Characterization of a Wheat Heme Oxygenase-1 Gene and Its Responses to Different Abiotic Stresses
Abstract
:1. Introduction
2. Results and Discussion
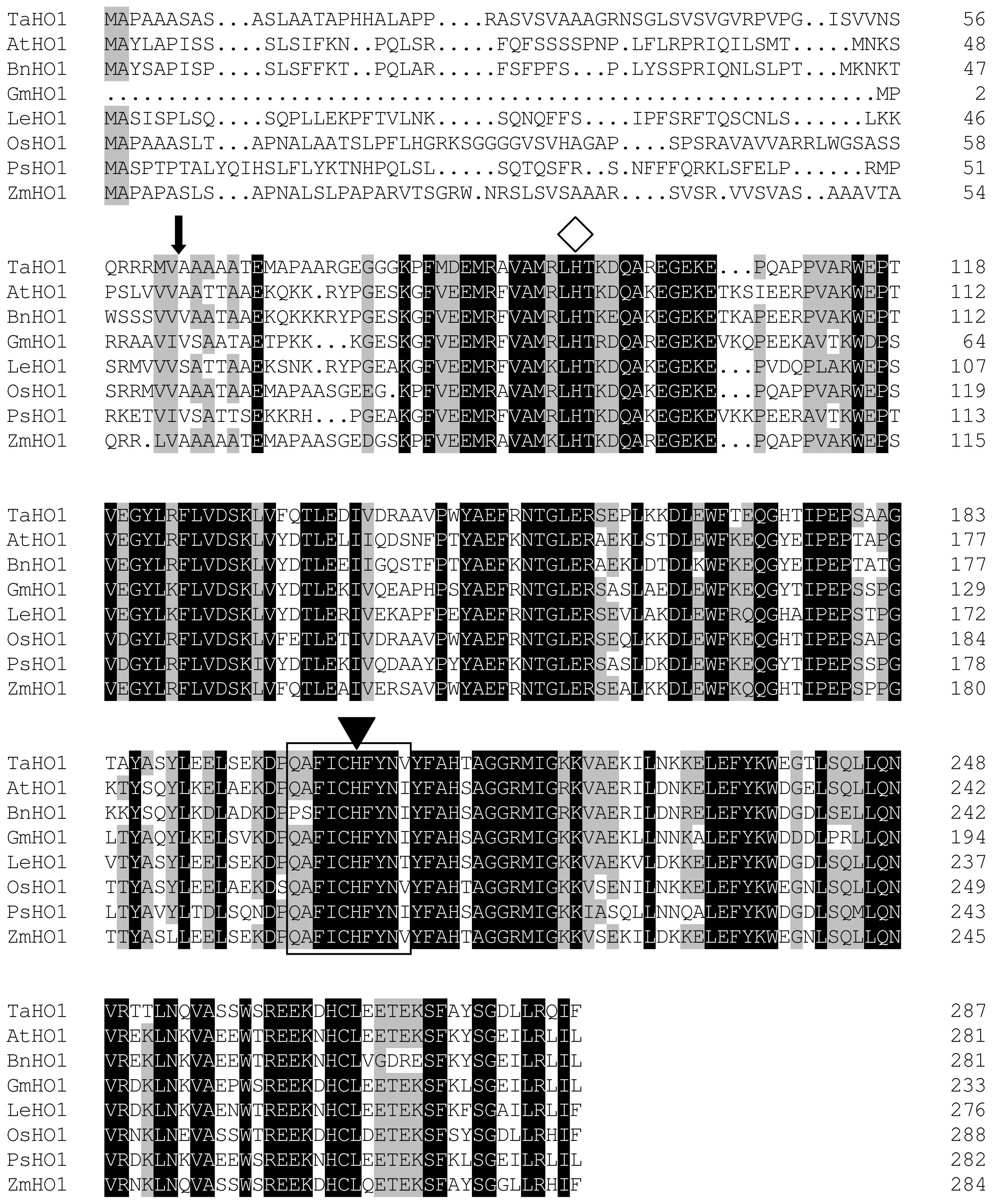
2.1. Identification and Cloning of a Wheat HO1 Gene
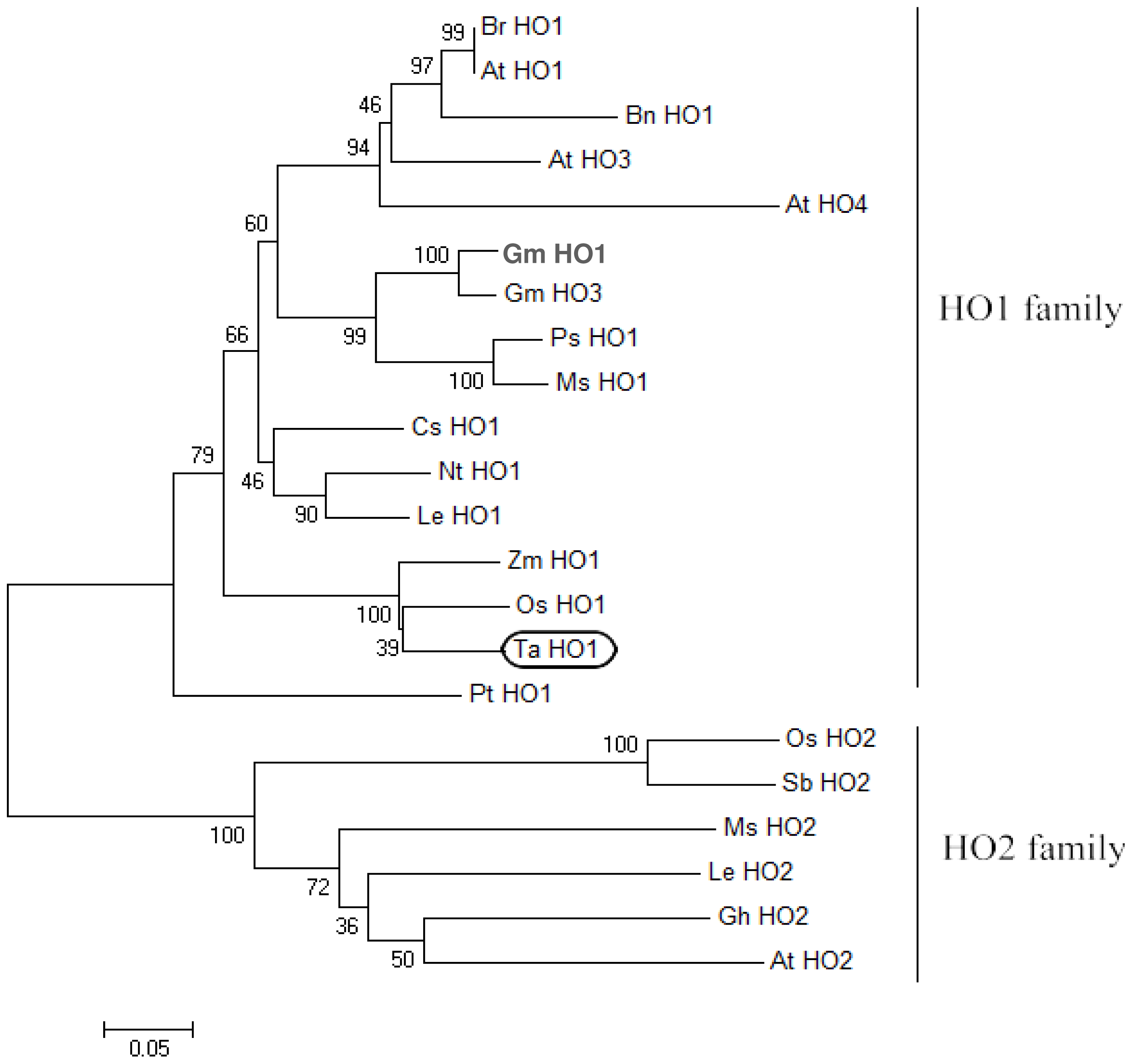
2.2. Phylogenetic Analysis of TaHO1
2.3. Expression and Purification of TaHO1
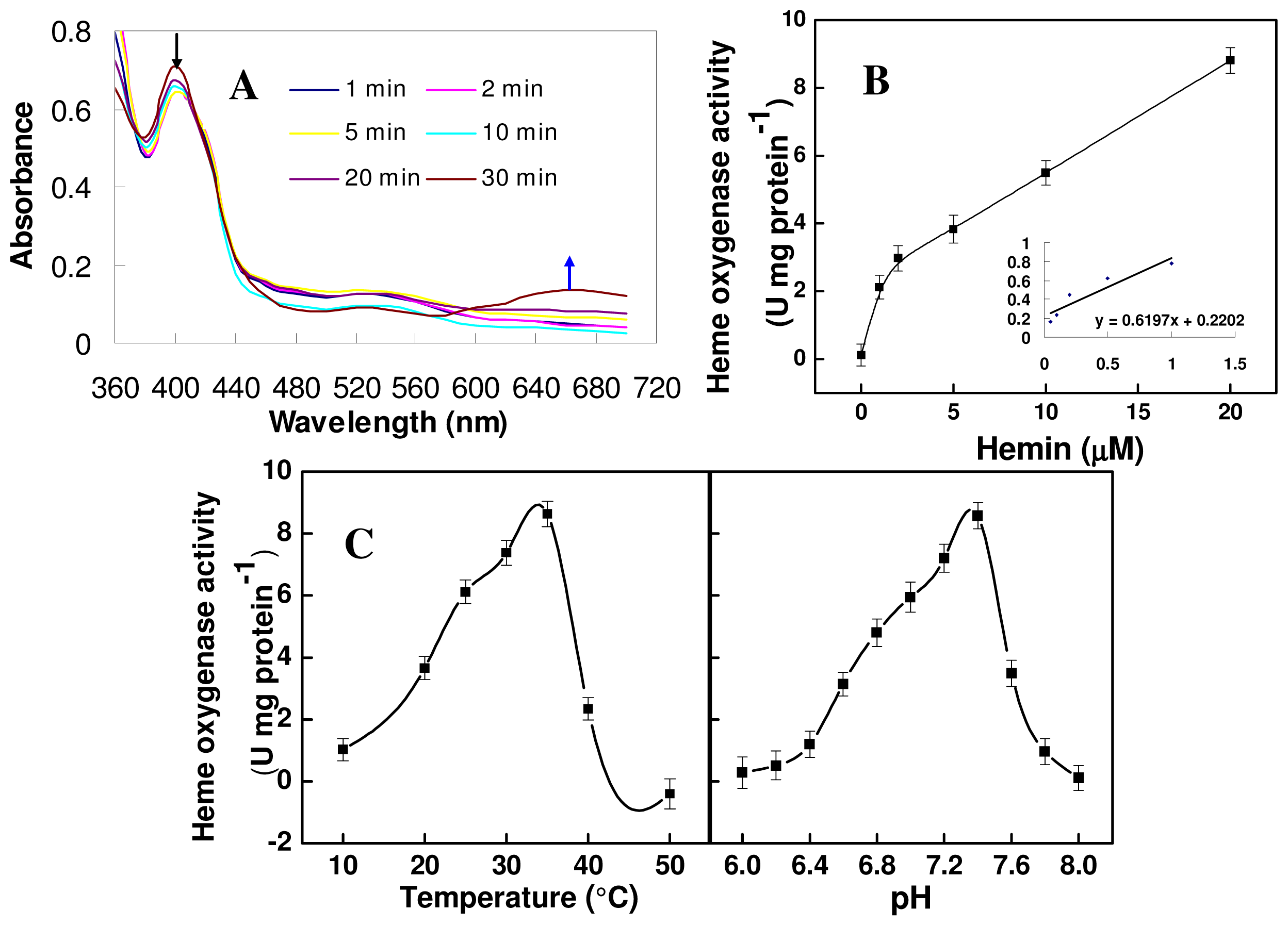
2.4. Biochemical Activity of the Mature TaHO1 Protein Expressed in Escherichia coli
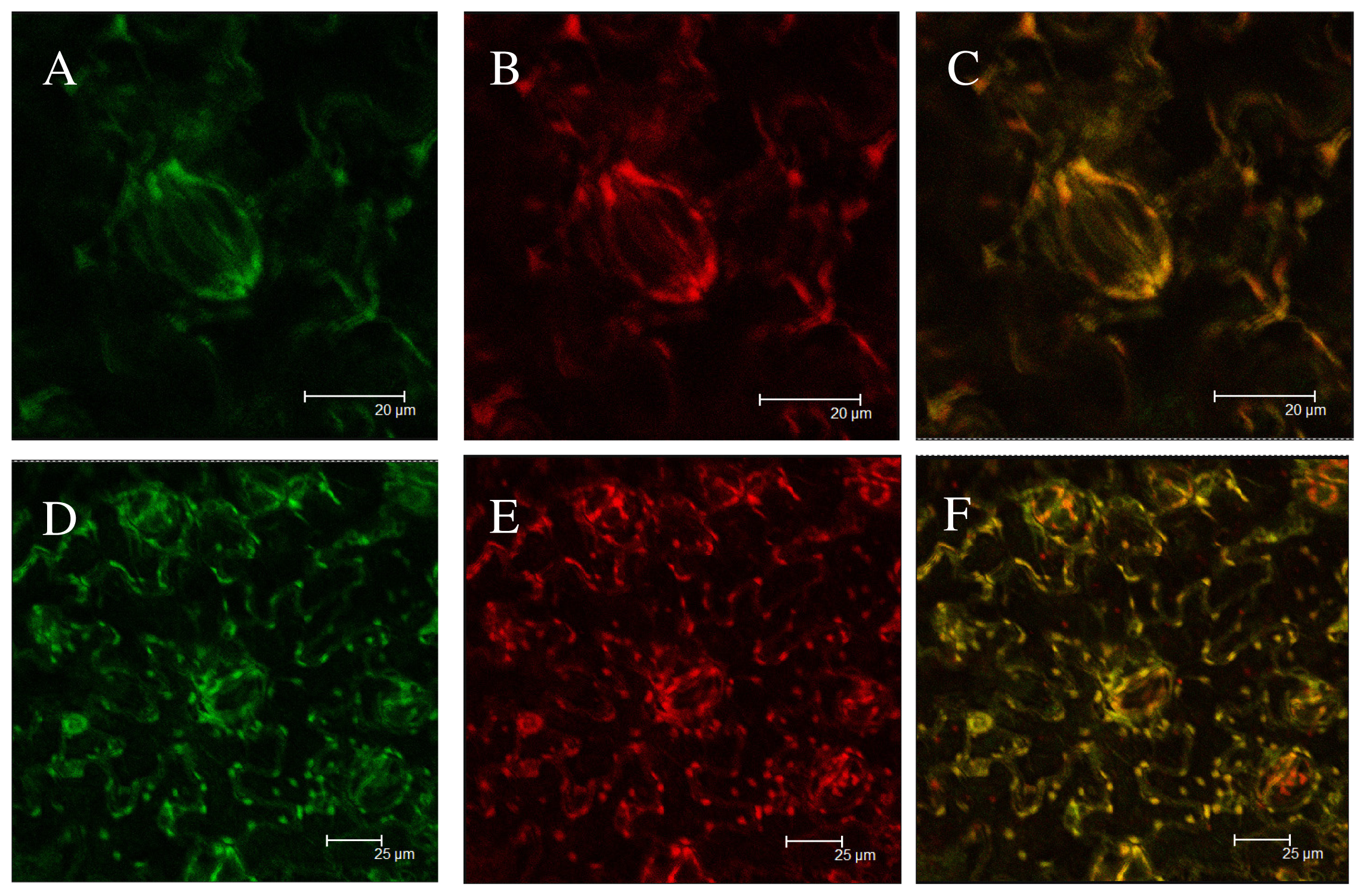
2.5. Subcellular Localization of Green Fluorescent Protein (GFP) Fusion Protein
2.6. Expression pattern Analysis of TaHO1 Protein
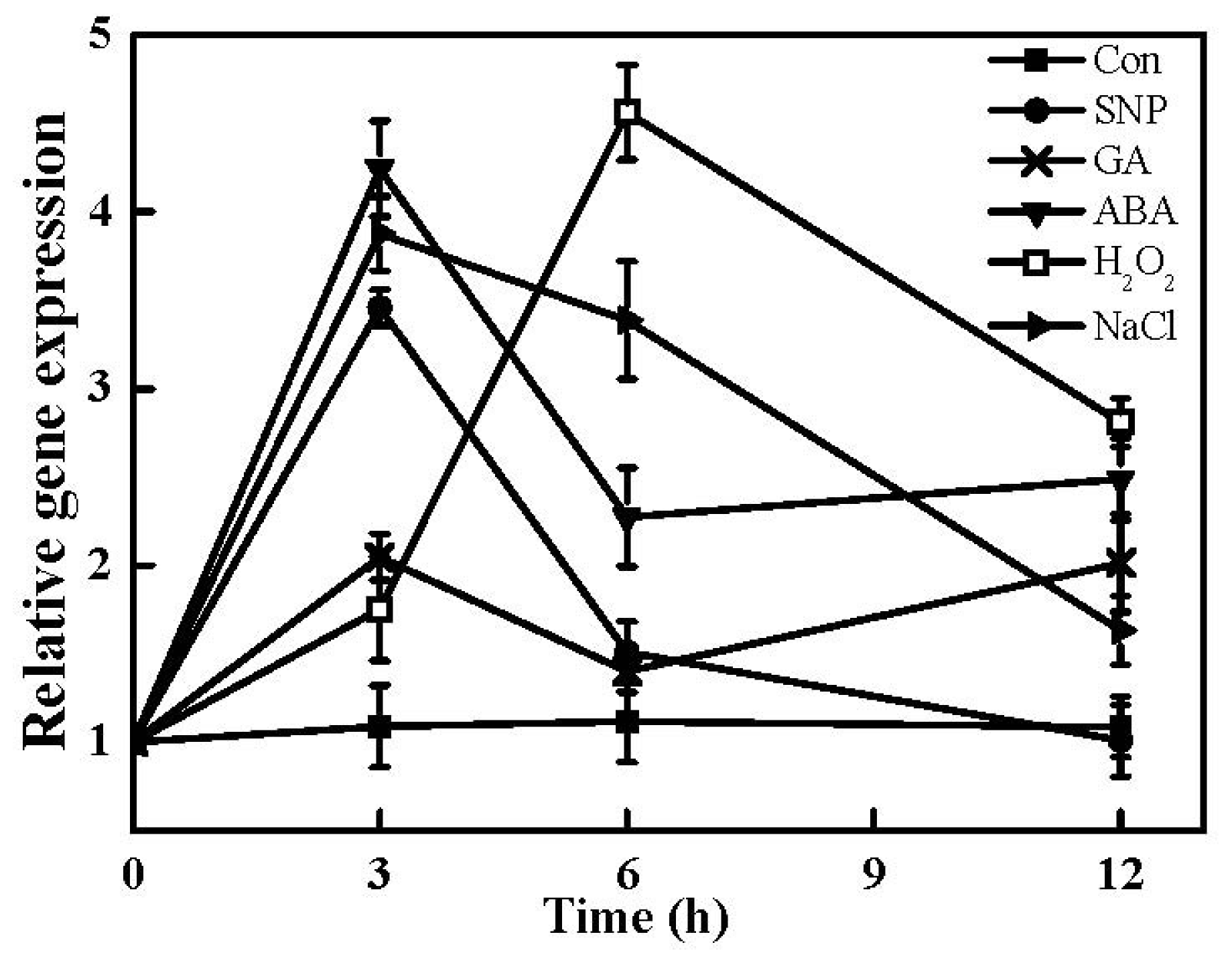
2.7. Expression of TaHO1 Gene in Responses to Different Exogenous Chemicals and Salinity Stress
3. Experimental Section
3.1. Plant Materials, Growth Condition and Treatments
3.2. Cloning of TaHO1
3.3. Multiple Sequence Alignment and Phylogenetic Analysis
3.4. Prokaryotic Expression and Purification of Recombinant His-Tagged TaHO1
3.5. HO Activity Determination
3.6. Subcellular Localization of TaHO1
3.7. Gel Blot Analysis for TaHO1
3.8. Real-Time RT-PCR Analysis
Acknowledgements
References
- Apel, K.; Hirt, H. Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol 2004, 55, 373–399. [Google Scholar]
- Salin, M.L. Toxic oxygen species and protective systems of the chloroplast. Physiol. Plant 1988, 72, 681–689. [Google Scholar]
- Shekhawat, G.S.; Verma, K. Haem oxygenase (HO): An overlooked enzyme of plant metabolism and defence. J. Exp. Bot 2010, 61, 2255–2270. [Google Scholar]
- Stocker, R.; Yamamoto, Y.; McDonagh, A.F.; Glazer, A.N.; Ames, B.N. Bilirubin is an antioxidant of possible physiological importance. Science 1987, 235, 1043–1046. [Google Scholar]
- Maines, M.D. Heme oxygenase: Function, multiplicity, regulatory mechanisms, and clinical applications. FASEB J 1988, 2, 2557–2568. [Google Scholar]
- Ewing, J.F.; Haber, S.N.; Maines, M.D. Normal and heat-induced patterns of expression of heme oxygenase-1 (HSP32) in rat brain: Hyperthermia causes rapid induction of mRNA and protein. J. Neurochem 1992, 58, 1140–1149. [Google Scholar]
- Yamaguchi, T.; Terakado, M.; Horio, F.; Aoki, K.; Tanaka, M.; Nakajima, H. Role of bilirubin as an antioxidant in an ischemia-reperfusion of rat liver and induction of heme oxygenase. Biochem. Biophys. Res. Commun 1996, 223, 129–135. [Google Scholar]
- Llesuy, S.F.; Tomaro, M.L. Heme oxygenase and oxidative stress. Evidence of involvement of bilirubin as physiological protector against oxidative damage. Biochim. Biophys. Acta 1994, 1223, 9–14. [Google Scholar]
- Platt, J.L.; Nath, K.A. Heme oxygenase: protective gene or Trojan horse. Nat. Med 1998, 4, 1364–1365. [Google Scholar]
- Emborg, T.J.; Walker, J.M.; Noh, B.; Vierstra, R.D. Multiple heme oxygenase family members contribute to the biosynthesis of the phytochrome chromophore in Arabidopsis. Plant Physiol 2006, 140, 856–868. [Google Scholar]
- Gisk, B.; Yasui, Y.; Kohchi, T.; Frankenberg-Dinkel, N. Characterization of the haem oxygenase protein family in Arabidopsis thaliana reveals a diversity of functions. Biochem. J 2010, 425, 425–434. [Google Scholar]
- Fu, G.Q.; Xu, S.; Xie, Y.J.; Han, B.; Nie, L.; Shen, W.B.; Wang, R. Molecular cloning, characterization, and expression of an alfalfa (Medicago sativa L.) heme oxygenase-1 gene, MsHO1, which is pro-oxidants-regulated. Plant Physiol. Biochem 2011, 49, 792–799. [Google Scholar]
- Noriega, G.O.; Yannarelli, G.G.; Balestrasse, K.B.; Batlle, A.; Tomaro, M.L. The effect of nitric oxide on heme oxygenase gene expression in soybean leaves. Planta 2007, 226, 1155–1163. [Google Scholar]
- Han, Y.; Zhang, J.; Chen, X.; Gao, Z.Z.; Xuan, W.; Xu, S.; Ding, X.; Shen, W.B. Carbon monoxide alleviates cadmium-induced oxidative damage by modulating glutathione metabolism in the roots of Medicago sativa. New Phytol 2008, 177, 155–166. [Google Scholar]
- Cui, W.T.; Fu, G.Q.; Wu, H.H.; Shen, W.B. Cadmium-induced heme oxygenase-1 gene expression is associated with the depletion of glutathione in the roots of Medicago sativa. Biometals 2011, 24, 93–103. [Google Scholar]
- Yannarelli, G.G.; Noriega, G.O.; Batlle, A.; Tomaro, M.L. Heme oxygenase up-regulation in ultraviolet-B irradiated soybean plants involves reactive oxygen species. Planta 2006, 224, 1154–1162. [Google Scholar]
- Xie, Y.J.; Ling, T.F.; Han, Y.; Liu, K.L.; Zheng, Q.S.; Huang, L.Q.; Yuan, X.X.; He, Z.Y.; Hu, B.; Fang, L.; et al. Carbon monoxide enhances salt tolerance by nitric oxide-mediated maintenance of ion homeostasis and up-regulation of antioxidant defense in wheat seedling roots. Plant Cell Environ 2008, 31, 1864–1881. [Google Scholar]
- Xie, Y.J.; Xu, S.; Han, B.; Wu, M.; Yuan, X.; Han, Y.; Gu, Q.; Xu, D.K.; Yang, Q.; Shen, W.B. Evidence of Arabidopsis salt acclimation induced by up-regulation of HY1 and the regulatory role of RbohD-derived reactive oxygen species synthesis. Plant J 2011, 66, 280–292. [Google Scholar]
- Xie, Y.J.; Cui, W.T.; Yuan, X.X.; Shen, W.B.; Yang, Q. Haem oxygenase-1 is associated with wheat salinity acclimation by modulating reactive oxygen species homeostasis. J. Integr. Plant Biol 2011, 53, 653–670. [Google Scholar]
- Chen, X.X.; Ding, X.; Xu, S.; Wang, R.; Xuan, W.; Cao, Z.Y.; Chen, J.; Wu, H.H.; Ye, M.B.; Shen, W.B. Endogenous hydrogen peroxide plays a positive role in the upregulation of heme oxygenase and acclimation to oxidative stress in wheat seedling leaves. J. Integr. Plant Biol 2009, 51, 951–960. [Google Scholar]
- Xuan, W.; Zhu, F.Y.; Xu, S.; Huang, B.K.; Ling, T.F.; Qi, J.Y.; Ye, M.B.; Shen, W.B. The heme oxygenase/carbon monoxide system is involved in the auxin-induced cucumber adventitious rooting process. Plant Physiol 2008, 148, 881–893. [Google Scholar]
- Muramoto, T.; Kohchi, T.; Yokota, A.; Hwang, I.; Goodman, H.M. The Arabidopsis photomorphogenic mutant hy1 is deficient in phytochrome chromophore biosynthesis as a result of a mutation in a plastid heme oxygenase. Plant Cell 1999, 11, 335–348. [Google Scholar]
- Izawa, T.; Oikawa, T.; Tokutomi, S.; Okuno, K.; Shimamoto, K. Phytochromes confer the photoperiodic control of flowering in rice (a short-day plant). Plant J 2000, 22, 391–399. [Google Scholar]
- Jin, Q.J.; Yuan, X.X.; Cui, W.T.; Han, B.; Feng, J.F.; Xu, S.; Shen, W.B. Isolation and characterization of a heme oxygenase-1 gene from Chinese cabbage. Mol. Biotechnol 2011. [Google Scholar] [CrossRef]
- Matera, K.M.; Zhou, H.; Migita, C.T.; Hobert, S.E.; Ishikawa, K.; Katakura, K.; Maeshima, H.; Yoshida, T.; Ikeda-Saito, M. Histidine-132 does not stabilize a distal water ligand and is not an important residue for the enzyme activity in heme oxygenase-1. Biochemistry 1997, 36, 4909–4915. [Google Scholar]
- Sun, J.; Loehr, T.M.; Wilks, A.; Ortiz de Montellano, P.R. Identification of histidine 25 as the heme ligand in human liver heme oxygenase. Biochemistry 1994, 33, 13734–13740. [Google Scholar]
- Yoshida, T.; Kikuchi, G. Purification and properties of heme oxygenase from rat liver microsomes. J. Biol. Chem 1979, 254, 4487–4491. [Google Scholar]
- Muramoto, T.; Tsurui, N.; Terry, M.J.; Yokota, A.; Kohchi, T. Expression and biochemical properties of a ferredoxin-dependent heme oxygenase required for phytochrome chromophore synthesis. Plant Physiol 2002, 130, 1958–1966. [Google Scholar]
- Wilks, A.; Black, S.M.; Miller, W.L.; Ortiz de Montellano, P.R. Expression and characterization of truncated human heme oxygenase (hHO-1) and a fusion protein of hHO-1 with human cytochrome P450 reductase. Biochemistry 1995, 34, 4421–4427. [Google Scholar]
- Liu, Y.H.; Xu, S.; Ling, T.F.; Xu, L.L.; Shen, W.B. Heme oxygenase/carbon monoxide system participates in regulating wheat seed germination under osmotic stress involving the nitric oxide pathway. J. Plant Physiol 2010, 167, 1371–1379. [Google Scholar]
- Besson-Bard, A.; Pugin, A.; Wendehenne, D. New insights into nitric oxide signaling in plants. Annu. Rev. Plant Biol 2008, 59, 21–39. [Google Scholar]
- Zimmermann, P.; Hirsch-Hoffmann, M.; Hennig, L.; Gruissem, W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol 2004, 136, 2621–2632. [Google Scholar]
- Zilli, C.G.; Balestrasse, K.B.; Yannarelli, G.G.; Polizio, A.H.; Santa-Cruz, D.M.; Tomaro, M.L. Heme oxygenase up-regulation under salt stress protects nitrogen metabolism in nodules of soybean plants. Environ. Exp. Bot 2008, 64, 83–89. [Google Scholar]
- NCBI BLAST program. Available online: http://www.ncbi.nlm.nih.gov/blast accessed on 14 September 2009.
- Liu, K.L.; Xu, S.; Xuan, W.; Ling, T.F.; Cao, Z.Y.; Huang, B.K.; Sun, Y.G.; Fang, L.; Liu, Z.Y.; Zhao, N.; et al. Carbon monoxide counteracts the inhibition of seed germination and alleviates oxidative damage caused by salt stress in Oryza sativa. Plant Sci 2007, 172, 544–555. [Google Scholar]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem 1976, 72, 248–254. [Google Scholar]
- Voinnet, O.; Rivas, S.; Mestre, P.; Baulcombe, D. An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J 2003, 33, 949–956. [Google Scholar]
- Brandizzi, F.; Frangne, N.; Marc-Martin, S.; Hawes, C.; Neuhaus, J.M.; Paris, N. The destination for single-pass membrane proteins is influenced markedly by the length of the hydrophobic domain. Plant Cell 2002, 14, 1077–1092. [Google Scholar]


© 2011 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Xu, D.-k.; Jin, Q.-j.; Xie, Y.-j.; Liu, Y.-h.; Lin, Y.-t.; Shen, W.-b.; Zhou, Y.-j. Characterization of a Wheat Heme Oxygenase-1 Gene and Its Responses to Different Abiotic Stresses. Int. J. Mol. Sci. 2011, 12, 7692-7707. https://doi.org/10.3390/ijms12117692
Xu D-k, Jin Q-j, Xie Y-j, Liu Y-h, Lin Y-t, Shen W-b, Zhou Y-j. Characterization of a Wheat Heme Oxygenase-1 Gene and Its Responses to Different Abiotic Stresses. International Journal of Molecular Sciences. 2011; 12(11):7692-7707. https://doi.org/10.3390/ijms12117692
Chicago/Turabian StyleXu, Dao-kun, Qi-jiang Jin, Yan-jie Xie, Ya-hui Liu, Yu-ting Lin, Wen-biao Shen, and Yi-jun Zhou. 2011. "Characterization of a Wheat Heme Oxygenase-1 Gene and Its Responses to Different Abiotic Stresses" International Journal of Molecular Sciences 12, no. 11: 7692-7707. https://doi.org/10.3390/ijms12117692
APA StyleXu, D.-k., Jin, Q.-j., Xie, Y.-j., Liu, Y.-h., Lin, Y.-t., Shen, W.-b., & Zhou, Y.-j. (2011). Characterization of a Wheat Heme Oxygenase-1 Gene and Its Responses to Different Abiotic Stresses. International Journal of Molecular Sciences, 12(11), 7692-7707. https://doi.org/10.3390/ijms12117692





