miRNAs as Biomolecular Markers for Food Safety, Quality, and Traceability in Poultry Meat—A Preliminary Study
Abstract
:1. Introduction
2. Results
2.1. Validation of the Reference Gene to Measure miRNA Expression
2.2. Analysis and Quantification of the Presence of Two Specific miRNAs
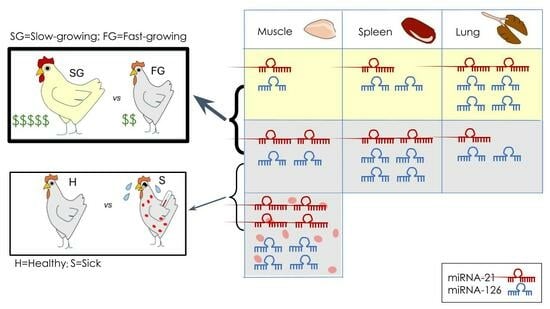
2.3. Determination of Tissue Specificity for miRNA Biomarkers
2.4. Expression of miRNAs in Healthy and Diseased R308 Broilers
3. Discussion
4. Materials and Methods
4.1. Animals and Tissues
4.2. miRNA Extractions
4.3. Quality Control small RNA
4.4. Reverse Transcription Quantitative Real-Time PCR (RT-qPCR)
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Petracci, M.; Mudalal, S.; Soglia, F.; Cavani, C. Meat Quality in Fast-Growing Broiler Chickens. World’s Poult. Sci. J. 2015, 71, 363–374. [Google Scholar] [CrossRef]
- UNAITALIA. Global and National Meat Consumption Data. 2021. Available online: https://www.unaitalia.com/filiera-avicola/ (accessed on 9 December 2023).
- Whitton, C.; Bogueva, D.; Marinova, D.; Phillips, C.J.C. Are We Approaching Peak Meat Consumption? Analysis of Meat Consumption from 2000 to 2019 in 35 Countries and Its Relationship to Gross Domestic Product. Animals 2021, 11, 3466. [Google Scholar] [CrossRef]
- Havenstein, G.; Ferket, P.; Qureshi, M. Growth, Livability, and Feed Conversion of 1957 versus 2001 Broilers When Fed Representative 1957 and 2001 Broiler Diets. Poult. Sci. 2003, 82, 1500–1508. [Google Scholar] [CrossRef] [PubMed]
- Welfare Quality R © Consortium, Lelystad N. Welfare Quality Protocols. 2009. Available online: http://www.welfarequalitynetwork.net/en-us/reports/assessment-protocols/ (accessed on 20 January 2024).
- Miretti, S.; Lecchi, C.; Ceciliani, F.; Baratta, M. MicroRNAs as Biomarkers for Animal Health and Welfare in Livestock. Front. Vet. Sci. 2020, 7, 578193. [Google Scholar] [CrossRef] [PubMed]
- Zootecnica, 2020: Increased Interest in the Slower-Growing Broiler Ranger Gold. Available online: https://zootecnicainternational.com/featured/increased-interest-slower-growing-broiler-Ranger-gold/ (accessed on 9 December 2023).
- Louton, H.; Keppler, C.; Erhard, M.; van Tuijl, O.; Bachmeier, J.; Damme, K.; Reese, S.; Rauch, E. Animal-Based Welfare Indicators of 4 Slow-Growing Broiler Genotypes for the Approval in an Animal Welfare Label Program. Poult. Sci. 2019, 98, 2326–2337. [Google Scholar] [CrossRef]
- Kestin, S.C.; Gordon, S.; Su, G.; Sørensen, P. Relationships in Broiler Chickens between Lameness, Liveweight, Growth Rate and Age. Vet. Rec. 2001, 148, 195–197. [Google Scholar] [CrossRef]
- Fanatico, A.C.; Pillai, P.B.; Hester, P.Y.; Falcone, C.; Mench, J.A.; Owens, C.M.; Emmert, J.L. Performance, Livability, and Carcass Yield of Slow- and Fast-Growing Chicken Genotypes Fed Low-Nutrient or Standard Diets and Raised Indoors or with Outdoor Access. Poult. Sci. 2008, 87, 1012–1021. [Google Scholar] [CrossRef]
- Kjaer, J.B.; Su, G.; Nielsen, B.L.; Sørensen, P. Foot Pad Dermatitis and Hock Burn in Broiler Chickens and Degree of Inheritance. Poult. Sci. 2006, 85, 1342–1348. [Google Scholar] [CrossRef] [PubMed]
- Bessei, W. Welfare of Broilers: A Review. World’s Poult. Sci. J. 2006, 62, 455–466. [Google Scholar] [CrossRef]
- Fleming, D.S.; Miller, L.C. Differentially Expressed MiRNAs and TRNA Genes Affect Host Homeostasis during Highly Pathogenic Porcine Reproductive and Respiratory Syndrome Virus Infections in Young Pigs. Front. Genet. 2019, 10, 691. [Google Scholar] [CrossRef]
- Wright, K.; Plain, K.; Purdie, A.; Saunders, B.M.; de Silva, K. Biomarkers for Detecting Resilience against Mycobacterial Disease in Animals. Infect. Immun. 2019, 88, e00401-19. [Google Scholar] [CrossRef]
- Miretti, S.; Volpe, M.G.; Martignani, E.; Accornero, P.; Baratta, M. Temporal Correlation between Differentiation Factor Expression and MicroRNAs in Holstein Bovine Skeletal Muscle. Animal 2017, 11, 227–235. [Google Scholar] [CrossRef]
- Duan, X.; Wang, L.; Sun, G.; Yan, W.; Yang, Y. Understanding the Cross-Talk between Host and Virus in Poultry from the Perspectives of MicroRNA. Poult. Sci. 2020, 99, 1838–1846. [Google Scholar] [CrossRef]
- Zhang, Y.; Wan, N.; Pan, T.; Hu, X.; Liu, Q.; Li, S. MicroRNA-33-3p Regulates Vein Endothelial Cell Apoptosis in Selenium-Deficient Broilers by Targeting E4F1. Oxidative Med. Cell. Longev. 2019, 2019, 6274010. [Google Scholar] [CrossRef]
- Rani, P.; Yenuganti, V.R.; Shandilya, S.; Onteru, S.K.; Singh, D. MiRNAs: The Hidden Bioactive Component of Milk. Trends Food Sci. Technol. 2017, 65, 94–102. [Google Scholar] [CrossRef]
- Chen, X.; Gao, C.; Li, H.; Huang, L.; Sun, Q.; Dong, Y.; Tian, C.; Gao, S.; Dong, H.; Guan, D.; et al. Identification and Characterization of MicroRNAs in Raw Milk during Different Periods of Lactation, Commercial Fluid, and Powdered Milk Products. Cell Res. 2010, 20, 1128–1137. [Google Scholar] [CrossRef] [PubMed]
- Medina, S.; Pereira, J.A.; Silva, P.; Perestrelo, R.; Câmara, J.S. Food Fingerprints—A Valuable Tool to Monitor Food Authenticity and Safety. Food Chem. 2019, 278, 144–162. [Google Scholar] [CrossRef]
- Valdés, A.; Álvarez-Rivera, G.; Socas-Rodríguez, B.; Herrero, M.; Ibáñez, E.; Cifuentes, A. Foodomics: Analytical Opportunities and Challenges. Anal. Chem. 2022, 94, 366–381. [Google Scholar] [CrossRef]
- Vishnuraj, M.R.; Devatkal, S.; Vaithiyanathan, S.; Uday Kumar, R.; Mendiratta, S.K. Development and Validation of MiRNA Based Method for Rapid Identification of Offal Meats in Processed Chicken Meat Products. Food Control 2021, 121, 107593. [Google Scholar] [CrossRef]
- Grundy, H.H.; Brown, L.C.; Romero, M.R.; Donarski, J.A. Review: Methods to Determine Offal Adulteration in Meat Products to Support Enforcement and Food Security. Food Chem. 2023, 399, 133818. [Google Scholar] [CrossRef]
- Selcuklu, S.D.; Donoghue, M.T.A.; Spillane, C. MiR-21 as a Key Regulator of Oncogenic Processes. Biochem. Soc. Trans. 2009, 37, 918–925. [Google Scholar] [CrossRef]
- Zhang, D.; Ran, J.; Li, J.; Yu, C.; Cui, Z.; Amevor, F.K.; Wang, Y.; Jiang, X.; Qiu, M.; Du, H.; et al. MiR-21-5p Regulates the Proliferation and Differentiation of Skeletal Muscle Satellite Cells by Targeting KLF3 in Chicken. Genes 2021, 12, 814. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, J.; Gou, D.; Turaka, P.; Viktorova, E.; Ramchandran, R.; Raj, J.U. MicroRNA-21 Plays a Role in Hypoxia-Mediated Pulmonary Artery Smooth Muscle Cell Proliferation and Migration. Am. J. Physiol.-Lung Cell. Mol. Physiol. 2010, 299, L861–L871. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.-B.; Li, X.; Li, Z.-Y.; Zhao, J.; Yuan, X.-B.; Ren, Y.; Cui, Z.-D.; Liu, Y.-D.; Yang, X.-J. MicroRNA-21 Promotes Osteogenic Differentiation of Mesenchymal Stem Cells by the PI3K/β-Catenin Pathway: MiR-21, OSTEOGENESIS, PI3K/AKT/β-CATENIN. J. Orthop. Res. 2015, 33, 957–964. [Google Scholar] [CrossRef] [PubMed]
- Novellino, L.; Rossi, R.L.; Bonino, F.; Cavallone, D.; Abrignani, S.; Pagani, M.; Brunetto, M.R. Circulating Hepatitis B Surface Antigen Particles Carry Hepatocellular MicroRNAs. PLoS ONE 2012, 7, e31952. [Google Scholar] [CrossRef]
- Long, G.; Wang, F.; Duan, Q.; Chen, F.; Yang, S.; Gong, W.; Wang, Y.; Chen, C.; Wang, D.W. Human Circulating MicroRNA-1 and MicroRNA-126 as Potential Novel Indicators for Acute Myocardial Infarction. Int. J. Biol. Sci. 2012, 8, 811–818. [Google Scholar] [CrossRef]
- Guan, P.; Yin, Z.; Li, X.; Wu, W.; Zhou, B. Meta-Analysis of Human Lung Cancer MicroRNA Expression Profiling Studies Comparing Cancer Tissues with Normal Tissues. J. Exp. Clin. Cancer Res. 2012, 31, 54. [Google Scholar] [CrossRef]
- Grabher, C.; Payne, E.M.; Johnston, A.B.; Bolli, N.; Lechman, E.; Dick, J.E.; Kanki, J.P.; Look, A.T. Zebrafish MicroRNA-126 Determines Hematopoietic Cell Fate through c-Myb. Leukemia 2011, 25, 506–514. [Google Scholar] [CrossRef]
- Wang, S.; Aurora, A.B.; Johnson, B.A.; Qi, X.; McAnally, J.; Hill, J.A.; Richardson, J.A.; Bassel-Duby, R.; Olson, E.N. The Endothelial-Specific MicroRNA MiR-126 Governs Vascular Integrity and Angiogenesis. Dev. Cell 2008, 15, 261–271. [Google Scholar] [CrossRef]
- Wang, X.-G.; Shao, F.; Wang, H.-J.; Yang, L.; Yu, J.-F.; Gong, D.-Q.; Gu, Z.-L. MicroRNA-126 Expression Is Decreased in Cultured Primary Chicken Hepatocytes and Targets the Sprouty-Related EVH1 Domain Containing 1 MRNA. Poult. Sci. 2013, 92, 1888–1896. [Google Scholar] [CrossRef]
- Yanaihara, N.; Caplen, N.; Bowman, E.; Seike, M.; Kumamoto, K.; Yi, M.; Stephens, R.M.; Okamoto, A.; Yokota, J.; Tanaka, T.; et al. Unique MicroRNA Molecular Profiles in Lung Cancer Diagnosis and Prognosis. Cancer Cell 2006, 9, 189–198. [Google Scholar] [CrossRef]
- Valente, V.; Teixeira, S.A.; Neder, L.; Okamoto, O.K.; Oba-Shinjo, S.M.; Marie, S.K.; Scrideli, C.A.; Paçó-Larson, M.L.; Carlotti, C.G., Jr. Selection of Suitable Housekeeping Genes for Expression Analysis in Glioblastoma Using Quantitative RT-PCR. ANS 2014, 21, 17. [Google Scholar] [CrossRef] [PubMed]
- Lim, Q.E.; Zhou, L.; Ho, Y.K.; Wan, G.; Too, H.P. SnoU6 and 5S RNAs Are Not Reliable MiRNA Reference Genes in Neuronal Differentiation. Neuroscience 2011, 199, 32–43. [Google Scholar] [CrossRef]
- Schwarzenbach, H.; da Silva, A.M.; Calin, G.; Pantel, K. Data Normalization Strategies for MicroRNA Quantification. Clin. Chem. 2015, 61, 1333–1342. [Google Scholar] [CrossRef] [PubMed]
- Meyer, S.U.; Pfaffl, M.W.; Ulbrich, S.E. Normalization Strategies for MicroRNA Profiling Experiments: A ‘Normal’ Way to a Hidden Layer of Complexity? Biotechnol. Lett. 2010, 32, 1777–1788. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Muñoz, J.J.; Anauate, A.C.; Amaral, A.G.; Ferreira, F.M.; Meca, R.; Ormanji, M.S.; Boim, M.A.; Onuchic, L.F.; Heilberg, I.P. Identification of Housekeeping Genes for MicroRNA Expression Analysis in Kidney Tissues of Pkd1 Deficient Mouse Models. Sci. Rep. 2020, 10, 231. [Google Scholar] [CrossRef]
- Lamba, V.; Ghodke-Puranik, Y.; Guan, W.; Lamba, J.K. Identification of Suitable Reference Genes for Hepatic MicroRNA Quantitation. BMC Res. Notes 2014, 7, 129. [Google Scholar] [CrossRef] [PubMed]
- Zou, X.; Wang, J.; Qu, H.; Lv, X.H.; Shu, D.M.; Wang, Y.; Ji, J.; He, Y.H.; Luo, C.L.; Liu, D.W. Comprehensive Analysis of MiRNAs, LncRNAs, and MRNAs Reveals Potential Players of Sexually Dimorphic and Left-Right Asymmetry in Chicken Gonad during Gonadal Differentiation. Poult. Sci. 2020, 99, 2696–2707. [Google Scholar] [CrossRef]
- Warnefors, M.; Mössinger, K.; Halbert, J.; Studer, T.; VandeBerg, J.L.; Lindgren, I.; Fallahshahroudi, A.; Jensen, P.; Kaessmann, H. Sex-Biased MicroRNA Expression in Mammals and Birds Reveals Underlying Regulatory Mechanisms and a Role in Dosage Compensation. Genome Res. 2017, 27, 1961–1973. [Google Scholar] [CrossRef]
- Sharma, S.; Eghbali, M. Influence of Sex Differences on MicroRNA Gene Regulation in Disease. Biol. Sex Differ. 2014, 5, 3. [Google Scholar] [CrossRef]
- Nikolic, I.; Plate, K.-H.; Schmidt, M.H. EGFL7 Meets MiRNA-126: An Angiogenesis Alliance. J. Angiogenesis Res. 2010, 2, 9. [Google Scholar] [CrossRef] [PubMed]
- Liao, L.; Yao, Z.; Kong, J.; Zhang, X.; Li, H.; Chen, W.; Xie, Q. Exploring the Role of MiRNAs in Early Chicken Embryonic Development and Their Significance. Poult. Sci. 2023, 102, 103105. [Google Scholar] [CrossRef] [PubMed]
- Darnell, D.K.; Kaur, S.; Stanislaw, S.; Konieczka, J.K.; Yatskievych, T.A.; Antin, P.B. MicroRNA Expression during Chick Embryo Development. Dev. Dyn. 2006, 235, 3156–3165. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Cheng, Y.; Wang, L.; Sun, A.; Lin, Z.; Zhu, W.; Wang, Z.; Ma, J.; Wang, H.; Yan, Y.; et al. Chicken MiR-126-5p Negatively Regulates Antiviral Innate Immunity by Targeting TRAF3. Vet. Res. 2022, 53, 82. [Google Scholar] [CrossRef] [PubMed]
- Ishizaki, T.; Tamiya, T.; Taniguchi, K.; Morita, R.; Kato, R.; Okamoto, F.; Saeki, K.; Nomura, M.; Nojima, Y.; Yoshimura, A. MiR126 Positively Regulates Mast Cell Proliferation and Cytokine Production through Suppressing Spred1: MiR126 and Spred1 in Mast Cells. Genes Cells 2011, 16, 803–814. [Google Scholar] [CrossRef] [PubMed]
- Rivas, D.A.; Lessard, S.J.; Rice, N.P.; Lustgarten, M.S.; So, K.; Goodyear, L.J.; Parnell, L.D.; Fielding, R.A. Diminished Skeletal Muscle MicroRNA Expression with Aging Is Associated with Attenuated Muscle Plasticity and Inhibition of IGF-1 Signaling. FASEB J. 2014, 28, 4133–4147. [Google Scholar] [CrossRef] [PubMed]
- Khatri, B.; Seo, D.; Shouse, S.; Pan, J.H.; Hudson, N.J.; Kim, J.K.; Bottje, W.; Kong, B.C. MicroRNA Profiling Associated with Muscle Growth in Modern Broilers Compared to an Unselected Chicken Breed. BMC Genom. 2018, 19, 683. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Riart, B.; Lorda-Diez, C.I.; Marin-Llera, J.C.; Garcia-Porrero, J.A.; Hurle, J.M.; Montero, J.A. Interdigital Tissue Remodelling in the Embryonic Limb Involves Dynamic Regulation of the miRNA Profiles. J. Anat. 2017, 231, 275–286. [Google Scholar] [CrossRef]
- Thum, T.; Gross, C.; Fiedler, J.; Fischer, T.; Kissler, S.; Bussen, M.; Galuppo, P.; Just, S.; Rottbauer, W.; Frantz, S.; et al. MicroRNA-21 Contributes to Myocardial Disease by Stimulating MAP Kinase Signaling in Fibroblasts. Nature 2008, 456, 980–984. [Google Scholar] [CrossRef]
- Papagiannakopoulos, T.; Shapiro, A.; Kosik, K.S. MicroRNA-21 Targets a Network of Key Tumor-Suppressive Pathways in Glioblastoma Cells. Cancer Res. 2008, 68, 8164–8172. [Google Scholar] [CrossRef]
- Ouyang, H.; Yu, J.; Chen, X.; Wang, Z.; Nie, Q. A Novel Transcript of MEF2D Promotes Myoblast Differentiation and Its Variations Associated with Growth Traits in Chicken. PeerJ 2020, 8, e8351. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Li, H.; Mu, H.; Luo, W.; Li, Y.; Jia, X.; Wang, S.; Jia, X.; Nie, Q.; Li, Y.; et al. Let-7b Regulates the Expression of the Growth Hormone Receptor Gene in Deletion-Type Dwarf Chickens. BMC Genom. 2012, 13, 306. [Google Scholar] [CrossRef]
- Kang, L.; Cui, X.; Zhang, Y.; Yang, C.; Jiang, Y. Identification of MiRNAs Associated with Sexual Maturity in Chicken Ovary by Illumina Small RNA Deep Sequencing. BMC Genom. 2013, 14, 352. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Gan, H.; Zhang, H.; Tang, W.; Sun, Y.; Tang, X.; Kong, D.; Zhou, J.; Wang, Y.; Zhu, Y. MicroRNA-21 Inhibits SMAD7 Expression through a Target Sequence in the 3′ Untranslated Region and Inhibits Proliferation of Renal Tubular Epithelial Cells. Mol. Med. Rep. 2014, 10, 707–712. [Google Scholar] [CrossRef]
- Ouyang, H.; He, X.; Li, G.; Xu, H.; Jia, X.; Nie, Q.; Zhang, X. Deep Sequencing Analysis of MiRNA Expression in Breast Muscle of Fast-Growing and Slow-Growing Broilers. Int. J. Mol. Sci. 2015, 16, 16242–16262. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]



| ID | Breed | Average Weight | Sex | Tissue | Notes | RNA Extraction Date | RNA Concentration (ng/μL) | OD 260/280 |
|---|---|---|---|---|---|---|---|---|
| FIRST ROUND SAMPLING * | ||||||||
| R308_1 | Ross 308 | unk | unk | Muscle (breast) | n/a | 14 July 2021 | 230.04 | 2.13 |
| R308_1 | Ross 308 | unk | unk | Liver | n/a | 20 July 2021 | 141.74 | 2.14 |
| R308_1 | Ross 308 | unk | unk | Spleen | n/a | 21 July 2021 | 516.37 | 2.04 |
| R308_1 | Ross 308 | unk | unk | Kidney | n/a | 22 July 2021 | 1303.69 | 2.14 |
| R308_1 | Ross 308 | unk | unk | Lung | n/a | 27 July 2021 | 1764.78 | 2.12 |
| RG_1 | Ranger Gold | unk | unk | Muscle (breast) | n/a | 16 July 2021 | 372.76 | 2.16 |
| RG_1 | Ranger Gold | unk | unk | Liver | n/a | 20 July 2021 | 1573.03 | 2.17 |
| RG_1 | Ranger Gold | unk | unk | Spleen | n/a | 21 July 2021 | 825.62 | 2.16 |
| RG_1 | Ranger Gold | unk | unk | Kidney | n/a | 22 July 2021 | 962.51 | 2.14 |
| RG_1 | Ranger Gold | unk | unk | Lung | n/a | 22 July 2021 | 567.03 | 2.09 |
| SECOND ROUND SAMPLING ** | ||||||||
| R308_1-B2 | Ross 308 | 3.56 | male | Muscle (breast) | hepatic steatosis | 26 April 2023 | 220.77 | 2 |
| R308_1-L2 | Ross 308 | 3.56 | male | Lung | hepatic steatosis | 26 April 2023 | 198.83 | 2.07 |
| R308_1-S2 | Ross 308 | 3.56 | male | Spleen | hepatic steatosis | 29 April 2023 | 188.17 | 2 |
| R308_2-B1 | Ross 308 | 3.56 | female | Muscle (breast) | n/a | 20 March 2023 | 222.33 | 2.11 |
| R308_2-L1 | Ross 308 | 3.56 | female | Lung | n/a | 20 March 2023 | 166.05 | 2.01 |
| R308_2-S1 | Ross 308 | 3.56 | female | Spleen | n/a | 20 March 2023 | 234.68 | 2.05 |
| R308_3-B1 | Ross 308 | 3.56 | male | Muscle (breast) | enlarged spleen | 20 March 2023 | 122.03 | 2.26 |
| R308_3-L1 | Ross 308 | 3.56 | male | Lung | enlarged spleen | 20 March 2023 | 228.21 | 2.01 |
| R308_3-S2 | Ross 308 | 3.56 | male | Spleen | enlarged spleen | 29 March 2023 | 306.64 | 2 |
| R308_4-B1 | Ross 308 | 3.56 | male | Muscle (breast) | airsacculitis | 21 March 2023 | 161.47 | 2.12 |
| R308_4-L1 | Ross 308 | 3.56 | male | Lung | airsacculitis | 20 March 2023 | 201.68 | 2.04 |
| R308_4-S1 | Ross 308 | 3.56 | male | Spleen | airsacculitis | 21 March 2023 | 202.3 | 2.07 |
| R308_5-B1 | Ross 308 | 3.56 | male | Muscle (breast) | airsacculitis | 21 March 2023 | 235.38 | 2.11 |
| R308_5-L1 | Ross 308 | 3.56 | male | Lung | airsacculitis | 21 March 2023 | 145.54 | 2.05 |
| R308_5-S1 | Ross 308 | 3.56 | male | Spleen | airsacculitis | 21 March 2023 | 236.91 | 2.03 |
| R308_6-B1 | Ross 308 | 3.56 | female | Muscle (breast) | n/a | 21 March 2023 | 138.56 | 2.13 |
| R308_6-L1 | Ross 308 | 3.56 | female | Lung | n/a | 21 March 2023 | 132.3 | 2.1 |
| R308_6-S1 | Ross 308 | 3.56 | female | Spleen | n/a | 21 March 2023 | 253.44 | 2.04 |
| RG_1-B1 | Ranger Gold | 3.14 | female | Muscle (breast) | n/a | 22 March 2023 | 175.81 | 2.08 |
| RG_1-L1 | Ranger Gold | 3.14 | female | Lung | n/a | 21 March 2023 | 152.04 | 2 |
| RG_1-S1 | Ranger Gold | 3.14 | female | Spleen | n/a | 21 March 2023 | 302.63 | 2.04 |
| RG_2-B1 | Ranger Gold | 3.14 | female | Muscle (breast) | n/a | 22 March 2023 | 165.86 | 2.09 |
| RG_2-L1 | Ranger Gold | 3.14 | female | Lung | n/a | 22 March 2023 | 159.73 | 2.03 |
| RG_2-S1 | Ranger Gold | 3.14 | female | Spleen | n/a | 22 March 2023 | 182.07 | 2.09 |
| RG_3-B1 | Ranger Gold | 3.14 | female | Muscle (breast) | n/a | 23 March 2023 | 151.64 | 2.22 |
| RG_3-L1 | Ranger Gold | 3.14 | female | Lung | n/a | 22 March 2023 | 166.03 | 2.04 |
| RG_3-S1 | Ranger Gold | 3.14 | female | Spleen | n/a | 23 March 2023 | 212.33 | 2.01 |
| RG_4-B1 | Ranger Gold | 3.14 | female | Muscle (breast) | n/a | 23 March 2023 | 214.42 | 2.13 |
| RG_4-L1 | Ranger Gold | 3.14 | female | Lung | n/a | 23 March 2023 | 233.54 | 2 |
| RG_4-S1 | Ranger Gold | 3.14 | female | Spleen | n/a | 23 March 2023 | 154.55 | 2.04 |
| RG_5-B1 | Ranger Gold | 3.14 | female | Muscle (breast) | n/a | 24 March 2023 | 146.81 | 2.17 |
| RG_5-L1 | Ranger Gold | 3.14 | female | Lung | n/a | 23 March 2023 | 177.1 | 2.05 |
| RG_5-S1 | Ranger Gold | 3.14 | female | Spleen | n/a | 24 March 2023 | 178.55 | 2.04 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baraldo, N.; Buzzoni, L.; Pasti, L.; Cavazzini, A.; Marchetti, N.; Mancia, A. miRNAs as Biomolecular Markers for Food Safety, Quality, and Traceability in Poultry Meat—A Preliminary Study. Molecules 2024, 29, 748. https://doi.org/10.3390/molecules29040748
Baraldo N, Buzzoni L, Pasti L, Cavazzini A, Marchetti N, Mancia A. miRNAs as Biomolecular Markers for Food Safety, Quality, and Traceability in Poultry Meat—A Preliminary Study. Molecules. 2024; 29(4):748. https://doi.org/10.3390/molecules29040748
Chicago/Turabian StyleBaraldo, Nada, Luna Buzzoni, Luisa Pasti, Alberto Cavazzini, Nicola Marchetti, and Annalaura Mancia. 2024. "miRNAs as Biomolecular Markers for Food Safety, Quality, and Traceability in Poultry Meat—A Preliminary Study" Molecules 29, no. 4: 748. https://doi.org/10.3390/molecules29040748