanotubes of size
n = 4 was first carried out. The two nanotubes were positioned relative to each other, so as to favor O-H· · ·O=C interactions between the tubes.
Figure 5 shows that the geometry optimization successfully culminated in an elongated nanotube about 54 Å long (slightly more than twice the length of the parent nanotube, which is about 26 Å). The corresponding geometry optimization of two nanotubes of size
n = 6 also resulted in tubular elongation through the O-H· · ·O=C hydrogen bonds (
Figure 5). BSSE-corrected dimer binding energies were calculated at the B3LYP/6-31G(d) and wB97XD/DGDZVP levels for each of the nanotube dimers. The B3LYP results were 67.71 and 84.34 kcal/mol for the smaller and larger nanotube dimer systems, respectively. Dividing these dimer binding energies by
n gives an average dimer binding energy of 16.93 and 14.06 kcal/mol for
n = 4 and
n = 6, respectively. The wB97XD/DGDZVP method resulted in binding energies of 72.83 and 93.86 kcal/mol and related average binding energies of 18.21 and 15.64 kcal/mol for
n = 4 and
n = 6, respectively. Although B3LYP somewhat underestimates the H-bond binding energies when compared with wB97XD, both methods agree that the intertubular O-H· · ·O=C hydrogen bonds provide a significant contribution to the stability of the elongated nanotubes. Intertubular growth is then expected to continue, which would progressively extend the length of the nanotubular structures. Because of the large computational cost, it becomes impractical to examine whether the decrease in the average dimer binding energies seen as
n increases from 4 to 6 will continue for larger values of
n.
To estimate the extent to which the O-H· · ·O=C hydrogen bond binding energies change with the size of the nanotubes upon dimer elongation, geometry optimization for smaller systems was performed. Specifically, only the region containing the intertubular H-bonds and the closest intratubular N-H· · ·O=C hydrogen bonds was considered. This approach significantly reduces the size of each system. Accordingly, the geometries of systems of size
n = 4, 6, 10, and 16 were optimized. In particular, the B3LYP/6-31G(d) optimized geometry of the
n = 16 system is displayed in
Figure 6.
Frequency calculations consistently show no imaginary frequencies, which confirms the minimum energy structure nature of the optimized dimer geometries for all
n values considered. BSSE-corrected dimer binding (and average binding) energies calculated at the B3LYP/6-31G(d) and wB97XD/DGDZVP levels for each of the modified nanotube dimers are listed in
Table 9. It is worth noting that the results listed in
Table 9 for the smaller
n = 4 and
n = 6 size dimer systems are close to those obtained for the larger (P-(CH
2)
7-P)
n dimer counterparts discussed previously. The results listed in
Table 9 indicate that, as
n increases, the average dimer binding energy does indeed decrease. However, this decrease in dimer binding energy tends towards some plateau value near 13.35 or 14.91 kcal/mol, depending on the DFT method used.