A LEA Gene Regulates Cadmium Tolerance by Mediating Physiological Responses
Abstract
:1. Introduction
2. Results
2.1. Time-Course Analysis of TaLEA1 Expression
2.2. Generation of Transgenic Poplar
2.3. Comparison of the Growth between TaLEA-Transformed and WT Poplar Plants after CdCl2 Exposure
2.4. Analyses of POD Activity in TaLEA1-Overexpressing Plants
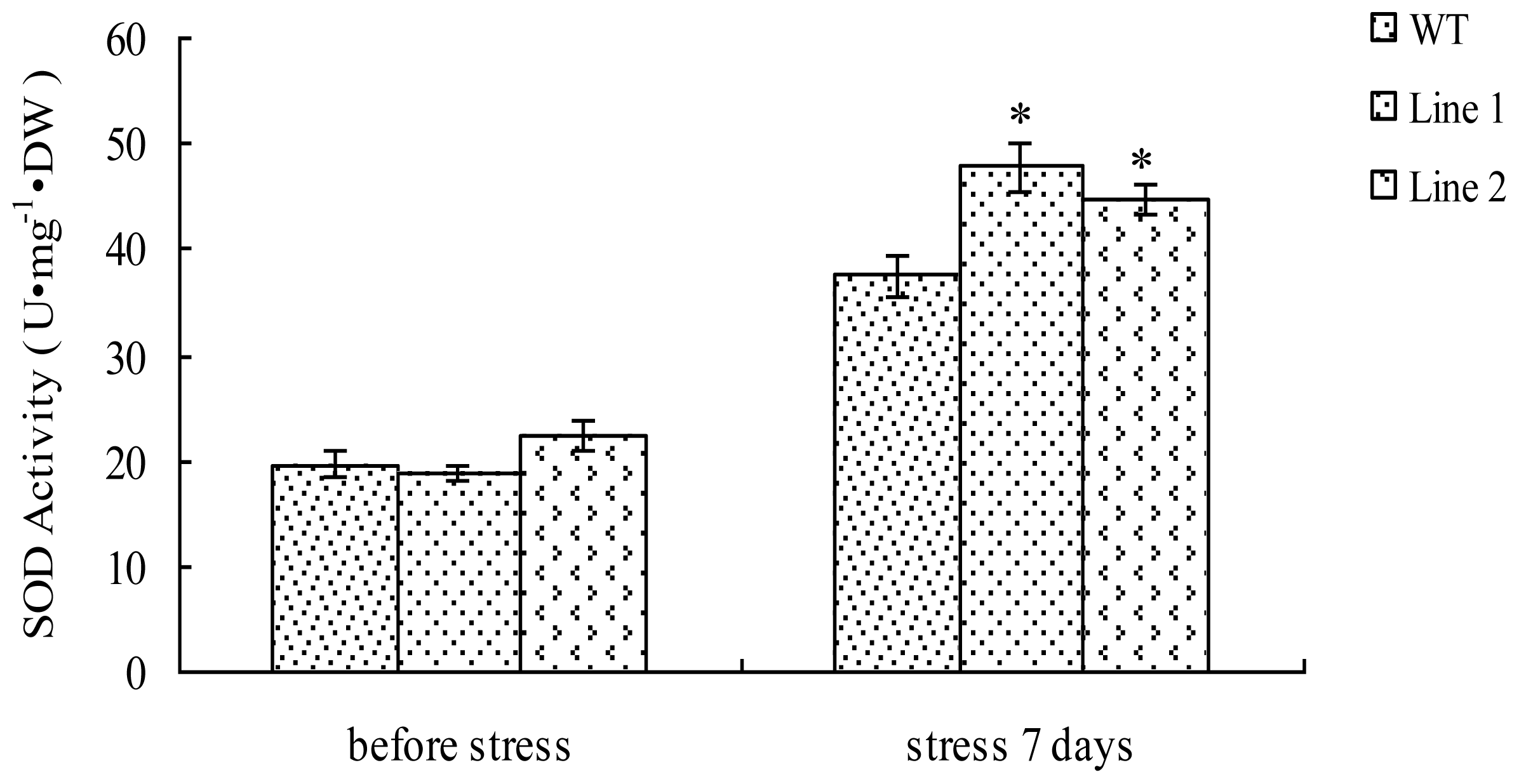
2.5. Analysis of the SOD Activity in TaLEA1-Overexpressing Plants
2.6. Analysis of MDA Content in TaLEA1-Overexpressing Plants
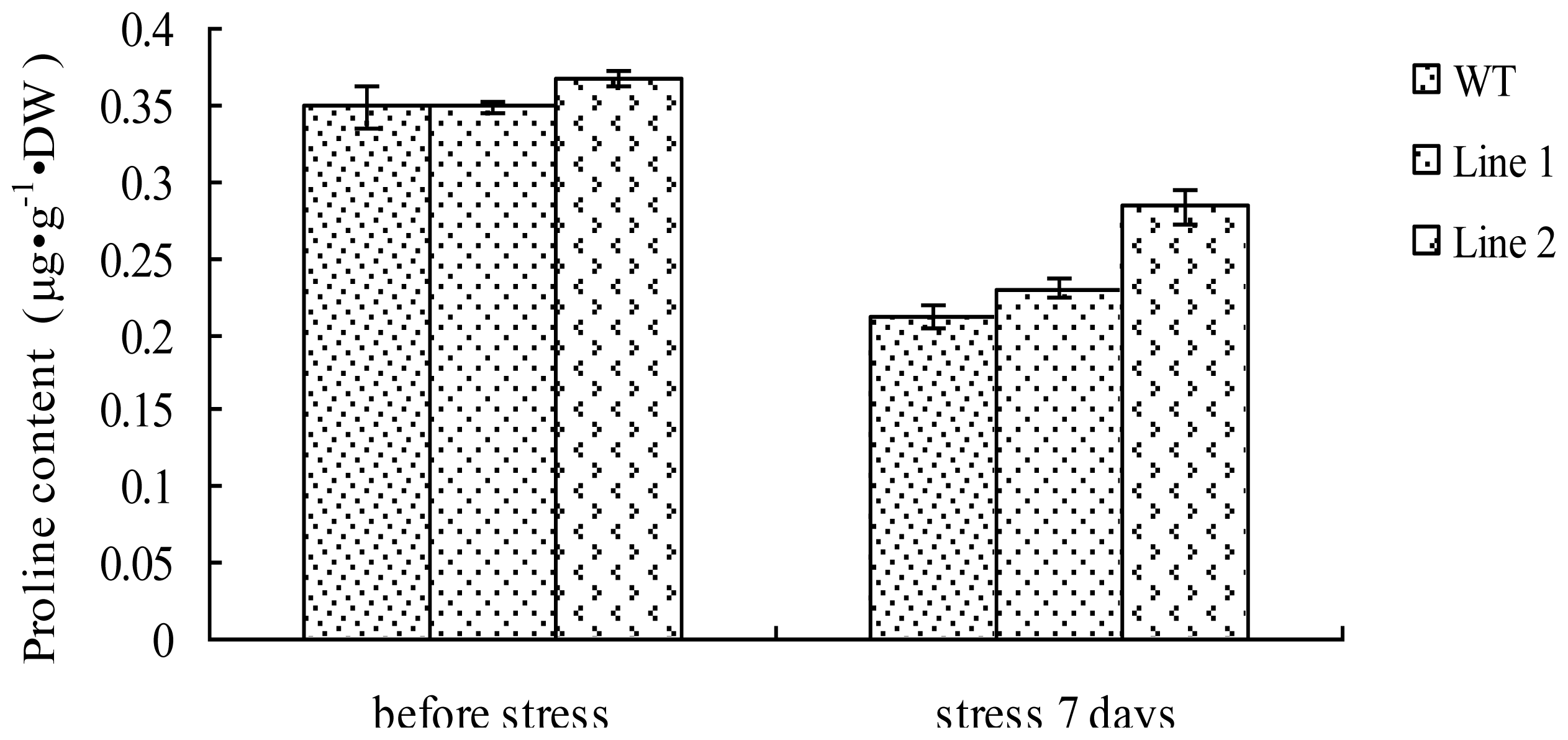
2.7. Analysis of the Proline Content in TaLEA1-Overexpressing Plants
2.8. Subcellular Localization of TaLEA1
3. Discussion
4. Experimental Section
4.1. Plant Culture and Treatments
4.2. Expression Analysis of TaLEA1 in Response to Different Stresses
4.3. Plant Transformation
4.4. Growth Analysis of Transgenic and WT Poplar Cultured in CdCl2 Media
4.5. Determination of Heavy Metal Tolerance of the TaLEA1 Transgenic Poplar Plants
4.6. Measurement of the Physiological Parameters in Transgenic Poplar Plants
4.7. Subcellular Localization of the TaLEA Gene
4.8. Data Analyses
5. Conclusions
Acknowledgments
References
- Blackman, S.A.; Wettlaufar, S.H.; Obendorf, R.L.; Leopold, A.C. Maturation proteins associated with desiccation tolerance in soybean. Plant Physiol 1991, 96, 868–874. [Google Scholar]
- Ali-Benali, M.A.; Alary, R.; Joudrier, P.; Gautier, M.F. Comparative expression of five Lea genes during wheat seed development and in response to abiotic stresses by real-time quantitative RT-PCR. Biochim. Biophys. Acta 2005, 1730, 56–65. [Google Scholar]
- Cuming, A.C.; Cho, S.H.; Kamisugi, Y.; Graham, H.; Quatrano, R.S. Microarray analysis of transcriptional responses to abscisic acid and osmotic, salt, and drought stress in the moss, Physcomitrella patens. New Phytol 2007, 176, 275–287. [Google Scholar]
- Serrano, R.; Montesinos, C. Molecular bases of desiccation tolerance in plant cells and potential applications in food dehydration. Food Sci. Technol. Int 2003, 9, 157–161. [Google Scholar]
- Goyal, K.; Walton, L.J.; Tunnacliffe, A. LEA proteins prevent protein aggregation due to water stress. Biochem. J 2005, 388, 151–157. [Google Scholar]
- Tunnacliffe, A.; Wise, M.J. The continuing conundrum of the LEA proteins. Naturwissenschaften 2007, 94, 791–812. [Google Scholar]
- Soulages, J.L.; Kim, K.; Walters, C.; Cushman, J.C. Temperature induced extended helix/random coil transitions in a group 1 late embryogenesis-abundant protein from soybean. Plant Physiol 2002, 128, 822–832. [Google Scholar]
- Xiao, B.; Huang, Y.; Tang, N.; Xiong, L. Over-expression of a LEA gene in rice improves drought resistance under the field conditions. Theor. Appl. Genet 2007, 115, 35–46. [Google Scholar]
- Shimamura, C.; Ohno, R.; Nakamura, C.; Takumi, S. Improvement of freezing tolerance in tobacco plants expressing a cold-responsive and chloroplast-targeting protein WCOR15 of wheat. J. Plant Physiol 2006, 163, 213–219. [Google Scholar]
- Wang, L.; Li, X.; Chen, S.; Liu, G. Enhanced drought tolerance in transgenic Leymus chinensis plants with constitutively expressed wheat TaLEA3. Biotechnol. Lett 2009, 31, 313–319. [Google Scholar]
- Gianazza, E.; Wait, R.; Sozzi, A.; Regondi, S.; Saco, D.; Labra, M.; Agradi, E. Growth and protein profile changes in Lepidium sativum L. plantlets exposed to cadmium. Environ. Exp. Bot 2007, 59, 179–187. [Google Scholar]
- Leshem, Y.; Seri, L.; Levine, A. Induction of phosphatidylinositol 3-kinase-mediated endocytosis by salt stress leads to intracellular production of reactive oxygen species and salt tolerance. Plant J 2007, 51, 185–197. [Google Scholar]
- Ostergaard, L.; Pedersen, A.G.; Jespersen, H.M.; Brunak, S.; Welinder, K.G. Computational analyses and annotations of the Arabidopsis peroxidase gene family. FEBS Lett 1998, 433, 98–102. [Google Scholar]
- Lin, J.N.; Kao, C.H. Effect of oxidative stress caused by hydrogen peroxide on senescence of rice leaves. Bot. Bull. Acad. Sin 1998, 39, 161–165. [Google Scholar]
- Kim, Y.H.; Kim, C.Y.; Song, W.K.; Park, D.S.; Kwon, S.Y.; Lee, H.S.; Bang, J.W.; Kwak, S.S. Overexpression of sweetpotato swpa4 peroxidase results in increased hydrogen peroxide production and enhances stress tolerance in tobacco. Planta 2008, 227, 867–881. [Google Scholar]
- Koussevitzky, S.; Suzuki, N.; Huntington, S.; Armijo, L.; Sha, W.; Cortes, D.; Shulaev, V.; Mittler, R. Ascorbate peroxidase 1 plays a key role in the response of Arabidopsis thaliana to stress combination. J. Biol. Chem 2008, 283, 34197–34203. [Google Scholar]
- Sunkar, R.; Bartels, D.; Kirch, H.H. Overexpression of a stress inducible aldehyde dehydrogenase gene from Arabidopsis thaliana in transgenic plants improves stress tolerance. Plant J 2003, 35, 452–464. [Google Scholar]
- Kishor, P.; Hong, Z.; Miao, G.H.; Hu, C.; Verma, D. Overexpression of [delta]-Pyrroline-5-carboxylate synthetase increases proline production and confers osmotolerance in transgenic plants. Plant Physiol 1995, 108, 1387–1394. [Google Scholar]
- Silva-Ortega, C.O.; Ochoa-Alfaro, A.E.; Reyes-Agüero, J.A.; Aguado-Santacruz, G.A.; Jiménez-Bremont, J.F. Salt stress increases the expression of p5cs gene and induces proline accumulation in cactus pear. Plant Physiol. Biochem 2008, 46, 82–92. [Google Scholar]
- Ban, Q.; Liu, G.; Wang, Y. A DREB gene from Limonium bicolor mediates molecular and physiological responses to copper stress in transgenic tobacco. J. Plant Physiol 2011, 168, 449–458. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔC(T) method. Methods 2001, 25, 402–408. [Google Scholar]
- Tzfira, T.; Jensen, C.S.; Wang, W.X.; Zuker, A.; Vinocur, B.; Altman, A.; Vainstein, A. Transgenic Populus tremula: A step-by-step protocol for its Agrobacterium-mediated transformation. Plant Mol. Biol. Rep 1997, 15, 219–235. [Google Scholar]
- Wang, Y.; Gao, C.; Liang, Y.; Wang, C.; Yang, C.; Liu, G. A novel bZIP gene from Tamarix hispida mediates physiological responses to salt stress in tobacco plants. J. Plant Physiol 2009, 167, 222–230. [Google Scholar]
- Bates, L.S.; Waldron, R.P.; Teare, I.D. Rapid determination of free proline for water stress studies. Plant Soil 1973, 39, 205–208. [Google Scholar]






| Roots Number | Leaf Length (cm) | |||
|---|---|---|---|---|
| Plants | Normal growth condition | Stress condition | Normal growth condition | Stress condition |
| WT | 41 ± 0.52 | 11 ± 0.65 | 2.67 ± 0.13 | 1.65 ± 0.06 |
| Line 1 | 47.5 ± 0.91 * | 57 ± 1.03 * | 2.39 ± 0.05 | 2.33 ± 0.01 * |
| Line 2 | 32.5 ± 0.84 * | 56.5 ± 0.98 * | 2.40 ± 0.08 | 2.43 ± 0.07 * |
| Gene | Forward and Reverse Primers (5′–3′) |
|---|---|
| TaLEA | 5-TAAATCACGAGGCGGCGAAACA-3 5-AACTCCAGCAACGTCATGCGAAGA-3 |
| α-actin | 5-AAACAATGGCTGATGCTG-3 5-ACAATACCGTGCTCAATAGG-3 |
| α-tubulin | 5-CACCCACCGTTGTTCCAG-3 5-ACCGTCGTCATCTTCACC-3 |
© 2012 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Gao, C.; Wang, C.; Zheng, L.; Wang, L.; Wang, Y. A LEA Gene Regulates Cadmium Tolerance by Mediating Physiological Responses. Int. J. Mol. Sci. 2012, 13, 5468-5481. https://doi.org/10.3390/ijms13055468
Gao C, Wang C, Zheng L, Wang L, Wang Y. A LEA Gene Regulates Cadmium Tolerance by Mediating Physiological Responses. International Journal of Molecular Sciences. 2012; 13(5):5468-5481. https://doi.org/10.3390/ijms13055468
Chicago/Turabian StyleGao, Caiqiu, Chao Wang, Lei Zheng, Liuqiang Wang, and Yucheng Wang. 2012. "A LEA Gene Regulates Cadmium Tolerance by Mediating Physiological Responses" International Journal of Molecular Sciences 13, no. 5: 5468-5481. https://doi.org/10.3390/ijms13055468
APA StyleGao, C., Wang, C., Zheng, L., Wang, L., & Wang, Y. (2012). A LEA Gene Regulates Cadmium Tolerance by Mediating Physiological Responses. International Journal of Molecular Sciences, 13(5), 5468-5481. https://doi.org/10.3390/ijms13055468




