Cell-Biomaterial Mechanical Interaction in the Framework of Tissue Engineering: Insights, Computational Modeling and Perspectives
Abstract
:1. Introduction
1.1. Structures and Functions of Living Cells
1.2. Mechanical Aspects of Cell-Biomaterial Interaction
1.3. Tissue Engineering Science
- Cell recruitment: isolation and expansion. Tissues and organs are made of cells and the specific function of a tissue or organ can only be developed by those cells. Potential limitation of cell therapy is bounded by restricted cell niches. For this, adult stem cells can be isolated and after expanded and differentiated in vitro. In order to improve tissue regeneration, signal molecules, such as growth factors, are used in combination with cell therapy;
- Biomaterial interaction. Recruited cells are usually seeded in vitro onto artificial biomaterial matrices, i.e., scaffolds. The aim of these matrices is to give support to the cells to develop its specific function during the healing or regenerative process. Cell-biomaterial interaction is critical and the biomaterial should mimic the extracellular space that it replaces. This issue is the focus of this review;
- Implantation. Seeded matrices, performed ex vivo, are implanted in vivo for tissue repairing. Here, biocompatibility must be assured in order to prevent an immunological rejection. Moreover, the vascularization of the surrounded tissue should occur within the artificial matrix.
2. Mechanisms for Cell-ECM and Artificial Substrate Interaction
2.1. Plasma Membrane
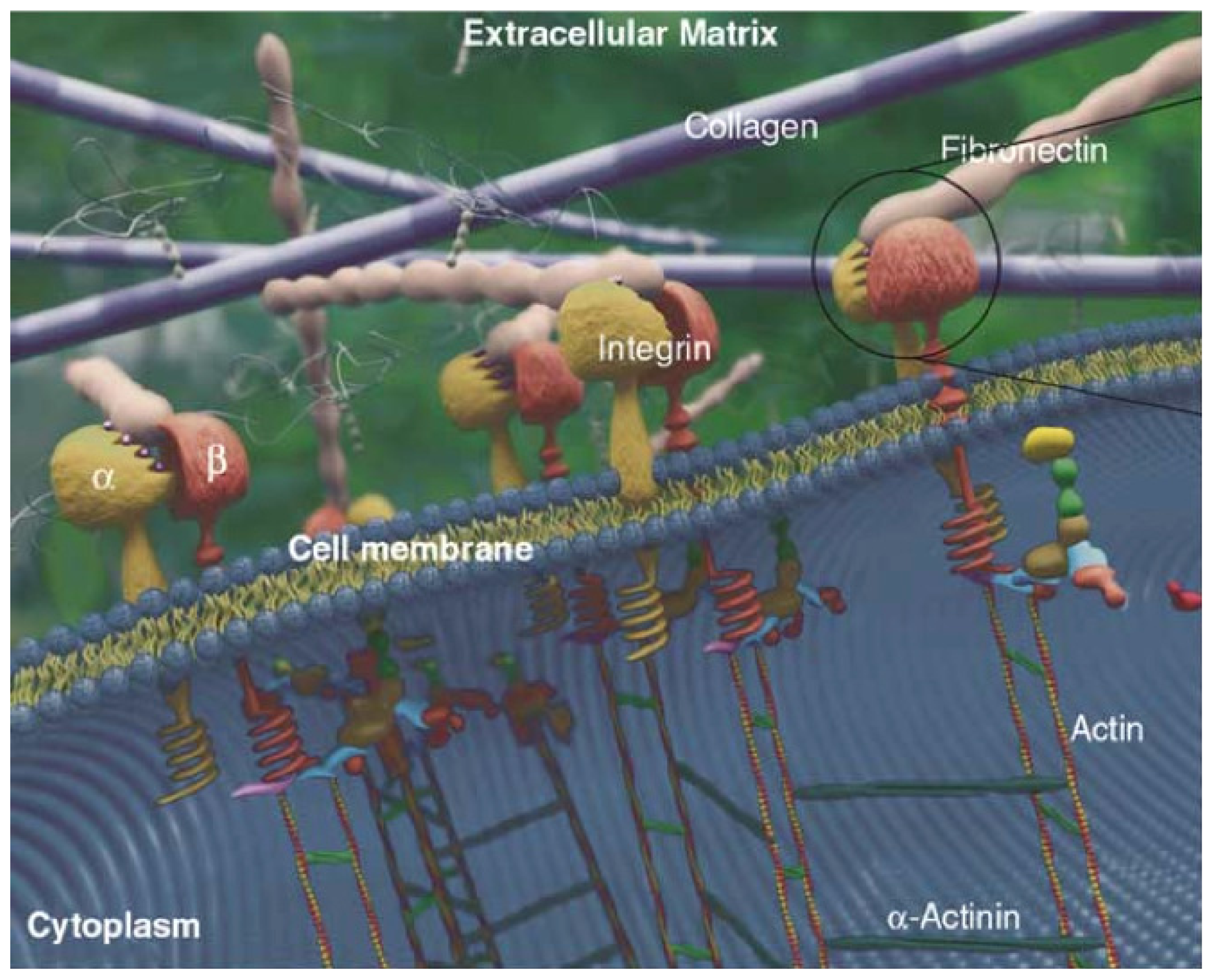
2.2. ECM
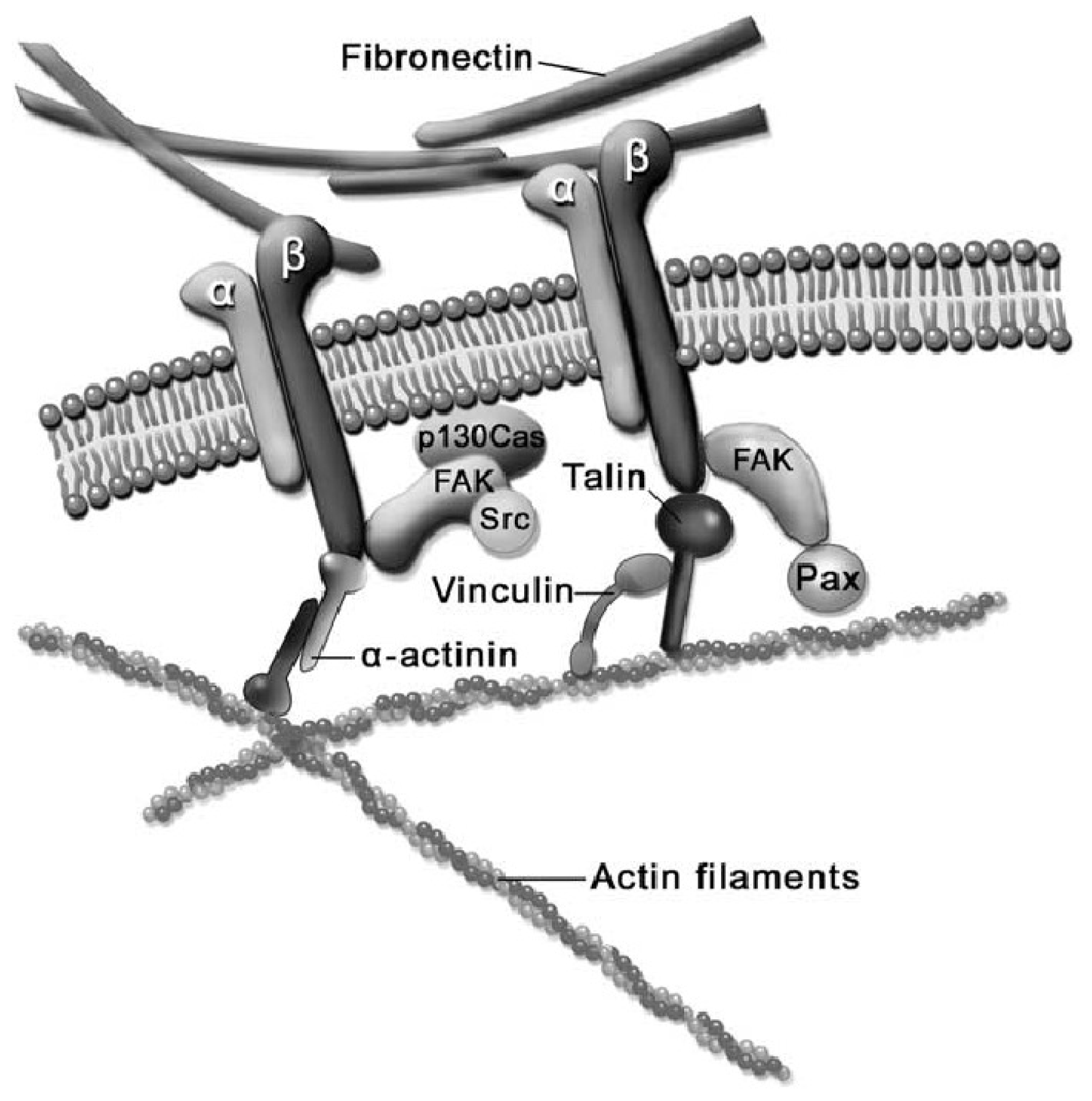
2.3. Cell-ECM Interaction
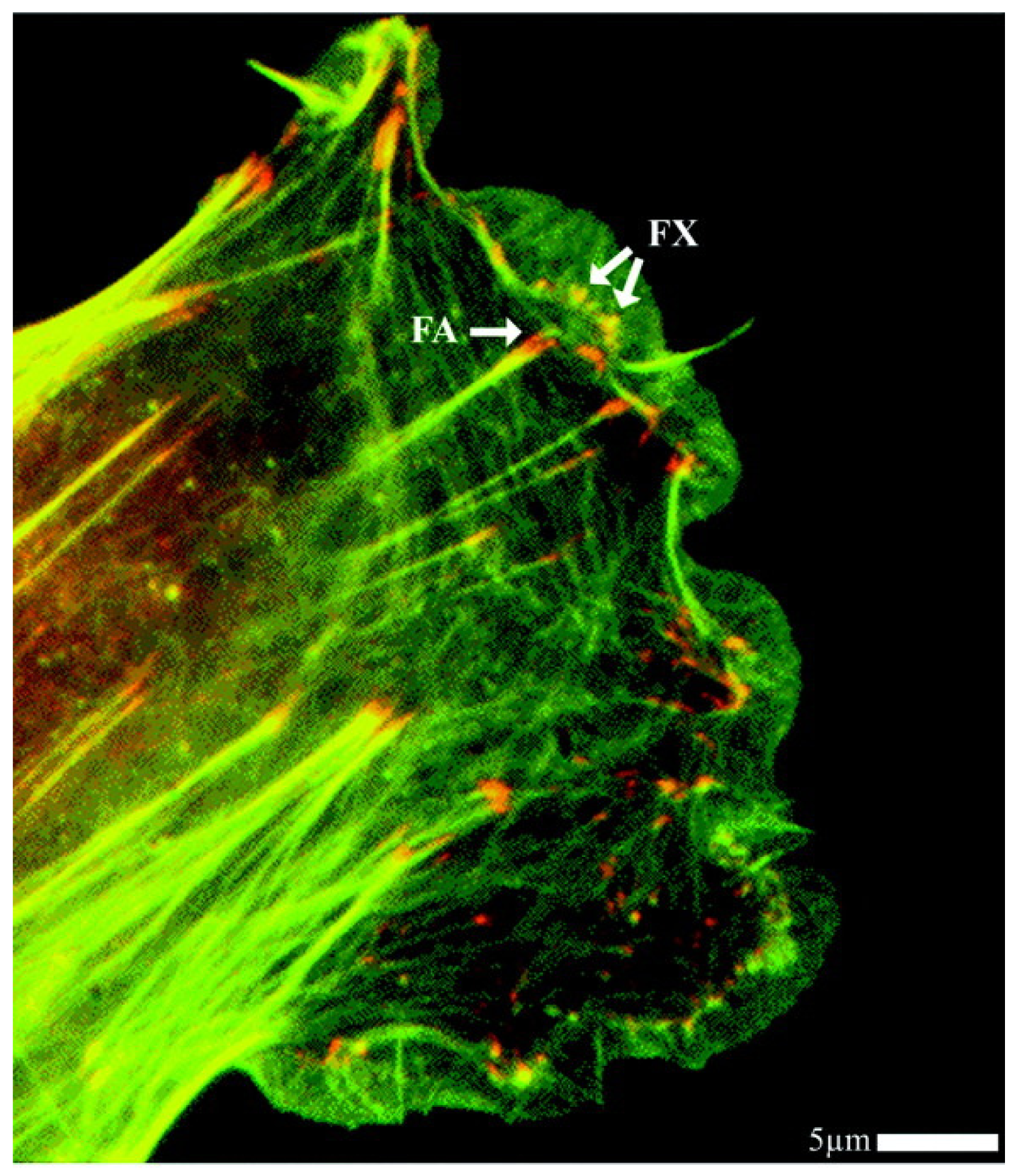
2.4. Cell-Biomaterial Interaction on Artificial Substrates
3. Cell-Biomaterial Interaction Models
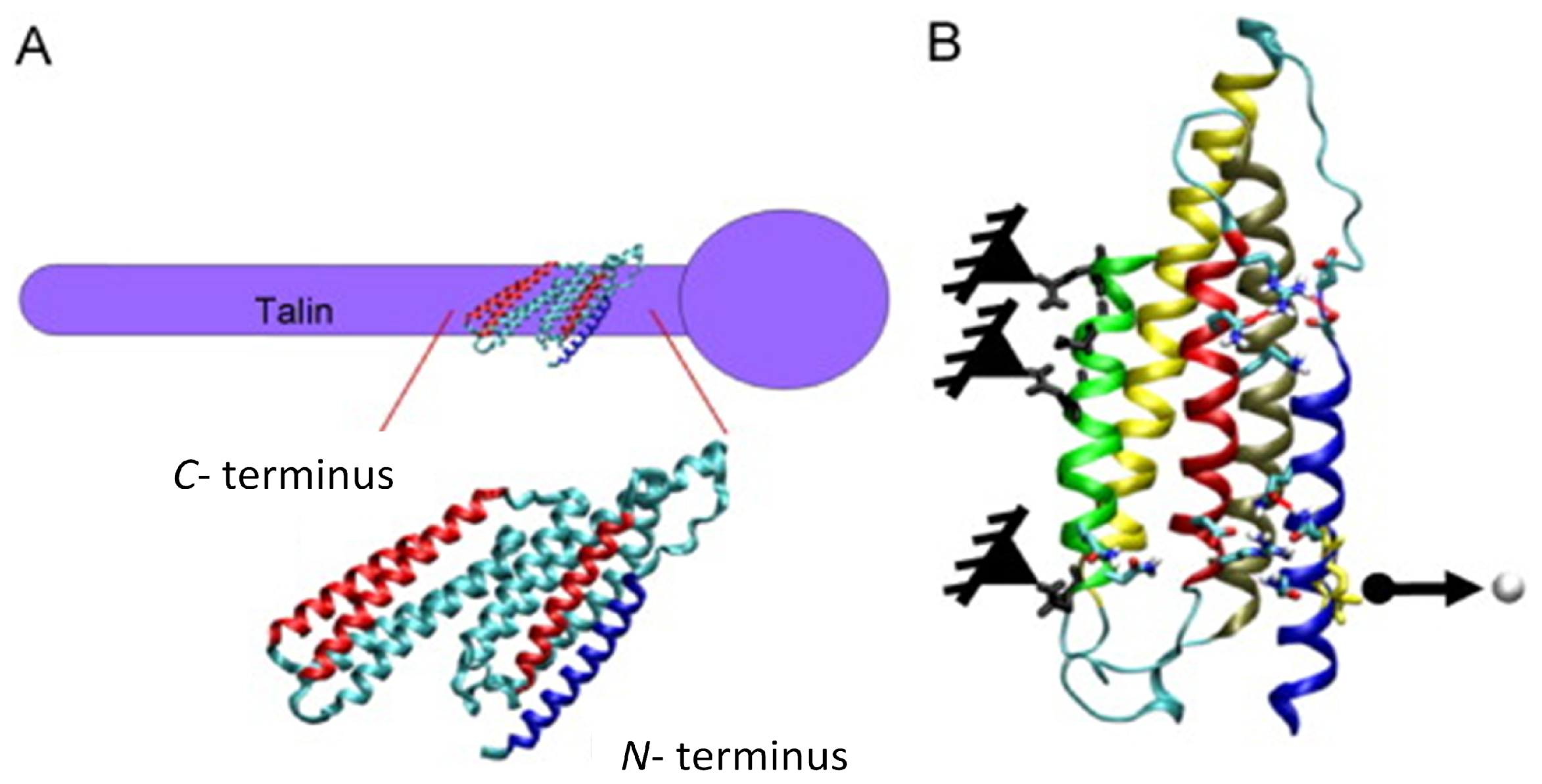
3.1. Molecular (Protein-Based) Models
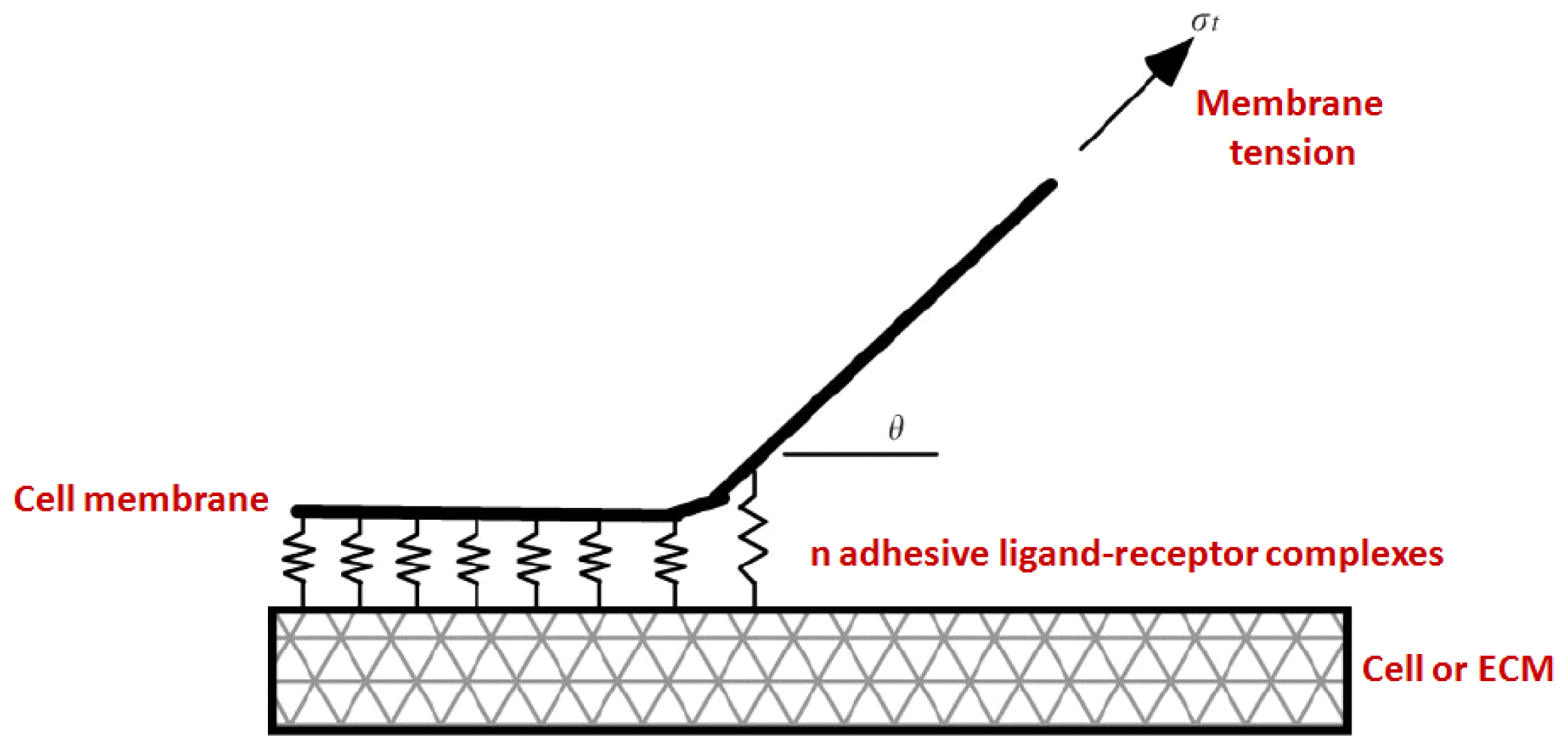
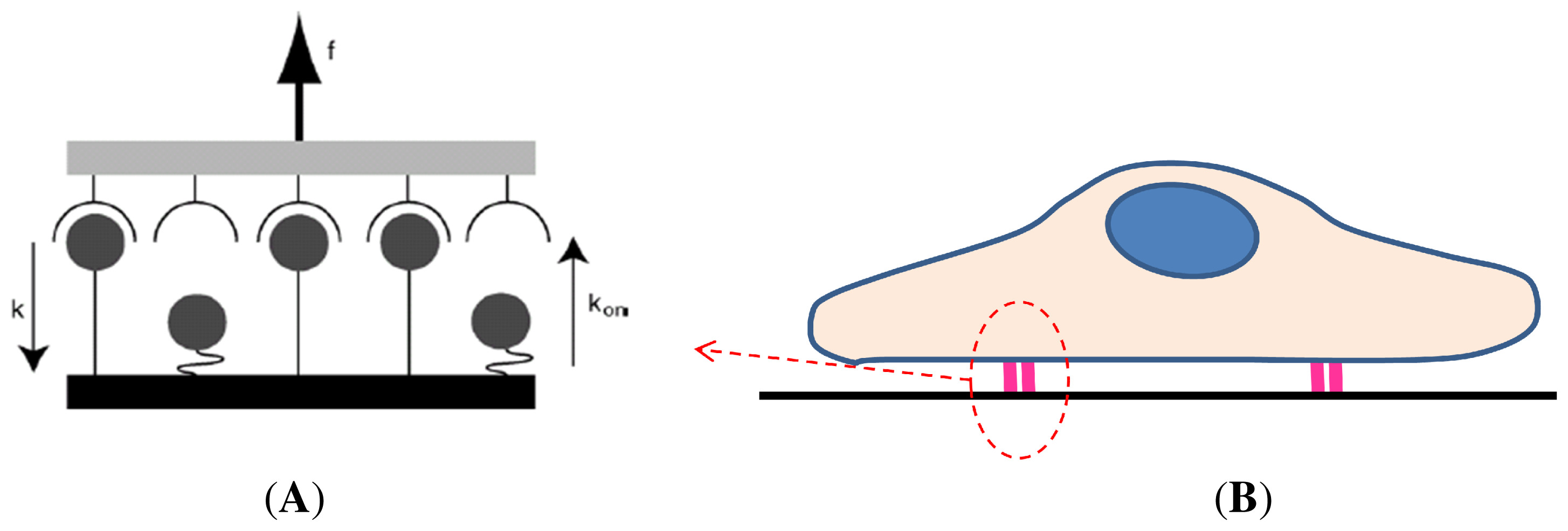
3.2. Kinetics and Chemomechanical Adhesion Models
3.3. Continuum Approaches
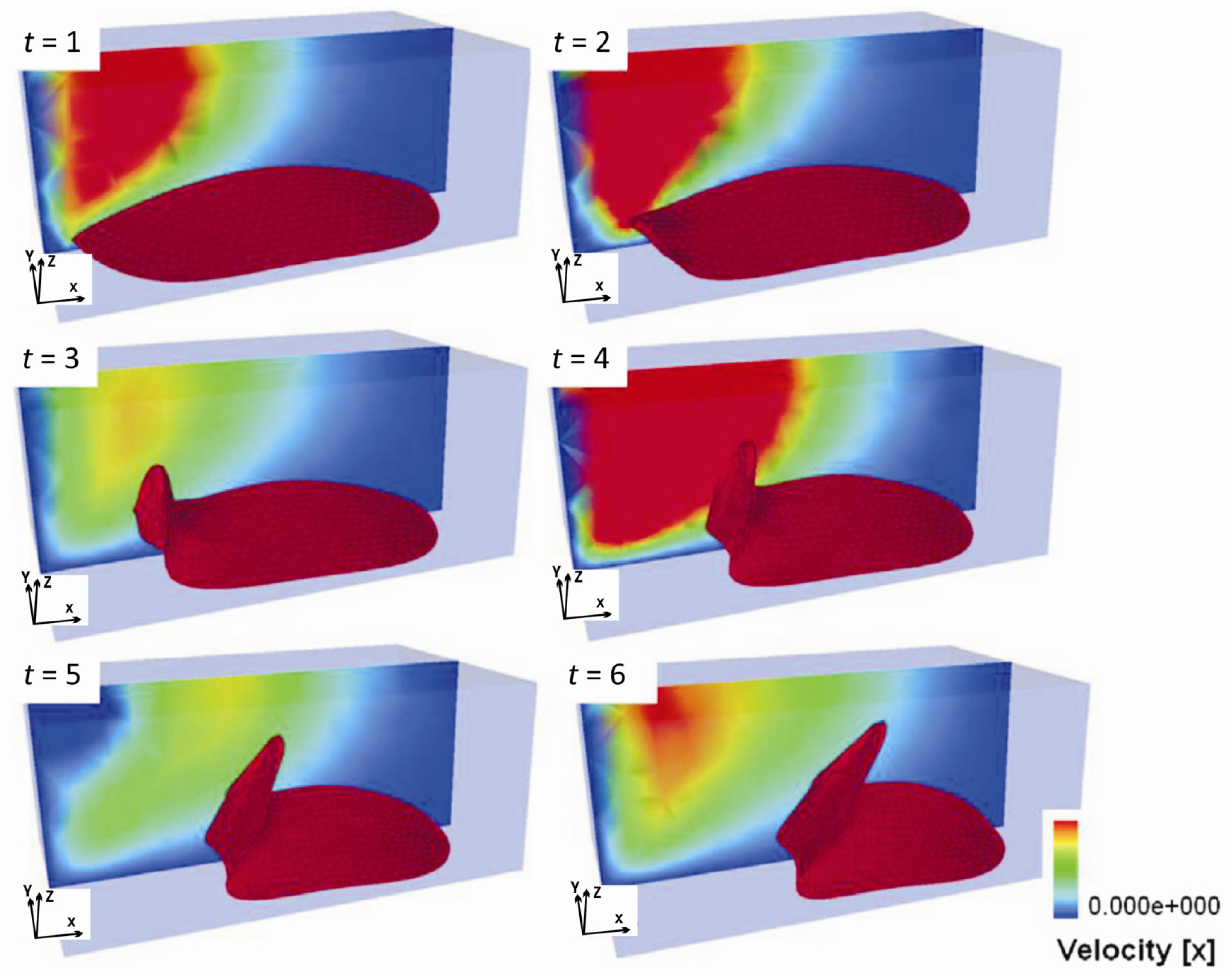
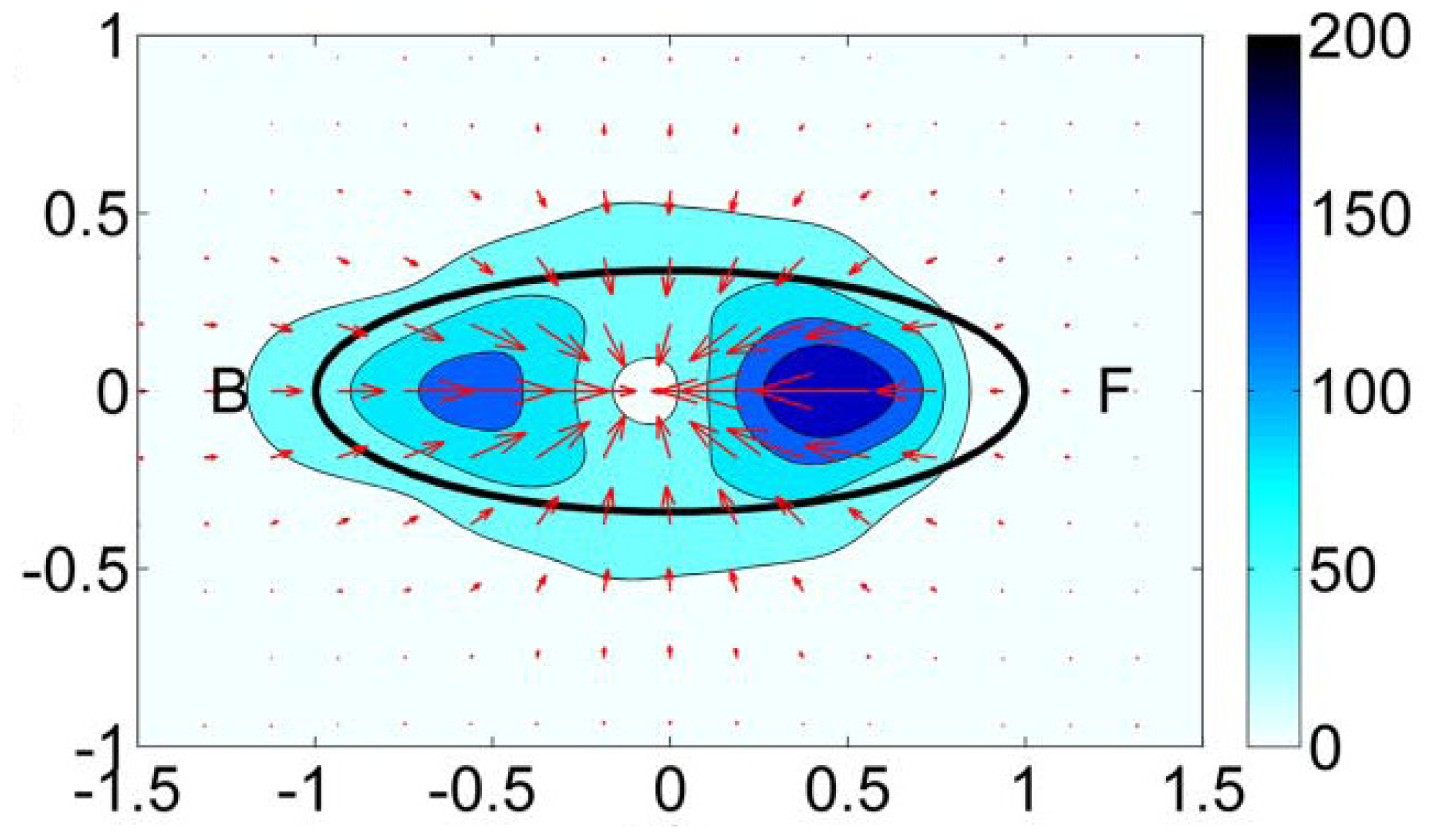
- Single-cell-biomaterial interaction models: This kind of approaches attempts to model in one hand the mechanical behavior of the CSK for a single cell, the mechanical behavior of the biomaterial on the other hand and the interaction between both by incorporating some of the characteristics of the previously discussed models. Cytoskeleton has been modeled as a continuum deformable body, as a soft glassy material [109], viscoelastic and continuum elastic [110,111], multiphasic model [112], as a gel [113], or based on the tensegrity theory [114]. So far, there is no consensus as to whether cytoskeleton behavior is closer to a fluid or to a solid, since it shows features of both. Likely, both perspectives should be accounted for in an overall global model for CSK (see Mofrad & Kamm [4] for a discussion on cell mechanics modeling). Then, the mechanical behavior of the biomaterial may be assumed under a simple viscoelastic or elastic behavior as a first approach in order to analyze its mechanical influence on cell attachment. Gracheva & Othmer [115] presented a 1D model which assumes the cell as a linear viscoelastic material interacting with the substrate through a drag (friction) coefficient. The substrate was considered as a rigid material. The main interest of that model was to analyze cell motility, so adhesion was considered only in phenomenological terms. This model was further extended to 2D in [116] accounting also for the matrix elasticity. In both models, contractile force generation and focal adhesion forces at the cell front during migration were phenomenologically modeled according to experimental evidences. In a similar model, Kuusela & Alt [117] phenomenologically modeled the force at cell-substrate interface as a drag force. However, this model considers motility of ligand and receptors adhesion proteins using diffusion-advection equations. On the other hand, Deshpande et al. [118] considered a contractile model for the CSK reminiscent to the Hill’s equations for muscle contraction [119]. The cell (considered as a square) is attached in the corners to four elastic elements (deflection posts) in order to analyze the effect of posts rigidity as cell contracts, as a measurement of focal adhesion forces. This model was extended in Deshpande et al. [120] where focal adhesion dynamics were considered in greater detail, similarly to the models discussed in the previous section, although here in a continuum framework. Kopacz et al. [110] simulated endothelial cell adhesion on arterial constructs under a blood flow. The cell is modeled as a viscoelastic material whereas the effect of the ECM was neglected. Focal adhesions were modeled using the classical Bell’s model [80]. Nevertheless, the focus of this work was the computational aspects of fluid-cell interaction (see Figure 7). Other works analyze cell-biomaterial interaction from another perspective rather than adhesion. For example, Sanz-Herrera et al. [121] investigated on the effect of substrate curvature on internal CSK forces redistribution in 3D matrices.
- Cell-populations-biomaterial interaction models: The main difference of this approach with the models discussed in (i) is the fact that a material point of analysis accounts for both cell and substrate, so the interaction between both cannot be easily incorporated in these models. However, cell and biomaterial mechanical behavior is modeled similar than in models presented above. Examples of these models may be found in Oster et al. [122]; Namy et al. [123]; Murray [124] and many others. Interestingly, Moreo et al. [125] used this approach to model the cell body as a contractile material [119] and a viscoelastic susbtrate, in an attempt to predict the experimentally observed phenomena of durotaxis and tensotaxis. Results showed an increasing concentration of cells in areas of strained and stiffer substrates.
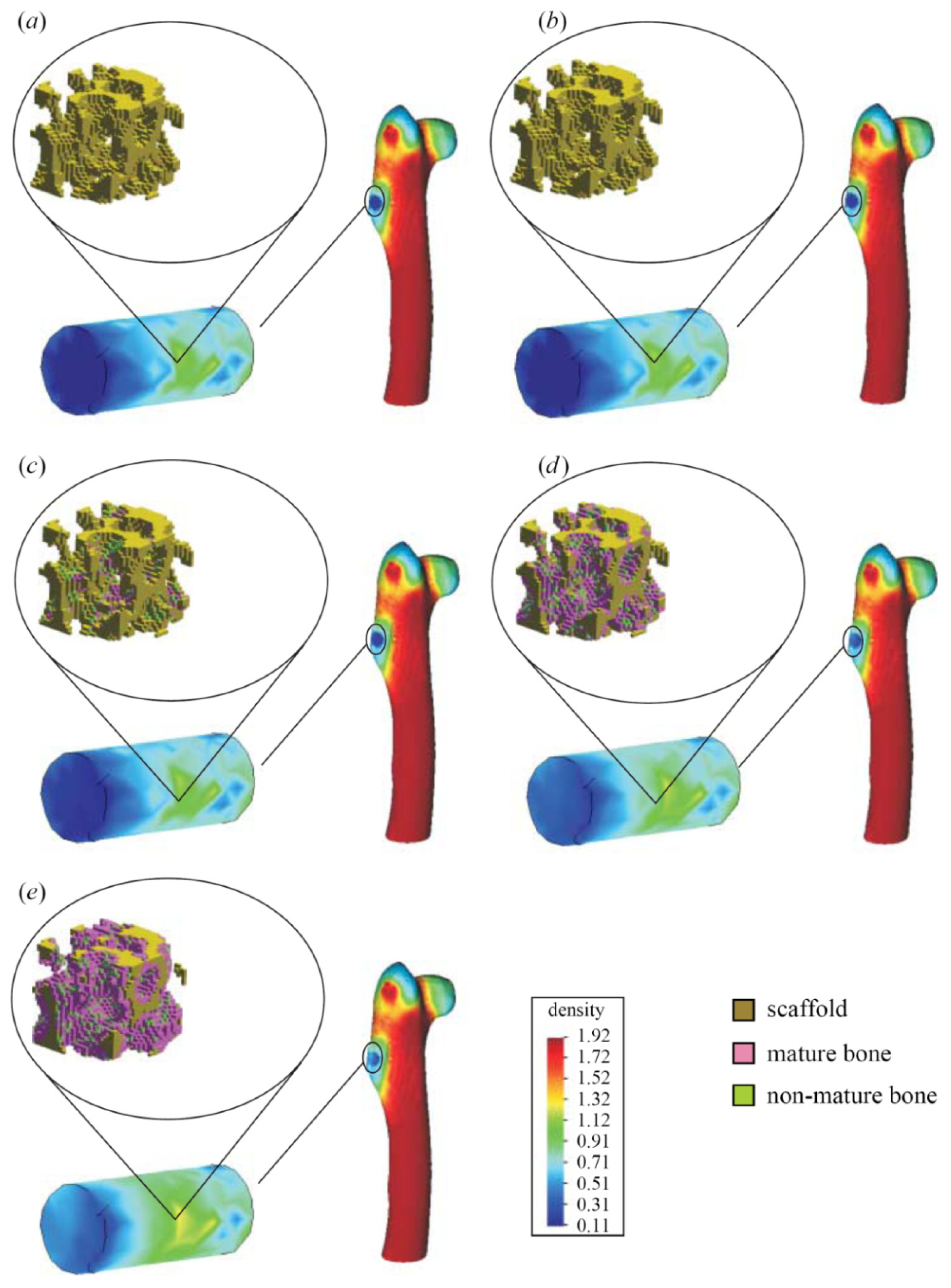
- Indirect mechanistic models in tissue engineering: Some examples of continuum studies that are based on cell-center approaches are within the tissue engineering framework. These models use lattice-based methods to discretize the spatial domain into a structured grid. The random-walk model introduced by Pérez & Prendergast [126] using a lattice based approach has been successfully applied to bone tissue engineering [127–130]. Byrne et al. [127] and Sanz-Herrera et al. [131] simulated tissue growth in a scaffold to investigate various design parameters (e.g., scaffold porosity, Young’s modulus, and dissolution rate) and Khayyeri et al. [128] studied tissue differentiation in an in vivo bone chamber. The random walk model early proposed by Pérez & Prendergast [126] has been extended further to account for the vascular network formation [132] and applied to different scaffold geometries for bone tissue engineering [129,130]. Other studies within the tissue engineering field have shown how mechanical loads and fluid flow applied on the scaffolds cause different levels of mechanical stimuli on cells at a microscopic level within the samples according to the morphology of the materials [133]. Other models to analyze cell activity on scaffolds [134–140] are also available for the macroscopic design of scaffolds in terms of porosity, permeability, apparent mechanical behavior, etc., which should be similar to properties of the natural tissue (see Figure 8).
4. Perspectives
5. Conclusions
Acknowledgments
References
- Alberts, B.; Bray, D.; Hopkin, K.; Johnson, A.; Lewis, J.; Raff, M.; Roberts, K.; Walter, P. Essential Cell Biology, 2nd ed; Garland Science/Taylor & Francis Group: New York, NY, USA, 2003. [Google Scholar]
- Belk, C.; Maier, V.B. Biology: Science for Life with Physiology, 2nd ed; Pearson Prentice Hall: Upper Saddle River, NJ, USA, 2007. [Google Scholar]
- Phillips, R.; Kondev, J.; Theriot, J. Physical Biology of the Cell; Garland Science: New York, NY, USA, 2009. [Google Scholar]
- Mofrad, M.R.K.; Kamm, R.D. Cytoskeletal Mechanics; Cambridge University Press: Cambridge, UK, 2006. [Google Scholar]
- Boal, D.H. Mechanics of the Cell; Cambridge University Press: Cambridge, UK, 2002. [Google Scholar]
- Discher, D.E.; Janmey, P.; Wang, Y.L. Tissue cells feel and respond to the stiffness of their substrate. Science 2005, 310, 1139–1143. [Google Scholar]
- Welch, D.R.; Sakamaki, T.; Pioquinto, R.; Leonard, T.O.; Goldberg, S.F.; Hon, Q.; Erikson, R.L.; Rieber, M.; Rieber, M.S.; Hicks, D.J.; et al. Transfection of constitutively active mitogen-activated protein/extracellular signal-regulated kinase kinase confers tumorigenic and metastatic potentials to NIH3T3 cells. Cancer Res 2000, 60, 1552–1556. [Google Scholar]
- Engler, A.J.; Sen, S.; Sweeney, H.L.; Discher, D.E. Matrix elasticity directs stem cell lineage specification. Cell 2006, 126, 677–689. [Google Scholar]
- Pelham, R.J.; Wang, Y. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc. Natl. Acad. Sci. USA 1997, 94, 13661–13665. [Google Scholar]
- Chrzanowska-Wodnicka, M.; Burridge, K. Rho-stimulated contractility drives the formation of stress fibers and focal adhesions. J. Cell Biol 1996, 133, 1403–1415. [Google Scholar]
- Griffin, M.A.; Sen, S.; Sweeney, H.L.; Discher, D.E. Adhesion-contractile balance in myocyte differentiation. J. Cell Sci 2004, 117, 5855–5863. [Google Scholar]
- Leader, W.M.; Stopak, D.; Harris, A.K. Increased contractile strength and tightened adhesions to the substratum result from reverse transformation of CHO cells by dibutyryl cyclic adenosine monophosphate. J. Cell Sci 1983, 64, 1–11. [Google Scholar]
- Engler, A.J.; Griffin, M.A.; Sen, S.; Bönnemann, C.G.; Sweeney, H.L.; Discher, D.E. Myotubes differentiate optimally on substrates with tissue-like stiffness: Pathological implications for soft or stiff microenvironments. J. Cell Biol 2004, 166, 877–887. [Google Scholar]
- Bridgman, P.C.; Dave, S.; Asnes, C.F.; Tullio, A.N.; Adelstein, R.S. Myosin IIB is required for growth cone motility. J. Neurosci 2001, 21, 6159–6169. [Google Scholar]
- Lauffenburger, D.A.; Horwitz, A.F. Cell migration: A physically integrated molecular process. Cell 1996, 84, 359–369. [Google Scholar]
- Bray, D. Cell Movements: From Molecules to Motility; Garland Publishing: New York, NY, USA, 2001. [Google Scholar]
- Lo, C.M.; Wang, H.B.; Dembo, M.; Wang, Y.L. Cell movement is guided by the rigidity of the substrate. Biophys. J 2000, 79, 144–152. [Google Scholar]
- Peyton, S.R.; Putnam, A.J. Extracellular matrix rigidity governs smooth muscle cell motility in a biphasic fashion. J. Cell. Physiol 2005, 204, 198–209. [Google Scholar]
- Zaari, N.; Rajagopalan, P.; Kim, S.K.; Engler, A.J.; Wong, J.Y. Photopolymerization in microfluidic gradient generators: Microscale control of substrate compliance to manipulate cell response. Adv. Mater 2004, 16, 2133–2137. [Google Scholar]
- Schwarz, U.S.; Bischofs, I.B. Physical determinants of cell organization in soft media. Med. Eng. Phys 2005, 7, 43–47. [Google Scholar]
- Riveline, D.; Zamir, E.; Balaban, N.Q.; Schwarz, U.S.; Ishizaki, T.; Narumiya, S.; Kam, Z.; Geiger, B.; Bershadsky, A.D. Focal contacts as mechanosensors: Externally applied local mechanical force induces growth of focal contacts by an mDia1-dependent and ROCK-independent mechanism. J. Cell Biol 2001, 153, 1175–1186. [Google Scholar]
- Tamada, M.; Sheetz, M.P.; Sawada, Y. Activation of a signaling cascade by cytoskeleton stretch. Dev. Cell 2004, 7, 709–718. [Google Scholar]
- Fillingham, I.; Gingras, A.R.; Papagrigoriou, E.; Patel, B.; Emsley, J.; Critchley, D.R.; Roberts, G.C.; Barsukov, I.L. A vinculin binding domain from the talin rod unfolds to form a complex with the vinculin head. Structure 2005, 13, 65–74. [Google Scholar]
- Law, R.; Liao, G.; Harper, S.; Yang, G.; Speicher, D.W.; Discher, D.E. Pathway shifts and thermal softening in temperature-coupled forced unfolding of spectrin domains. Biophys. J 2003, 85, 3286–3293. [Google Scholar]
- Langer, R.; Vacanti, J.P. Tissue engineering. Science 1993, 260, 920–926. [Google Scholar]
- Skalak, R.; Fox, C.F. Tissue Engineering; Wiley-Liss: New York, NY, USA, 1988. [Google Scholar]
- Landowne, D. Cell Physiology; McGraw-Hill Medical Publishing Division: Blacklick, OH, USA, 2006; pp. 17–47. [Google Scholar]
- Kumar, V.; Abbas, A.; Fausto, N. Robbins and Cotran: Pathologic Basis of Disease, 7th ed; Elsevier: Philadelphia, PA, USA, 2004. [Google Scholar]
- Tirrell, M.; Kokkoli, E.; Biesalski, M. The role of surface science in bioengineered materials. Surf. Sci 2002, 500, 61–83. [Google Scholar]
- Geiger, B.; Bershadsky, A. Exploring the neighborhood: Adhesion-coupled cell mechanosensors. Cell 2002, 110, 139–142. [Google Scholar]
- Lodish, H.; Berk, A.; Zipursky, L.; Matsudaira, P.; Baltimore, D.; Darnell, J. Molecular Cell Biology, 4th ed; W.H. Freeman: New York, NY, USA, 2000. [Google Scholar]
- Critchley, D.R. Focal adhesions—the cytoskeletal connection. Curr. Opin. Cell Biol 2000, 12, 133–139. [Google Scholar]
- Calderwood, D.A. Talin controls integrin activation. Biochem. Soc. Trans 2004, 32, 434–437. [Google Scholar]
- Hemmings, L.; Rees, D.J.G.; Ohanian, V.; Bolton, S.J.; Gilmore, A.P.; Patel, B.; Priddle, H.; Trevithick, J.E.; Hynes, R.O.; Critchley, D.R. Talin contains three actin-binding sites each of which is adjacent to a vinculin-binding site. J. Cell Sci 1996, 109, 2715–2726. [Google Scholar]
- Lee, S.E.; Kamm, R.D.; Mofrad, M.R.K. Force-induced activation of Talin and its possible role in focal adhesion mechanotransduction. J. Biomech 2007, 40, 2096–2106. [Google Scholar]
- Kamm, R.D.; Mofrad, M.R.K. Cellular Mechanotransduction; Mofrad, M.R.K., Kamm, R.D., Eds.; Cambridge University Press: Cambridge, UK, 2010; Volume Chapter 1, p. 1. [Google Scholar]
- Winkler, J.; Lunsdorf, H.; Jockusch, B.M. The ultrastructure of chicken gizzard vinculin as visualized by high-resolution electron microscopy. J. Struct. Biol 1996, 116, 270–277. [Google Scholar]
- Bakolitsa, C.; de Pereda, J.M.; Bagshaw, C.R.; Critchley, D.R.; Liddington, R.C. Crystal structure of the vinculin tail suggests a pathway for activation. Cell 1999, 99, 603–613. [Google Scholar]
- Xu, W.M.; Coll, J.L.; Adamson, E.D. Rescue of the mutant phenotype by reexpression of full-length vinculin in null F9 cells; effects on cell locomotion by domain deleted vinculin. J. Cell Sci 1998, 111, 1535–1544. [Google Scholar]
- Priddle, H.; Hemmings, L.; Monkley, S.; Woods, A.; Patel, B.; Sutton, D.; Dunn, G.A.; Zicha, D.; Critchley, D.R. Disruption of the talin gene compromises focal adhesion assembly in undifferentiated but not differentiated embryonic stem cells. J. Cell Biol 1998, 142, 1121–1133. [Google Scholar]
- Kaverina, I.; Krylyshkina, O.; Small, J.V. Regulation of substrate adhesion dynamics during cell motility. Int. J. Biochem. Cell Biol 2002, 34, 746–761. [Google Scholar]
- Geiger, B.; Bershadsky, A. Assembly and mechanosensory function of focal contacts. Curr. Opin. Cell Biol 2001, 13, 584–592. [Google Scholar]
- Wang, N.; Butler, J.P.; Ingber, D.E. Mechanotransduction across the cell surface and through the cytoskeleton. Science 1993, 260, 1124–1127. [Google Scholar]
- Choquet, D.; Felsenfeld, D.P.; Sheetz, M.P. Extracellular matrix rigidity causes strengthening of integrin-cytoskeleton linkages. Cell 1997, 88, 39–48. [Google Scholar]
- Koo, L.Y.; Irvine, D.J.; Mayes, A.M.; Lauffenburger, D.A.; Griffith, L.G. Co-regulation of cell adhesion by nanoscale RGD organization and mechanical stimulus. J. Cell Sci 2002, 115, 1423–1433. [Google Scholar]
- Wang, H.B.; Dembo, M.; Hanks, S.K.; Wang, Y. Focal adhesion kinase is involved in mechanosensing during fibroblast migration. Proc. Natl. Acad. Sci. USA 2001, 98, 11295–11300. [Google Scholar]
- Suska, F.; Emanuelsson, L.; Johansson, A.; Tengvall, P.; Thomsen, P. Fibrous capsule formation around titanium and copper. J. Biomed. Mater. Res. A 2007, 85, 888–896. [Google Scholar]
- Andersson, M.; Suska, F.; Johansson, A.; Berglin, M.; Emanuelsson, L.; Elwing, H.; Thomsen, P. Effect of molecular mobility of polymeric implants on soft tissue reactions: an in vivo study in rats. J. Biomed. Mater. Res. A 2007, 84, 652–660. [Google Scholar]
- Biggs, M.J.; Richards, R.G.; Dalby, M.J. Nanotopographical modification: A regulator of cellular function through focal adhesions. Nanomedicine 2010, 6, 619–633. [Google Scholar]
- Cuvelier, D.; Théry, M.; Chu, Y.S.; Dufour, S.; Thiéry, J.P.; Bornens, M.; Nassoy, P.; Mahadevan, L. The universal dynamics of cell spreading. Curr. Biol 2007, 17, 694–699. [Google Scholar]
- Oleschuk, R.D.; Mccomb, M.E.; Chow, A.; Ens, W.; Standing, K.G.; Perreault, H.; Marois, Y.; King, M. Characterization of plasma proteins adsorbed onto biomaterials. By MALDI-TOFMS. Biomaterials 2000, 21, 1701–1710. [Google Scholar]
- Suh, C.W.; Kim, M.Y.; Choo, J.B.; Kim, J.K.; Kim, H.K.; Lee, E.K. Analysis of protein adsorption characteristics to nano-pore silica particles by using confocal laser scanning microscopy. J. Biotechnol 2004, 112, 267–277. [Google Scholar]
- Dettin, M.; Conconi, M.T.; Gambaretto, R.; Bagno, A.; Di Bello, C.; Menti, A.M.; Grandi, C.; Parnigotto, P.P. Effect of synthetic peptides on osteoblast adhesion. Biomaterials 2005, 26, 4507–4515. [Google Scholar]
- Keselowsky, B.G.; Collard, D.M.; Garcia, A.J. Surface chemistry modulates focal adhesion composition and signaling through changes in integrin binding. Biomaterials 2004, 25, 5947–5954. [Google Scholar]
- Hench, L.L.; Polak, J.M. Third-generation biomedical materials. Science 2002, 295, 1014–1017. [Google Scholar]
- Badylak, S.F. The extracellular matrix as a biologic scaffold material. Biomaterials 2007, 28, 3587–3593. [Google Scholar]
- Stevens, M.M.; George, J.H. Exploring and engineering the cell surface interface. Science 2005, 310, 1135–1138. [Google Scholar]
- Norman, J.J.; Desai, T.A. Methods for fabrication of nanoscale topography for tissue engineering scaffolds. Ann. Biomed. Eng 2006, 34, 89–101. [Google Scholar]
- Curtis, A.; Wilkinson, C. New depths in cell behavior: Reactions of cells to nanotopography. Biochem. Soc. Symp 1999, 65, 15–26. [Google Scholar]
- Pattison, M.A.; Wurster, S.; Webster, T.J.; Haberstroh, K.M. Three-dimensional, nano-structured PLGA scaffolds for bladder tissue replacement applications. Biomaterials 2005, 26, 2491–2500. [Google Scholar]
- Lee, J.; Chu, B.H.; Chen, K.H.; Ren, F.; Lele, T.P. Randomly oriented, upright SiO2 coated nanorods for reduced adhesion of mammalian cells. Biomaterials 2009, 30, 4488–4493. [Google Scholar]
- Ramalanjaona, G.; Kempczinski, R.F.; Rosenman, J.E.; Douville, E.C.; Silberstein, E.B. The effect of fibronectin coating on endothelial cell kinetics in polytetrafluoroethylene grafts. J. Vasc. Surg 1986, 3, 264–272. [Google Scholar]
- Kesler, K.A.; Herring, M.B.; Arnold, M.P.; Glover, J.L.; Park, H.M.; Helmus, M.N.; Bendick, P.J. Enhanced strength of endothelial attachment on polyester elastomer and polytetrafluoroethylene graft surfaces with fibronectin substrate. J. Vasc. Surg 1986, 3, 58–64. [Google Scholar]
- Pratt, K.J.; Jarrell, B.E.; Williams, S.K.; Carabasi, R.A.; Rupnick, M.A.; Hubbard, F.A. Kinetics of endothelial cell-surface attachment forces. J. Vasc. Surg. 1988, 7, 591–599. [Google Scholar]
- Cozens-Roberts, C.; Lauffenburger, D.A.; Quinn, J.A. Receptor-mediated cell attachment and detachment kinetics. I. Probabilistic model and analysis. Biophys. J 1990, 58, 841–856. [Google Scholar]
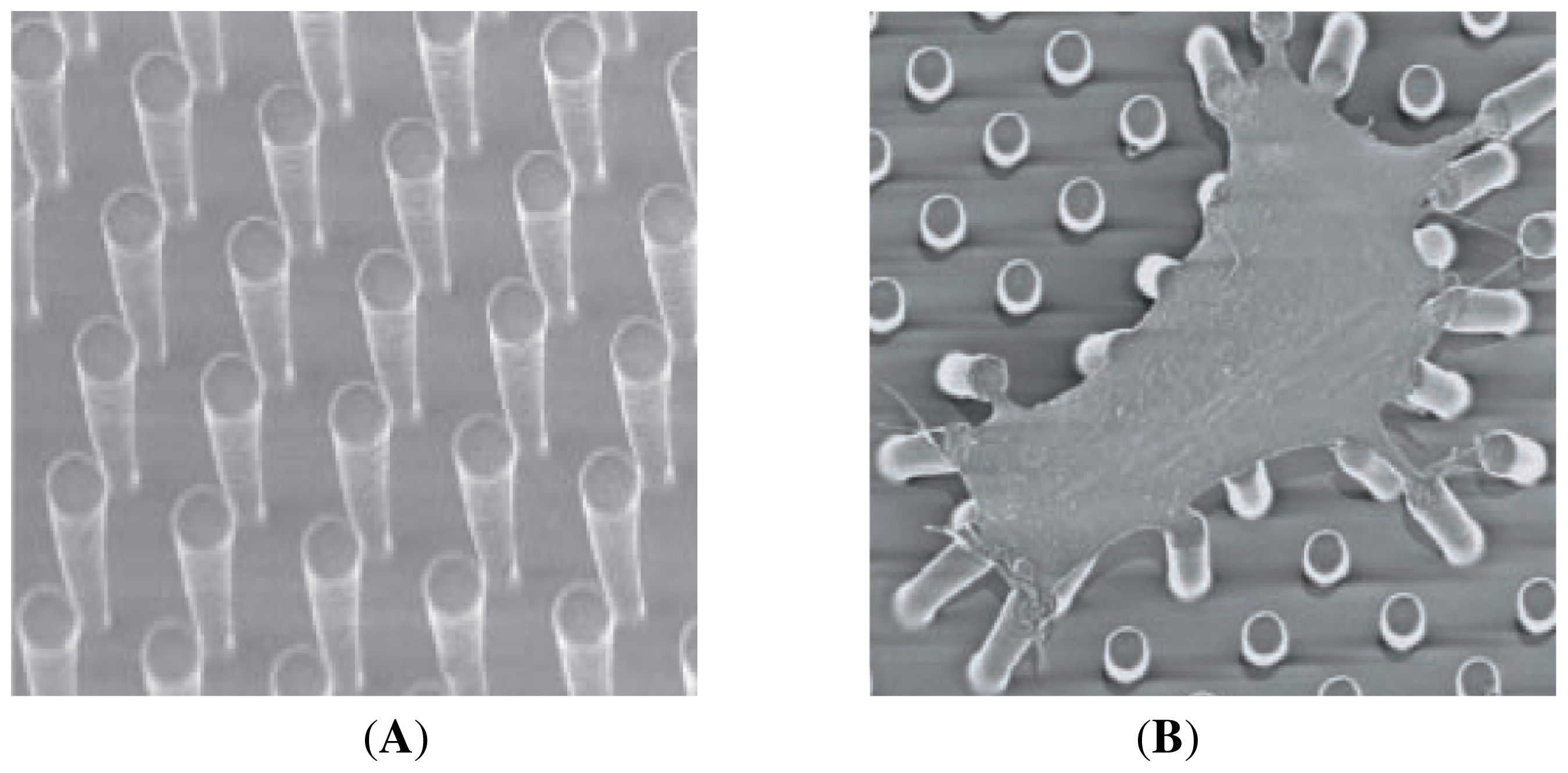
- Tan, J.L.; Tien, J.; Pirone, D.M.; Gray, D.S.; Bhadriraju, K.; Chen, C.S. Cells lying on a bed of microneedles: An approach to isolate mechanical force. Proc. Natl. Acad. Sci. USA 2003, 100, 1484–1489. [Google Scholar]
- Hammer, D.A.; Lauffenburger, D.A. A dynamical model for receptor-mediated cell adhesion to surfaces. Biophys. J 1987, 52, 475–487. [Google Scholar]
- Krishnan, R.; Oommen, B.; Walton, E.B.; Maloney, J.M.; van Vliet, K.J. Modeling and simulation of chemomechanics at the cell-matrix interface. Cell Adh. Migr 2008, 2, 83–94. [Google Scholar]
- Krammer, A.; Lu, H.; Isralewitz, B.; Schulten, K.; Vogel, V. Forced unfolding of the fibronectin type III module reveals a tensile molecular recognition switch. Proc. Natl. Acad. Sci. USA 1999, 96, 1351–1356. [Google Scholar]
- Krammer, A.; Craig, D.; Thomas, W.E.; Schulten, K.; Vogel, V. A structural model for force regulated integrin binding to fibronectin’s RGD-synergy site. Matrix Biol 2002, 21, 139–147. [Google Scholar]
- Gao, M.; Craig, D.; Vogel, V.; Schulten, K. Identifying unfolding intermediates of FN-III10 by steered molecular dynamics. J. Mol. Biol 2002, 323, 939–950. [Google Scholar]
- Grubmüller, H.; Heymann, B.; Tavan, P. Ligand binding: molecular mechanics calculation of the streptavidin-biotin rupture force. Science 1996, 271, 997–999. [Google Scholar]
- Hytönen, V.P.; Vogel, V. How force might activate talin’s vinculin binding sites: SMD reveals a structural mechanism. PLoS Comput. Biol 2008, 4. [Google Scholar] [CrossRef]
- Golji, J.; Mofrad, M.R. A molecular dynamics investigation of vinculin activation. Biophys. J 2010, 99, 1073–1081. [Google Scholar]
- Golji, J.; Lam, J.; Mofrad, M.R. Vinculin activation is necessary for complete talin binding. Biophys. J 2011, 100, 332–340. [Google Scholar]
- Skalak, R.; Zarda, P.R.; Jan, K.M.; Chien, S. Mechanics of rouleau formation. Biophys. J 1981, 35, 771–781. [Google Scholar]
- Evans, E.A. Detailed mechanics of membrane-membrane adhesion and separation. I. Continuum of molecular cross-bridges. Biophys. J 1985, 48, 175–183. [Google Scholar]
- Bell, G.I.; Dembo, M.; Bongrand, P. Cell adhesion. Competition between nonspecific repulsion and specific bonding. Biophys. J 1984, 45, 1051–1064. [Google Scholar]
- Zhu, C. Kinetics and mechanics of cell adhesion. J. Biomech 2000, 33, 23–33. [Google Scholar]
- Bell, G.I. Models for the specific adhesion of cells to cells. Science 1978, 200, 618–627. [Google Scholar]
- Dembo, M.; Torney, D.C.; Saxman, K.; Hammer, D.A. The reaction-limited kinetics of membrane-to-surface adhesion and detachment. Proc. R. Soc. London Ser. B 1988, 234, 55–83. [Google Scholar]
- Seifert, U. Rupture of multiple parallel molecular bonds under dynamic loading. Phys. Rev. Lett 2000, 84, 2750–2753. [Google Scholar]
- Seifert, U. Dynamic strength of adhesion molecules: Role of rebinding and self consistent rates. Europhys. Lett 2002, 58, 792–798. [Google Scholar]
- Chesla, S.E.; Selvaraj, P.; Zhu, C. Measuring two-dimensional receptor-ligand binding kinetics with micropipette. Biophys. J 1998, 75, 1553–1572. [Google Scholar]
- Piper, J.W.; Swerlick, R.A.; Zhu, C. Determining force dependence of two-dimensional receptor-ligand binding affinity by centrifugation. Biophys. J 1998, 74, 492–513. [Google Scholar]
- Kaplanski, G.; Farnarier, C.; Tissot, O.; Pierres, A.; Benoliel, A.M.; Alessi, M.C.; Kaplanski, S.; Bongrand, P. Granulocyte-endothelium initial adhesion. Analysis of transient binding events mediated by E-selectin in a laminar shear flow. Biophys. J 1993, 64, 1922–1933. [Google Scholar]
- Erdmann, T.; Schwarz, U.S. Stability of adhesion clusters under constant force. Phys. Rev. Lett 2004, 92, 102–108. [Google Scholar]
- Erdmann, T.; Schwarz, U.S. Stochastic dynamics of adhesion clusters under shared constant force and with rebinding. J. Chem. Phys 2004, 121, 8997–9017. [Google Scholar]
- Erdmann, T.; Schwarz, U.S. Adhesion clusters under shared linear loading: a stochastic analysis. Europhys. Lett 2004, 66, 603–609. [Google Scholar]
- Hammer, D.A.; Apte, S.M. Simulation of cell rolling and adhesion on surfaces in shear flow: General results and analysis of selectin-mediated neutrophil adhesion. Biophys. J 1992, 63, 35–57. [Google Scholar]
- Evans, E.; Ritchie, K. Dynamic strength of molecular adhesion bonds. Biophys. J 1997, 72, 1541–1555. [Google Scholar]
- Tees, D.F.J.; Woodward, J.T.; Hammer, D.A. Reliability theory for receptor-ligand bond dissociation. J. Chem. Phys 2001, 114, 7483–7496. [Google Scholar]
- Erdmann, T.; Schwarz, U.S. Impact of receptor-ligand distance on adhesion cluster stability. Eur. Phys. J. E 2007, 22, 123–137. [Google Scholar]
- Schwarz, U.S.; Erdmann, T.; Bischofs, I.B. Focal adhesions as mechanosensors: The two-spring model. Biosystems 2006, 83, 225–232. [Google Scholar]
- Nicolas, A.; Geiger, B.; Safran, S.A. Cell mechanosensitivity controls the anisotropy of focal adhesions. Proc. Natl. Acad. Sci. USA 2004, 101, 12520–12525. [Google Scholar]
- Besser, A.; Safran, S.A. Force-induced adsorption and anisotropic growth of focal adhesions. Biophys. J 2006, 90, 3469–3484. [Google Scholar]
- Nicolas, A.; Besser, A.; Safran, S.A. Dynamics of cellular focal adhesions on deformable substrates: consequences for cell force microscopy. Biophys. J 2008, 95, 527–539. [Google Scholar]
- Nicolas, A.; Safran, S.A. Limitation of cell adhesion by the elasticity of the extracellular matrix. Biophys. J 2006, 91, 61–73. [Google Scholar]
- Gov, N.S. Modeling the size distribution of focal adhesions. Biophys. J 2006, 91, 2844–2847. [Google Scholar]
- Bischofs, I.B.; Safran, S.A.; Schwarz, U.S. Elastic interactions of active cells with soft materials. Phys. Rev. E 2004, 69, 021911:1–021911:17. [Google Scholar]
- Stéphanou, A.; Mylona, E.; Chaplain, M.; Tracqui, P. A computational model of cell migration coupling the growth of focal adhesions with oscillatory cell protrusions. J. Theor. Biol 2008, 253, 701–716. [Google Scholar]
- Zaidel-Bar, R.; Cohen, M.; Addadi, L.; Geiger, B. Hierarchical assembly of cell-matrix adhesion complexes. Biochem. Soc. Trans 2004, 32, 416–420. [Google Scholar]
- Galbraith, C.G.; Yamada, K.M.; Sheetz, M.P. The relationship between force and focal complex development. J. Cell Biol 2002, 159, 695–705. [Google Scholar]
- Olberding, J.E.; Thouless, M.D.; Arruda, E.M.; Garikipati, K. The non-equilibrium thermodynamics and kinetics of focal adhesion dynamics. PLoS One 2010, 5, e12043. [Google Scholar]
- Freund, L.B.; Lin, Y. The role of binder mobility in spontaneous contact and implications for cell adhesion. J. Mech. Phys. Solids 2004, 52, 2455–2472. [Google Scholar]
- Wang, J.; Gao, H. Clustering instability in adhesive contact between elastic solids via diffusive molecular bonds. J. Mech. Phys. Solids 2008, 56, 251–266. [Google Scholar]
- Lin, Y. A model of cell motility leading to biphasic dependence of transport speed on adhesive strength. J. Mech. Phys. Solids 2010, 58, 502–514. [Google Scholar] [Green Version]
- Lin, Y.; Inamdar, M.; Freund, L.B. The competition between Brownian motion and adhesion in soft materials. J. Mech. Phys. Solids 2008, 56, 241–250. [Google Scholar]
- Gunst, S.J.; Fredberg, J.J. The first three minutes: Smooth muscle contraction, cytoskeletal events, and soft glasses. J. Appl. Physiol 2003, 95, 413–425. [Google Scholar]
- Kopacz, A.M; Liu, W.K.; Liu, S.Q. Simulation and prediction of endothelial cell adhesion modulated by molecular engineering. Comput. Methods Appl. Mech. Eng. 2008, 197, 2340–2352. [Google Scholar]
- Lim, C.T.; Zhou, E.H.; Quek, S.T. Mechanical models for living cells-a review. J. Biomech 2006, 39, 195–216. [Google Scholar]
- Lai, W.M.; Hou, J.S.; Mow, V.C. A triphasic theory for the swelling and deformation behaviors of articular caetilage. J. Biomech. Eng 1991, 113, 245–258. [Google Scholar]
- Janmey, P.A.; Shah, J.V.; Tang, J.X.; Stossel, T.P. Actin filament networks. Results Probl. Cell Differ 2001, 32, 181–199. [Google Scholar]
- Ingber, D.E. Cellular tensegrity: Defining new rules of biological design that govern the cytoskeleton. J. Cell Sci 1993, 104, 613–627. [Google Scholar]
- Gracheva, M.E.; Othmer, H.G. A continuum model of motility in ameboid cells. Bull. Math. Biol 2004, 66, 167–193. [Google Scholar]
- Dokukina, I.V.; Gracheva, M.E. A model of fibroblast motility on substrates with different rigidities. Biophys. J 2010, 98, 2794–2803. [Google Scholar]
- Kuusela, E.; Alt, W. Continuum model of cell adhesion and migration. J. Math. Biol 2009, 58, 135–161. [Google Scholar]
- Deshpande, V.S.; McMeeking, R.M.; Evans, A.G. A bio-chemo-mechanical model for cell contractility. Proc. Natl. Acad. Sci. USA 2006, 103, 14015–14020. [Google Scholar]
- Hill, A.V. The heat of shortening and the dynamic constants of muscle. Proc. R. Soc. Lond. Ser. B 1938, 126, 136–195. [Google Scholar]
- Deshpande, V.S.; Mrksich, M.; McMeeking, R.M.; Evans, A.G. A bio-mechanical modeling for coupling cell contractility with focal adhesion formation. J. Mech. Phys. Solids 2008, 56, 1484–1510. [Google Scholar]
- Sanz-Herrera, J.A.; Moreo, P.; García-Aznar, J.M.; Doblaré, M. On the effect of substrate curvature on cell mechanics. Biomaterials 2009, 30, 6674–6686. [Google Scholar]
- Oster, G.F.; Murray, J.D.; Harris, A.K. Mechanical aspects of mesenchymal morphogenesis. J. Embryol. Exp. Morphol 1983, 78, 83–125. [Google Scholar]
- Namy, P.; Ohayon, J.; Tracqui, P. Critical conditions for pattern formation and in vitro tubulogenesis driven by cellular traction fields. J. Theor. Biol 2004, 202, 103–120. [Google Scholar]
- Murray, J.D. Mathematical Biology, 3rd ed; Springer: New York, NY, USA, 2005. [Google Scholar]
- Moreo, P.; García-Aznar, J.M.; Doblaré, M. Modeling mechanosensing and its effect on the migration and proliferation of adherent cells. Acta Biomater 2008, 4, 613–621. [Google Scholar]
- Pérez, M.A.; Prendergast, P.J. Random-walk models of cell dispersal included in mechanobiological simulations of tissue differentiation. J. Biomech 2007, 40, 2244–2253. [Google Scholar]
- Byrne, D.P.; Lacroix, D.; Planell, J.A.; Kelly, D.J.; Prendergast, P.J. Simulation of tissue differentiation in a scaffold as a function of porosity, Young’s modulus and dissolution rate: Application of mechanobiological models in tissue engineering. Biomaterials 2007, 28, 5544–5554. [Google Scholar]
- Khayyeri, H.; Checa, S.; Tägil, M.; Prendergast, P.J. Corroboration of mechanobiological simulations of tissue differentiation in an in vivo bone chamber using a lattice-modeling approach. J. Orthop. Res 2009, 27, 1659–1666. [Google Scholar]
- Checa, S.; Prendergast, P.J. Effect of cell seeding and mechanical loading on vascularization and tissue formation inside a scaffold: Amechano-biological model using a lattice approach to simulate cell activity. J. Biomech 2010, 43, 961–968. [Google Scholar]
- Sandino, C.; Checa, S.; Prendergast, P.J.; Lacroix, D. Simulation of angiogenesis and cell differentiation in a CaP scaffold subjected to compressive strains using a lattice modeling approach. Biomaterials 2010, 31, 2446–2452. [Google Scholar]
- Sanz-Herrera, J.A.; García-Aznar, J.M.; Doblaré, M. On scaffold designing for bone regeneration: A computational multiscale approach. Acta Biomater 2009, 5, 219–229. [Google Scholar]
- Checa, S.; Prendergast, P.J. A mechanobiological model for tissue differentiation that includes angiogenesis: A lattice-based modeling approach. Ann. Biomed. Eng 2009, 37, 129–145. [Google Scholar]
- Sandino, C.; Planell, J.A.; Lacroix, D. A finite element study of mechanical stimuli in scaffolds for bone tissue engineering. J. Biomech 2008, 41, 1005–1014. [Google Scholar]
- Hollister, S.J.; Maddox, R.D.; Taboas, J.M. Optimal design and fabrication of scaffolds to mimic tissue properties and satisfy biological constraints. Biomaterials 2002, 23, 4095–4103. [Google Scholar]
- Taboas, J.M.; Maddox, R.D.; Krebsbach, P.H.; Hollister, S.J. Indirect solid free form fabrication of local and global porous, biomimetic and composite 3D polymer–ceramic scaffolds. Biomaterials 2003, 24, 181–194. [Google Scholar]
- Adachi, T.; Osako, Y.; Tanaka, M.; Hojo, M.; Hollister, S.J. Framework for optimal design of porous scaffold microstructure by computational simulation of bone regeneration. Biomaterials 2006, 27, 3964–3972. [Google Scholar]
- Sanz-Herrera, J.A.; Garcia-Aznar, J.M.; Doblare, M. Micro–macro numerical modeling of bone regeneration in tissue engineering. Comput. Methods Appl. Mech. Eng 2008, 197, 3092–3107. [Google Scholar]
- Sanz-Herrera, J.A.; Kasper, C.; van Griensven, M.; Garcia-Aznar, J.M.; Ochoa, I.; Doblaré, M. Mechanical and flow characterization of Sponceram carriers: Evaluation by homogenization theory and experimental validation. J. Biomed. Mater. Res. B 2008, 87, 42–48. [Google Scholar]
- Sanz-Herrera, J.A.; García-Aznar, J.M.; Doblaré, M. A mathematical approach to bone tissue engineering. Philos. Trans. R. Soc. Ser. A 2009, 367, 2055–2078. [Google Scholar]
- Carlier, A.; Chai, Y.C.; Moesen, M.; Theys, T.; Schrooten, J.; van Oosterwyck, H.; Geris, L. Designing optimal calcium phosphate scaffold-cell combinations using an integrative model-based approach. Acta Biomater 2011, 7, 3573–3585. [Google Scholar]
- Karcher, H.; Lee, S.E.; Kaazempur-Mofrad, M.R.; Kamm, R.D. A coarse-grained model for force-induced protein deformation and kinetics. Biophys. J 2006, 90, 2686–2697. [Google Scholar]
- Buehler, M.J. Atomistic and continuum modeling of mechanical properties of collagen: Elasticity, fracture, and self-assembly. J. Mater. Res 2006, 21, 1947–1961. [Google Scholar]
- Xiao, S.P.; Belytschko, T. A bridging domain method for coupling continua with molecular dynamics. Comput. Methods Appl. Mech. Eng 2004, 193, 1645–1669. [Google Scholar]
- Reina-Romo, E.; Sanz-Herrera, J.A. Multiscale simulation of particle-reinforced elastic–plastic adhesives at small strains. Comput. Methods Appl. Mech. Eng 2011, 200, 2211–2222. [Google Scholar]
- DiMilla, P.A.; Barbee, K.; Lauffenburger, D.A. Mathematical model for the effects of adhesion and mechanics on cell migration speed. Biophys. J 1991, 60, 15–37. [Google Scholar]
- Mogilner, A.; Verzi, D.W. A simple 1-D physical model for the crawling nematode sperm cell. J. Stat. Phys 2003, 110, 1169–1189. [Google Scholar]
- Marée, A.F.; Jilkine, A.; Dawes, A.; Grieneisen, V.A.; Edelstein-Keshet, L. Polarization and movement of keratocytes: A multiscale modeling approach. Bull. Math. Biol 2006, 68, 1169–1211. [Google Scholar]
- Satulovsky, J.; Lui, R.; Wang, Y.L. Exploring the control circuit of cell migration by mathematical modeling. Biophys. J 2008, 94, 3671–3683. [Google Scholar]
- Zaman, M.H.; Kamm, R.D.; Matsudaira, P.; Lauffenburger, D.A. Computational model for cell migration in three-dimensional matrices. Biophys. J 2005, 89, 1389–1397. [Google Scholar]
- Mogilner, A. Mathematics of cell motility: Have we got its number? J. Math. Biol 2009, 58, 105–134. [Google Scholar]
- Flaherty, B.; McGarry, J.P.; McHugh, P.E. Mathematical models of cell motility. Cell Biochem. Biophys 2007, 49, 14–28. [Google Scholar]
- Boulbitch, A.; Guttenberg, Z.; Sackmann, E. Kinetics of membrane adhesion mediated by ligand-receptor interaction studied with a biomimetic system. Biophys. J 2001, 81, 2743–2751. [Google Scholar]
- Balaban, N.Q.; Schwarz, U.S.; Riveline, D.; Goichberg, P.; Tzur, G.; Sabanay, I.; Mahalu, D.; Safran, S.; Bershadsky, A.; Addadi, L.; et al. Force and focal adhesion assembly: A close relationship studied using elastic micropatterned substrates. Nat. Cell Biol 2001, 3, 466–472. [Google Scholar]
- Sagvolden, G.; Giaever, I.; Pettersen, E.O.; Feder, J. Cell adhesion force microscopy. Proc. Natl. Acad. Sci. USA 1999, 96, 471–476. [Google Scholar]
- del Alamo, J.C.; Meili, R.; Alonso-Latorre, B.; Rodríguez-Rodríguez, J.; Aliseda, A.; Firtel, R.A.; Lasheras, J.C. Spatio-temporal analysis of eukaryotic cell motility by improved force cytometry. Proc. Natl. Acad. Sci. USA 2007, 104, 13343–13348. [Google Scholar]
© 2011 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Sanz-Herrera, J.A.; Reina-Romo, E. Cell-Biomaterial Mechanical Interaction in the Framework of Tissue Engineering: Insights, Computational Modeling and Perspectives. Int. J. Mol. Sci. 2011, 12, 8217-8244. https://doi.org/10.3390/ijms12118217
Sanz-Herrera JA, Reina-Romo E. Cell-Biomaterial Mechanical Interaction in the Framework of Tissue Engineering: Insights, Computational Modeling and Perspectives. International Journal of Molecular Sciences. 2011; 12(11):8217-8244. https://doi.org/10.3390/ijms12118217
Chicago/Turabian StyleSanz-Herrera, Jose A., and Esther Reina-Romo. 2011. "Cell-Biomaterial Mechanical Interaction in the Framework of Tissue Engineering: Insights, Computational Modeling and Perspectives" International Journal of Molecular Sciences 12, no. 11: 8217-8244. https://doi.org/10.3390/ijms12118217




