Identification of the Metabolites of Both Formononetin in Rat Hepatic S9 and Ononin in Rat Urine Samples and Preliminary Network Pharmacology Evaluation of Their Main Metabolites
Abstract
:1. Introduction
2. Results and Discussion
2.1. Isolation and Determination of the Structure of Formononetin Metabolites
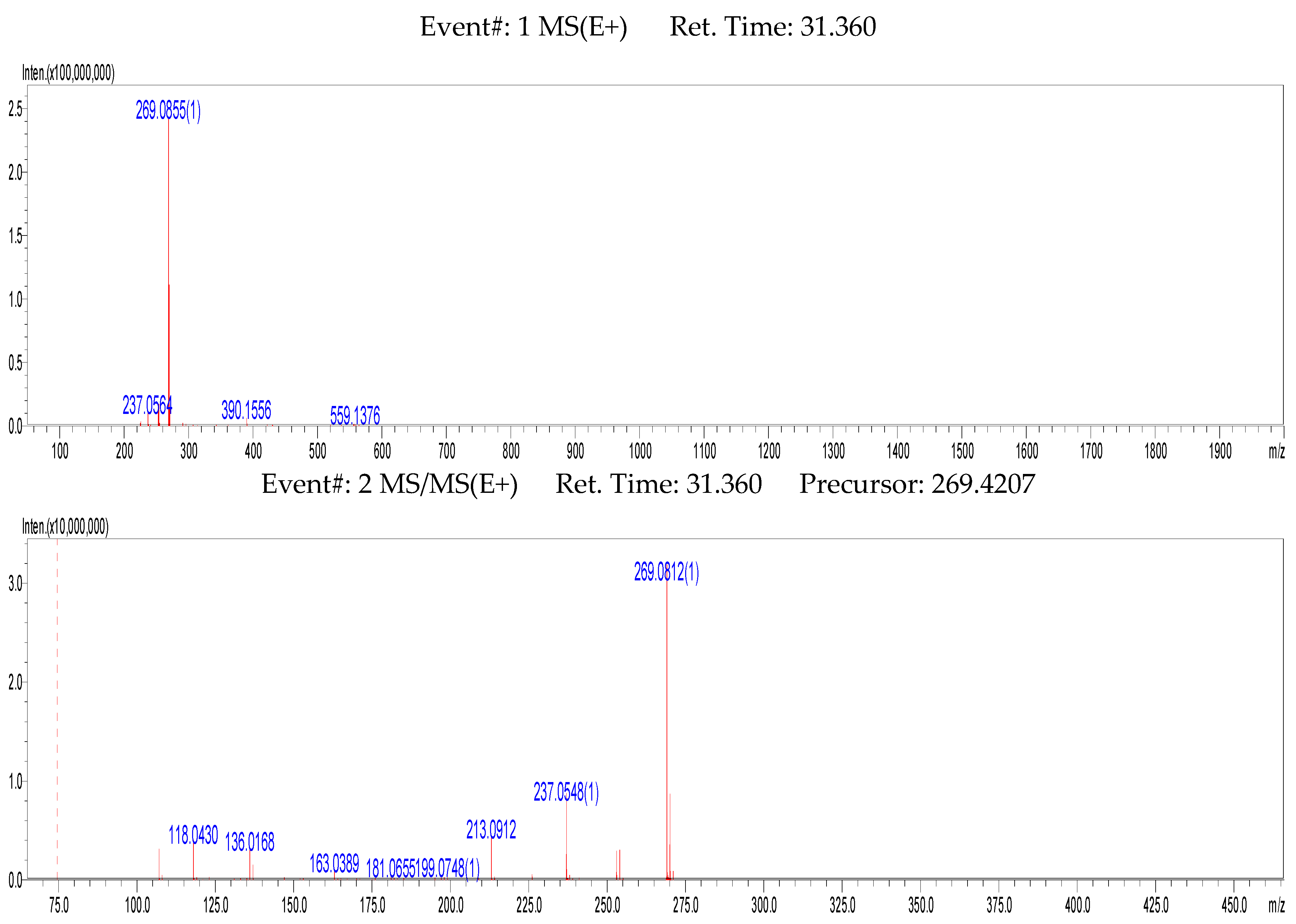
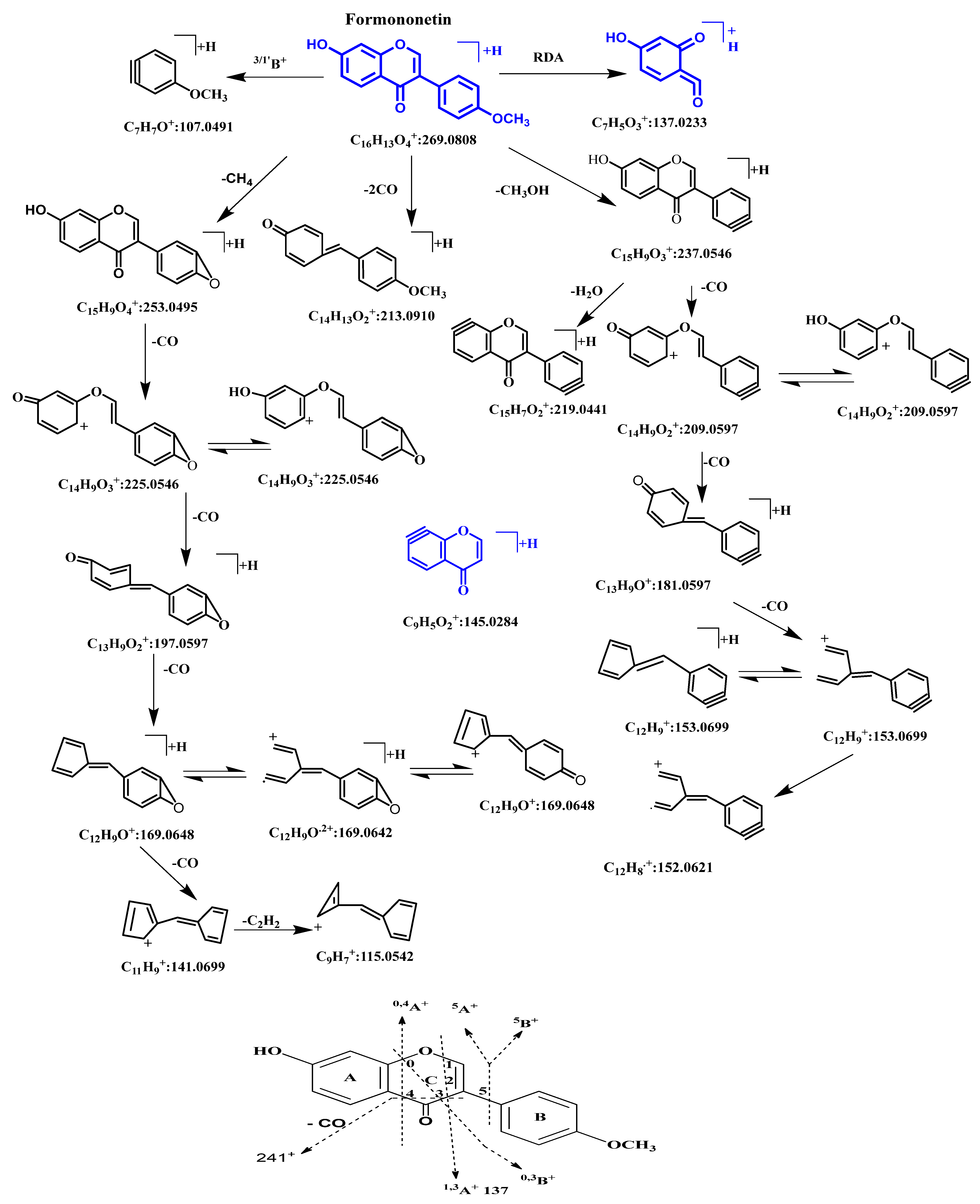
2.2. MSn Fragmentation Behavior of Formononetin
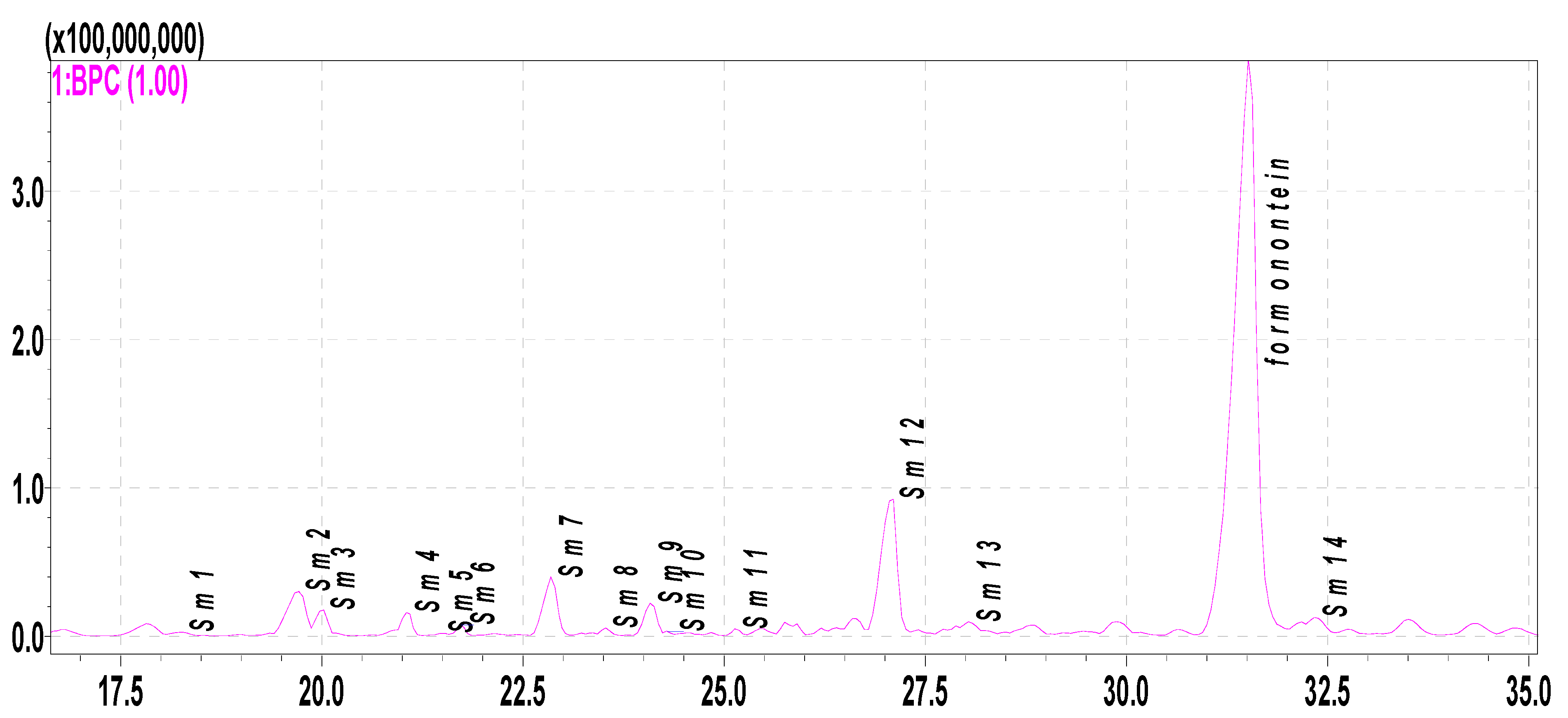
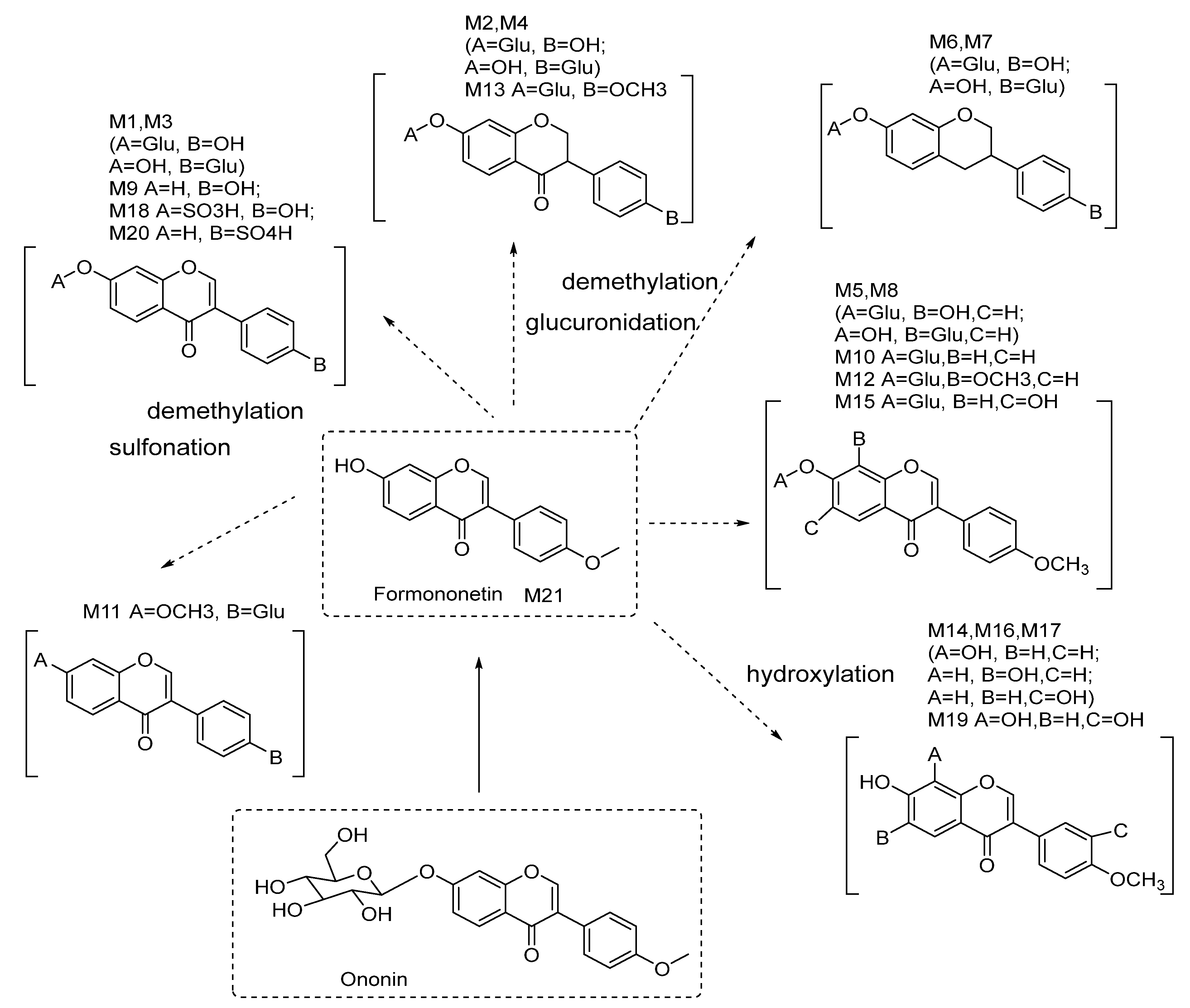
2.3. Metabolite Profiling of Formononetin Using Rat Hepatic S9 by HPLC-DAD-ESI-IT-TOF-MSn Analysis
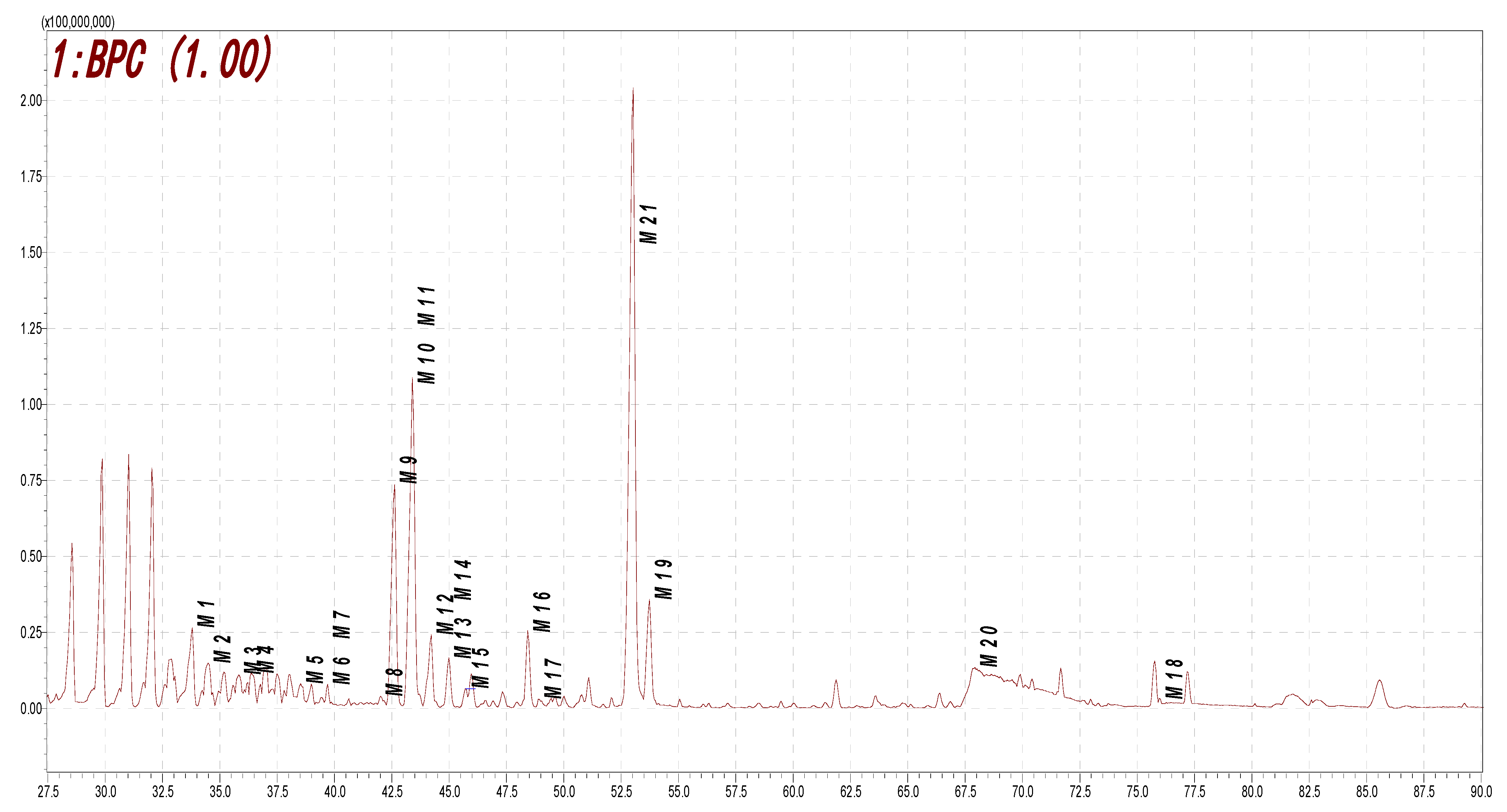
2.4. Metabolite Profiling of Rat Urine Samples after Oral Administration of Ononin via HPLC-DAD-ESI-IT-TOF-MSn Analysis
2.5. Results of Network Pharmacology and Macromolecular Docking
2.6. Discussion
3. Materials and Methods
3.1. Materials and Reagents
3.2. Ethical Approval of Animal Experiments
3.3. Preparation of the Rat Hepatic S9 Fraction
3.4. Rat Hepatic S9 Incubation for Metabolite Profiling
3.5. Enlarged Rat Hepatic S9 Incubation for Isolation of Metabolites
3.6. Rats’ Urine Samples Collected after Oral Administered with Ononin
3.7. The Instrument Parameters of HPLC-DAD-ESI-IT-TOF-MSn
3.8. Spectroscopic Methods
3.9. Research on Network Pharmacology and Macromolecular Docking
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fu, J.; Wang, Z.; Huang, L.; Zheng, S.; Wang, D.; Chen, S.; Zhang, H.; Yang, S. Review of the botanical characteristics, phytochemistry, and pharmacology of Astragalus membranaceus (Huangqi). Phytother. Res. 2014, 28, 1275–1283. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Sun, Y.; Huang, J.; Wang, B.; Gong, Y.; Fang, Y.; Liu, Y.; Wang, S.; Guo, Y. Anti-tumor effects and mechanisms of Astragalus membranaceus (AM) and its specific immunopotentiation: Status and prospect. J. Ethnopharmacol. 2020, 258, 112797. [Google Scholar] [CrossRef] [PubMed]
- Tay, K.C.; Tan, L.T.H.; Chan, C.K.; Hong, S.L.; Chan, K.G.; Yap, W.H.; Pusparajah, P.; Lee, L.-H.; Goh, B.H. Formononetin: A review of its anticancer potentials and mechanisms. Front. Pharmacol. 2019, 10, 820. [Google Scholar] [CrossRef] [PubMed]
- Dutra, J.M.; Espitia, P.J.; Batista, R.A. Formononetin: Biological effects and uses—A review. Food Chem. 2021, 359, 129975. [Google Scholar] [CrossRef] [PubMed]
- Mu, H.; Bai, Y.H.; Wang, S.T.; Zhu, Z.M.; Zhang, Y.W. Research on antioxidant effects and estrogenic effect of formononetin from Trifolium pratense (red clover). Phytomedicine 2009, 16, 314–319. [Google Scholar] [CrossRef]
- Dong, L.; Yin, L.; Zhang, Y.; Fu, X.; Lu, J. Anti-inflammatory effects of ononin on lipopolysaccharide-stimulated RAW 264.7 cells. Mol. Immunol. 2017, 83, 46–51. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, M.; Ahmed, M.; Surapaneni, K.M.; Veeraraghavan, V.P.; Arulselvan, P. Neuroprotective effects of ononin against the aluminium chloride-induced Alzheimer’s disease in rats. Saudi J. Biol. Sci. 2021, 28, 4232–4239. [Google Scholar] [CrossRef]
- Cao, J.; Li, T.; Liu, T.; Zheng, Y.; Liu, J.; Yang, Q.; Li, X.; Lu, W.; Wei, Y.; Li, W. A Study of the Mechanisms and Characteristics of Fluorescence Enhancement for the Detection of Formononetin and Ononin. Molecules 2023, 28, 1543. [Google Scholar] [CrossRef]
- Cayen, M.N.; Tang, G.; Common, R.H. Urinary conversion products of biochanin A and formononetin in the fowl. Biochim. Biophys. Acta Gen. Subj. 1965, 111, 349–357. [Google Scholar] [CrossRef]
- Shutt, D.A.; Braden, A.W.H. The significance of equol [7,4′-dihydroxyisoflavan] in relation to the estrogenic responses in sheep ingesting clover with a high formononetin content. Aust. J. Agr. Res. 1968, 19, 545–553. [Google Scholar] [CrossRef]
- Kulling, S.E.; Lehmann, L.; Metzler, M. Oxidative metabolism and genotoxic potential of major isoflavone phytoestrogens. J. Chromatogr. B 2002, 777, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Heinonen, S.; Waehaelae, K.; Adlercreutz, H. Identification of urinary metabolites of the red clover isoflavones formononetin and biochanin A in human subjects. J. Agric. Food Chem. 2004, 52, 6802–6809. [Google Scholar] [CrossRef] [PubMed]
- Jeong, E.J.; Jia, X.; Hu, M. Disposition of Formononetin via Enteric Recycling: Metabolism and Excretion in Mouse Intestinal Perfusion and Caco-2 Cell Models. Mol. Pharm. 2005, 2, 319–328. [Google Scholar] [CrossRef]
- Hendrich, S. Bioavailability of isoflavones. J. Chromatogr. B 2002, 777, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Cremoux, P.; Leclercq, G.; Jacquot, Y. Controversies concerning the use of phytoestrogens in menopause management: Bioavailability and metabolism. Maturitas 2010, 65, 334–339. [Google Scholar] [CrossRef] [PubMed]
- Hosoda, K.; Furuta, T.; Ishii, K. Simultaneous determination of glucuronic acid and sulfuric acid conjugated metabolites of daidzein and genistein in human plasma by high-performance liquid chromatography. J. Chromatogr. B 2010, 878, 628–636. [Google Scholar] [CrossRef]
- Wen, X.D.; Qi, L.W.; Li, B. Microsomal metabolism of calycosin, formononetin and drug–drug interactions by dynamic microdialysis sampling and HPLC–DAD–MS analysis. J. Pharmaceut. Biomed. 2009, 50, 100–105. [Google Scholar] [CrossRef]
- Wu, W.N.; Mckown, L.A.; Moyer, M.D.; Johannsen, T.B.; Takacs, A.R. In vitro metabolism of mifepristone (RU-486) in rat, monkey and human hepatic S9 fractions: Identification of three new mifepristone metabolites. Xenobiotica 1999, 29, 1089–1100. [Google Scholar] [CrossRef]
- Zhang, Y.Z.; Xu, F.; Dong, J.; Liang, J.; Hashi, Y.; Shang, M.Y.; Wang, X.; Cai, S.Q. Profiling the metabolites of astrapterocarpan in rat hepatic 9000 g supernatant. Chin. J. Nat. Med. 2019, 17, 842–857. [Google Scholar] [CrossRef]
- Li, S.; Zhang, B. Traditional Chinese medicine network pharmacology: Teory, methodology and application. Chin. J. Nat. Med. 2013, 11, 110–120. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissTargetPrediction: Updated data and new features for efcient prediction of protein targets of small molecules. Nucleic Acids Res. 2019, 47, 357–364. [Google Scholar] [CrossRef]
- Sherman, B.T.; Hao, M.; Qiu, J. A web server for functional enrichment analysis and functional annotation of gene list. Nucleic Acids Res. 2022, 23, 216–221. [Google Scholar] [CrossRef] [PubMed]
- Taylor, R.D.; Jewsbury, P.J.; Essex, J.W. A review of protein-small molecule docking methods. J. Comput.-Aided Mol. Des. 2002, 16, 151–166. [Google Scholar] [CrossRef] [PubMed]
- Patwardhan, B. Ethnopharmacology and drug discovery. J. Ethnopharmacol. 2005, 100, 50–52. [Google Scholar] [CrossRef] [PubMed]
- Dickinson, J.M.; Smith, G.R.; Randel, R.D.; Pemberton, I.J. In vitro metabolism of formononetin and biochanin A in bovine rumen fluid. J. Anim. Sci. 1988, 66, 1969–1973. [Google Scholar] [CrossRef]
- Chang, H.H.; Robinson, A.R.; Chan, A.H. Radioactive conversion products of intramuscularly injected [4-14C]-formononetin including sulfates in the urine of hens. Can. J. Biochem. 1977, 55, 50–55. [Google Scholar] [CrossRef]
- Nilsson, A.; Hill, J.L.; Davies, H.L. In vitro study of formononetin and biochanin A metabolism in rumen fluid from sheep. Biochim. Biophys. Acta. Gen. Subj. 1967, 148, 92–98. [Google Scholar] [CrossRef]
- Lindsay, D.R.; Kelly, R.W. Metabolism of phytoestrogens in sheep. Aust. Vet. J. 1970, 46, 219–222. [Google Scholar] [CrossRef]
- March, R.E.; Miao, X.S.; Metcalfe, C.D.; Stobiecki, M.; Marczak, L. A fragmentation study of an isoflavone glycoside, genistein-7-O-glucoside, using electrospray quadrupole time-of-flight mass spectrometry at high mass resolution. Int. J. Mass. Spectrom. 2004, 232, 171–183. [Google Scholar] [CrossRef]
- Heinonen, S.M.; Hoikkala, A.; Wähälä, K.; Adlercreutz, H. Metabolism of the soy isoflavones daidzein, genistein and glycitein in human subjects. Identification of new metabolites having an intact isoflavonoid skeleton. J. Steroid. Biochem. 2003, 87, 285–299. [Google Scholar] [CrossRef]
- Yu, J.; Wu, Q.; Qiao, S.L.; Yu, Z.P.; Jin, N.Z.; Yu, B.Y. Simultaneous determination of phytoestrogens and key metabolites in breast cancer patients’ urine by liquid chromatography–tandem mass spectrometry. J. Pharmaceut. Biomed. 2009, 50, 939–946. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.Z.; Xu, F.; Yi, T.; Zhang, J.Y.; Tang, Y.N.; Chen, H.B. Investigations of the fragmentation behavior of 11 isoflavones with ESI-IT-TOF-MSn. Chin. Pharm. J. Sci. 2014, 23, 631–641. [Google Scholar] [CrossRef]
- Zhang, Y.Z.; Xu, F.; Yi, T.; Zhang, J.Y.; Xu, J.; Tang, Y.N.; Chen, H.B. Chemical profile analysis and comparison of two versions of the classic TCM formula Danggui Buxue Tang by HPLC-DAD-ESI-IT-TOF-MSn. Molecules 2014, 19, 5650–5673. [Google Scholar] [CrossRef] [PubMed]
- Labow, R.S.; Layne, D.S. Formation of glucosides of isoflavones and of other phenols by rabbit liver microsomal fractions. Biochem. J. 1972, 128, 491–497. [Google Scholar] [CrossRef]
- Qi, L.W.; Yu, Q.T.; Li, P.; Li, S.L.; Wang, Y.X.; Sheng, L.H.; Yi, L. Quality evaluation of Radix Astragali through a simultaneous demermination of six major active isoflavonoids and four main saponins by high-performance liquid chromatography coupled with diode array and evaporative light scattering detectors. J. Chromatogr. A 2006, 1134, 162–169. [Google Scholar] [CrossRef] [PubMed]
- Li, H.F.; Li, T.; Yang, P.; Wang, Y.; Tang, X.J.; Liu, L.J.; Xu, F.; Cai, S.Q. Global profiling and structural characterization of metabolites of ononin using HPLC-ESI-IT-TOF-MSn after oral administration to rats. J. Agric. Food Chem. 2020, 68, 15164–15175. [Google Scholar] [CrossRef]
- Nestel, P.; Fujii, A.; Zhang, L. An isoflavone metabolite reduces arterial stiffness and blood pressure in overweight men and postmenopausal women. Atherosclerosis 2007, 192, 184–189. [Google Scholar] [CrossRef] [PubMed]
- Bednarek, P.; Frański, R.; Kerhoas, L.; Einhorn, J.; Wojtaszek, P. Profling changes in metabolism of isoflavonoids and their conjugates in Lupinus albus treated with biotic elicitor. Phytochemistry 2001, 56, 77–85. [Google Scholar] [CrossRef]
- Huh, J.E.; Seo, D.M.; Baek, Y.H.; Choi, D.Y.; Park, D.S.; Lee, J.D. Biphasic positive effect of formononetin on metabolic activity of human normal and osteoarthritic subchondral osteoblasts. Int. Immunopharmacol. 2010, 10, 500–507. [Google Scholar] [CrossRef]



| Formononetin | Sm7 | Sm5 | Sm10 | Sm9 | Sm2 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| δC | δH (J = Hz) | δC | δH (J = Hz) | δC | δH (J = Hz) | δC | δH (J = Hz) | δC | δH (J = Hz) | δC | δH (J = Hz) | |
| 1 | ||||||||||||
| 2 | 153.5 | 8.32 s | 153.3 | 8.30 s | 153.4 | 8.37 s | 153.1 | 8.27 s | 153.4 | 8.31 s | 152.7 | 8.22 s |
| 3 | 123.6 | 123.0 | 123.0 | 122.8 | 123.1 | 123.2 | ||||||
| 4 | 175.0 | 175.1 | 175.5 | 174.7 | 175.6 | 174.8 | ||||||
| 5 | 127.7 | 7.94d (9) | 127.7 | 7.94d (8) | 116.1 | 7.45d (8) | 108.5 | 7.37 s | 116.1 | 7.44d (8) | 108.6 | 7.36 s |
| 6 | 115.6 | 6.91dd (2,8) | 115.6 | 6.90dd (2,8) | 114.7 | 6.94d (8) | 145.1 | 9.77 s | 114.6 | 6.92d (8) | 145.1 | 9.48 s |
| 7 | 163.0 | 10.78 s | 162.9 | 10.76 s | 150.5 | 10.31 s | 151.3 | 10.41 s | 150.4 | 10.28 s | 151.3 | 10.38 s |
| 8 | 102.6 | 6.85d (2) | 102.5 | 6.85d (2) | 133.4 | 9.42 s | 103.2 | 6.89 s | 133.3 | 9.41 s | 103.2 | 6.87 s |
| 9 | 157.9 | 157.6 | 147.2 | 152.7 | 147.2 | 152.8 | ||||||
| 10 | 117.1 | 117.1 | 117.9 | 117.0 | 117.9 | 117.0 | ||||||
| 1′ | 124.7 | 123.9 | 124.8 | 125.0 | 123.4 | 123.4 | ||||||
| 2′ | 130.5 | 7.48d (8) | 130.5 | 7.35d (8) | 130.6 | 7.49d (8) | 130.5 | 7.47d (8) | 130.6 | 7.36d (8) | 130.5 | 7.34d (8) |
| 3′ | 114.1 | 6.96d (8) | 115.4 | 6.78 (8) | 114.0 | 6.96 (8) | 114.0 | 6.95 (8) | 115.4 | 6.78 (8) | 115.3 | 6.77d (8) |
| 4′ | 159.4 | 157.9 | 9.50 s | 159.4 | 159.3 | 157.6 | 9.50 s | 157.5 | 9.75 s | |||
| 5′ | 114.1 | 6.96d (8) | 115.4 | 6.78 (8) | 114.0 | 6.96 (8) | 114.0 | 6.95 (8) | 115.4 | 6.78 (8) | 115.3 | 6.77d (8) |
| 6′ | 130.5 | 7.48d (8) | 130.5 | 7.35d (8) | 130.6 | 7.49d (8) | 130.5 | 7.47d (8) | 130.6 | 7.36d (8) | 130.5 | 7.34d (8) |
| -OCH3 | 55.6 | 3.77 s | 55.6 | 3.77 s | 55.6 | 3.77 s | ||||||
| Metabolites | tR (min) | [M+H]+ (m/z) | [M−H]− (m/z) | Formula | Error (ppm) | Main Fragment in MS2 or MS3 | Metabolic Pathways |
|---|---|---|---|---|---|---|---|
| S0 | 31.205 | 269.0852 | 267.0669 | C16H12O4 | 4.49 | 269, 237, 213, 163, 118, 107 | Prototype (formononetin) |
| Sm1 | 18.568 | 447.1289 | 445.1134 | C22H22O10 | −0.45 | 447, 285(−162), 229, 152 | hydroxylation, glycosylation |
| Sm2 | 19.663 | 271.0597 | 269.0452 | C15H10O5 | −3.32 | 271, 253, 225, 215, 197, 169, 153 | hydroxylation, demethylation |
| Sm3 | 20.413 | 445.1115 | C22H22O10 | −4.49 | hydroxylation, glycosylation | ||
| Sm4 | 20.897 | 431.1337 | C22H22O9 | −1.16 | 431, 269(−162), 237, 213, 181, 152, 136 | glycosylation | |
| Sm5 | 21.483 | 301.0699 | 299.0560 | C16H12O6 | 1.34 | 301, 286, 269, 241, 229, 153 | dihydroxylation |
| Sm6 | 22.458 | 417.1179 | 415.1011 | C21H20O9 | −4.34 | 417, 285(−132), 268, 152, 124 | hydroxylation, glycosylation (pentose) |
| Sm7 | 22.847 | 255.0659 | 253.0493 | C15H10O4 | 0.78 | 255, 227, 199, 137, 152, 109 | demethylation |
| Sm8 | 23.227 | 417.1188 | 415.1025 | C21H20O9 | −0.96 | 417, 285(−132), 229, 152, | hydroxylation, glycosylation (pentose) |
| Sm9 | 23.425 | 271.0606 | C15H10O5 | −3.32 | 271, 253, 225, 215, 197, 169, 153 | hydroxylation, demethylation | |
| Sm10 | 24.080 | 285.0755 | 283.0599 | C16H12O5 | −2.81 | 285, 253, 225, 197, 141 | hydroxylation |
| Sm11 | 25.132 | 417.1193 | 415.1055 | C21H20O9 | 1.68 | 417, 285(−132), 253, 152, 123 | hydroxylation, glycosylation (pentose) |
| Sm12 | 27.053 | 285.0811 | 283.0611 | C16H12O5 | 1.77 | 285, 211, 183, 152 | hydroxylation |
| Sm13 | 27.518 | 285.0756 | 283.0604 | C16H12O5 | −2.46 | 285, 270, 229, 211, 197, 183, 152 | hydroxylation |
| Sm14 | 32.385 | 299.385 | C17H14O5 | 0.33 | 299, 270, 254, 237, 213, 181 | hydrolation, methylation |
| Metabolites | tR (min) | [M+H]+ (m/z) | [M−H]− (m/z) | Formula | Error (ppm) | Main Fragment in MS2 or MS3 | Metabolic Pathways |
|---|---|---|---|---|---|---|---|
| M1 | 33.707 | 431.0954 | 429.0815 | C21H18O10 | −1.63 | 429, 253(−176), 224, 208, 175, 135 | demethylation, glucuronidation |
| M2 | 34.397 | 431.0982 | C21H20O10 | 0.93 | 431, 255(−176), 175, 149 | hydrogenation, demethylation, glucuronidation | |
| M3 | 35.872 | 429.0833 | C21H18O10 | 2.56 | 429, 253(−176), 224, 175 | demethylation, glucuronidation | |
| M4 | 36.372 | 431.0967 | C21H20O10 | −2.55 | hydrogenation, demethylation, glucuronidation | ||
| M5 | 38.525 | 459.0938 | C22H20O11 | 2.40 | 459, 283(−176), 268 | hydroxylation, glucuronidation | |
| M6 | 39.447 | 417.1188 | C21H22O9 | 0.48 | hydrogenation, carbonyl reduction, glucuronidation | ||
| M7 | 39.705 | 417.1181 | C21H22O9 | −1.20 | hydrogenation, carbonyl reduction, glucuronidation | ||
| M8 | 42.007 | 461.1097 | 459.0923 | C22H20O11 | 2.82 | 461, 285(−176), 270, 152, 123 | hydroxylation, glucuronidation |
| M9 | 42.627 | 255.0668 | 253.0525 | C15H10O4 | 4.31 | 255, 199, 152, 137 | demethylation |
| M10 | 43.143 | 445.1127 | 443.0983 | C22H20O10 | −1.80 | 445, 269(−176), 237, 118 | glucuronidation |
| M11 | 43.445 | 445.1114 | 443.0988 | C22H20O10 | −4.72 | 443, 267(−176), 152, 175 | glucuronidation |
| M12 | 43.703 | 475.1224 | 473.1077 | C23H22O11 | −3.37 | methylation, hydroxylation, glucuronidation | |
| M13 | 44.168 | 447.1294 | 445.1158 | C22H22O10 | 0.67 | 445, 269(−176), 254, 175, 135 | hydrogenation, demethylation, glucuronidation |
| M14 | 44.988 | 285.0778 | 283.0611 | C16H12O5 | 5.26 | 283, 268, 224, 131 | hydroxylation |
| M15 | 45.652 | 461.1067 | 459.0924 | C22H20O11 | −0.65 | 461, 285(−176) | hydroxylation, glucuronidation |
| M16 | 48.427 | 285.0760 | 283.0617 | C16H12O5 | −1.05 | 283, 268, 224 | hydroxylation |
| M17 | 48.935 | 285.0779 | C16H12O5 | 5.61 | 285, 241 | hydroxylation | |
| M18 | 53.890 | 299.0905 | C 17H14O5 | −4.68 | 299, 284, 243, 166, 137 | hydroxylation, methylation | |
| M19 | 67.932 | 335.0203 | 333.0083 | C15H10O7S | 1.19 | 333, 253(−80), 224, 135 | demethylation, sulfonation |
| M20 | 76.412 | 335.0229 | 333.0092 | C15H10O7S | 4.20 | 333, 253(−80), 225, 211, 135 | demethylation, sulfonation |
| M21 | 52.977 | 269.0847 | 267.0654 | C16H12O4 | −1.12 | formononetin |
| No. | Names of the Receptors | Betweenness |
|---|---|---|
| 1 | HSP90AA1 | 4195.108 |
| 2 | SRC | 3675.1655 |
| 3 | ESR1 | 2125.8835 |
| 4 | HSPA8 | 1557.0778 |
| 5 | APP | 1246.3787 |
| 6 | AHR | 1209.1862 |
| 7 | EGFR | 1152.9003 |
| 8 | PLG | 1064.3462 |
| 9 | CYP1B1 | 1034.5195 |
| 10 | HDAC1 | 950.46075 |
| Six Isolated Metabolites | Binding Energy/kcal.mol−1 | |
|---|---|---|
| HSP90AA1 | SRC | |
| 6,7,4′-trihydroxy-isoflavonoid (Sm2) | −4.04 | −4.38 |
| 7,4′-dihydroxy-isoflavonoid (Sm7) | −4.76 | −5.01 |
| 7,8,4′-trihydroxy-isoflavonoid (Sm9) | −4.18 | −4.5 |
| 7,8-dihydroxy-4′-methoxy-isoflavonoid (Sm10) | −5.4 | −4.06 |
| 6,7-dihydroxy-4′-methoxy-isoflavonoid (Sm12) | −5.16 | −4.89 |
| Formononetin (prototype) | −5.37 | −4.37 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Y.-Z.; Wang, T.; Chen, Q.-L.; Chen, H.-B.; He, Q.-S.; Zhang, Y.-Z. Identification of the Metabolites of Both Formononetin in Rat Hepatic S9 and Ononin in Rat Urine Samples and Preliminary Network Pharmacology Evaluation of Their Main Metabolites. Molecules 2023, 28, 7451. https://doi.org/10.3390/molecules28217451
Yang Y-Z, Wang T, Chen Q-L, Chen H-B, He Q-S, Zhang Y-Z. Identification of the Metabolites of Both Formononetin in Rat Hepatic S9 and Ononin in Rat Urine Samples and Preliminary Network Pharmacology Evaluation of Their Main Metabolites. Molecules. 2023; 28(21):7451. https://doi.org/10.3390/molecules28217451
Chicago/Turabian StyleYang, Yu-Zhu, Tao Wang, Qi-Lei Chen, Hu-Biao Chen, Qian-Song He, and Ya-Zhou Zhang. 2023. "Identification of the Metabolites of Both Formononetin in Rat Hepatic S9 and Ononin in Rat Urine Samples and Preliminary Network Pharmacology Evaluation of Their Main Metabolites" Molecules 28, no. 21: 7451. https://doi.org/10.3390/molecules28217451




