Genetic Diversity of Pinus nigra Arn. Populations in Southern Spain and Northern Morocco Revealed By Inter-Simple Sequence Repeat Profiles †
Abstract
:1. Introduction
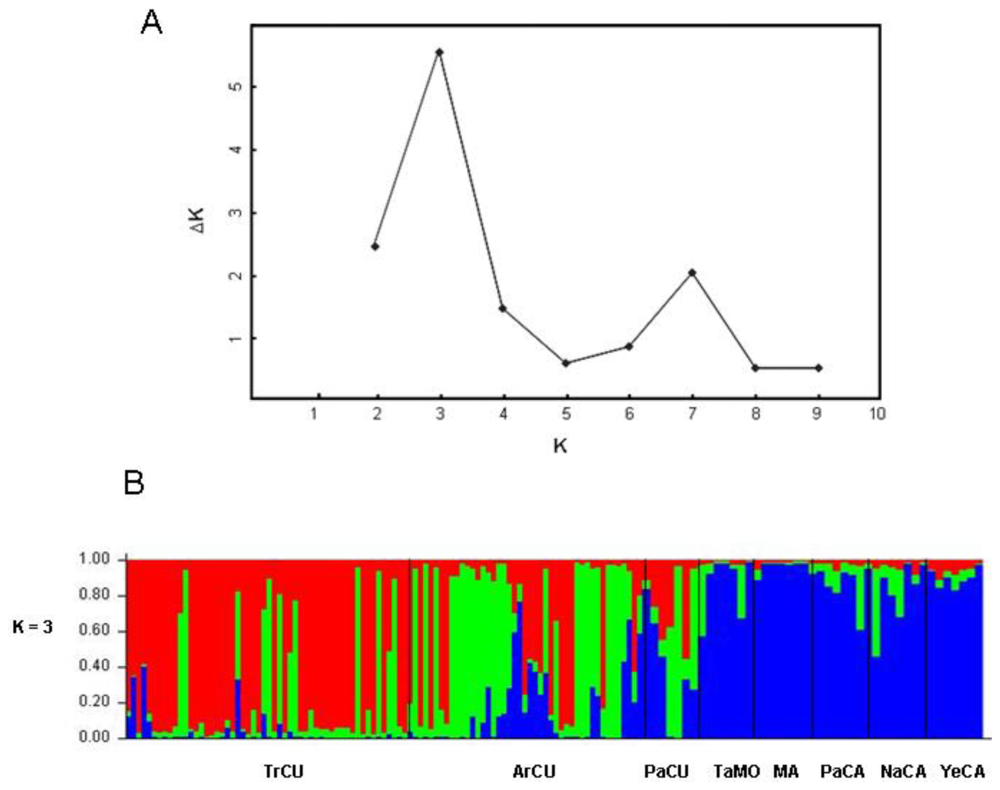
2. Results and Discussion
3. Experimental Section
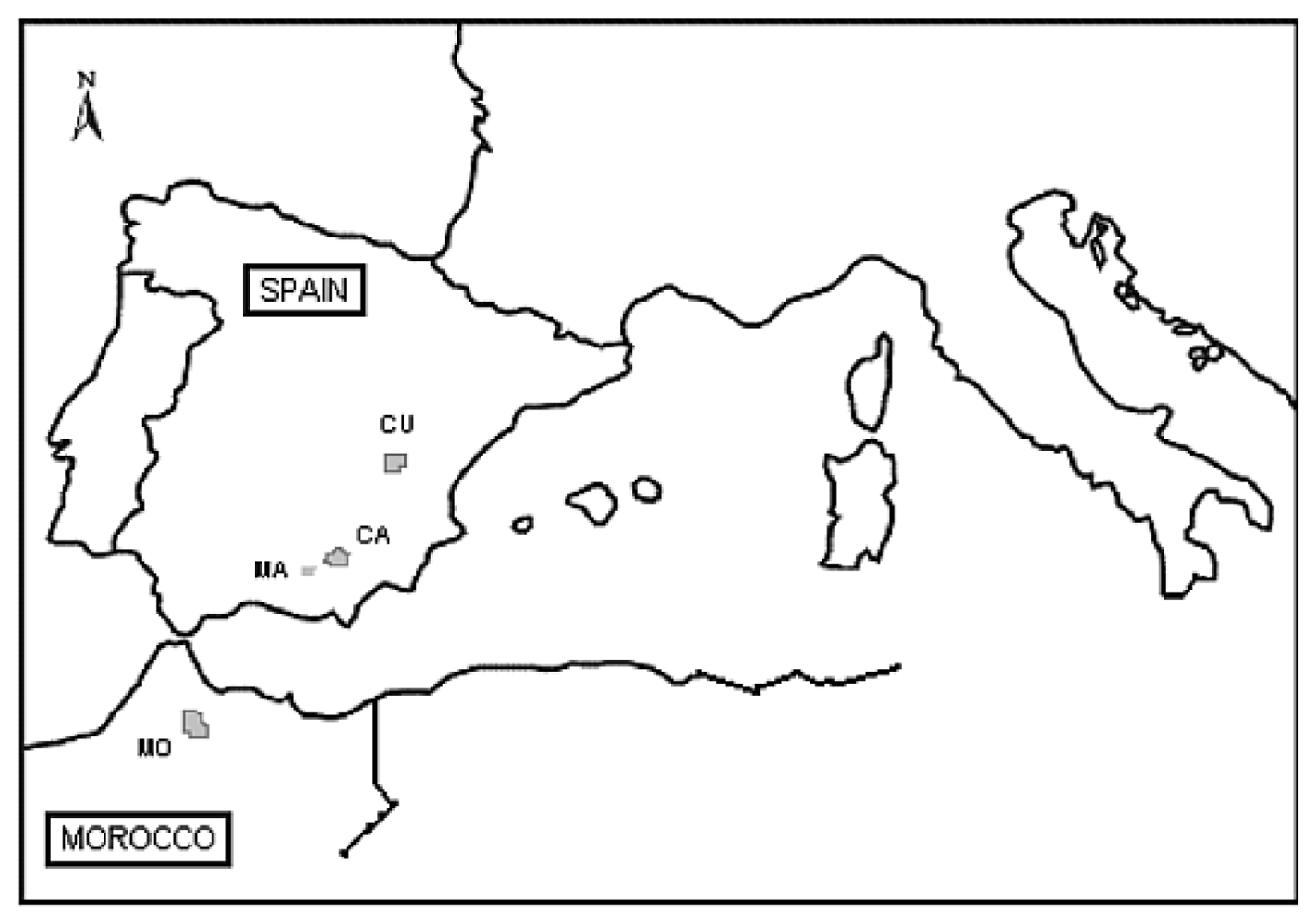
3.1. Plant Material and Population Selection
3.2. DNA Extraction
3.3. DNA Amplification
3.4. Data Analysis
4. Conclusions
Acknowledgments
- †This article is dedicated to the memory of Dr. Antonio Del Cerro Barja (1952–2010).
References
- Bogunic, F.; Muratovic, E.; Ballian, D.; Siljak-Yakovlev, S.; Brown, S. Genome size stability among five subspecies of Pinus nigra Arnold s.l. Environ. Exp. Bot 2007, 59, 354–360. [Google Scholar]
- Kerr, G. Natural regeneration of corsican pine (Pinus nigra subsp. laricio) in Great Britain. Forestry 2000, 73, 479–487. [Google Scholar]
- Lucas-Borja, M.E.; Fonseca, T.; Parresol, B.R.; Silva-Santos, P.; García-Morote, F.A.; Tíscar-Oliver, P.A. Modelling Spanish black pine seedling emergence: Establishing management strategies for endangered forest areas. For. Ecol. Manag 2011, 262, 195–202. [Google Scholar]
- Tíscar, P.A. Condicionantes y limitaciones de la regeneración natural en un pinar oromediterráneo de Pinus nigra subsp. salzmannii. Investig. Agrar. Sist. Recur. For 2003, 12, 55–64. [Google Scholar]
- Gernandt, D.S.; Magallón, S.; López, G.G.; Flores, O.Z.; Willyard, A.; Liston, A. Use of simultaneous analyses to guide fossil-based calibrations of pinaceae phylogeny. Int. J. Plant Sci 2008, 169, 1086–1099. [Google Scholar]
- Naydenov, K.D.; Tremblay, F.M.; Fenton, N.J.; Alexandrov, A. Structure of Pinus nigra Arn. populations in Bulgaria revealed by chloroplast microsatellites and terpenes analysis: Provenance tests. Biochem. Syst. Ecol 2006, 34, 562–574. [Google Scholar]
- Afzal-Rafii, Z.; Dodd, R.S. Chloroplast DNA supports a hypothesis of glacial refugia over postglacial recolonization in disjunct populations of black pine (Pinus nigra) in western Europe. Mol. Ecol 2007, 16, 723–736. [Google Scholar]
- Littell, J.S.; Oneil, E.E.; McKenzie, D.; Hicke, J.A.; Lutz, J.A.; Norheim, R.A.; Elsner, M.M. Forest ecosystems, disturbance, and climatic change in Washington State, USA. Clim. Change 2011, 102, 129–158. [Google Scholar]
- Oleksyn, J.; Tjoelker, M.G.; Reich, P.B. Adaptation to changing environment in Scots pine populations across a latitudinal gradient. Silva Fenn 1998, 32, 129–140. [Google Scholar]
- Aitken, S.N.; Yeaman, S.; Holliday, J.A.; Wang, T.; Curtis-McLane, S. Adaptation, migration or extirpation: Climate change outcomes for tree populations. Evol. Appl 2008, 1, 95–111. [Google Scholar]
- Thuiller, W.; Albert, C.; Araújo, M.B.; Berry, P.M.; Cabeza, M.; Guisan, A.; Hickler, T.; Midgley, G.F.; Paterson, J.; Schurr, F.M.; et al. Predicting global change impacts on plant species’ distributions: Future challenges. Perspect. Plant Ecol. Evol. Syst 2008, 9, 137–152. [Google Scholar]
- Chezhian, P.; Yasodha, R.; Ghosh, M. Genetic diversity analysis in a seed orchard of Eucalyptus tereticornis. New For 2010, 40, 85–99. [Google Scholar]
- Naik, D.; Singh, D.; Vartak, V.; Paranjpe, S.; Bhargava, S. Assessment of morphological and genetic diversity in Gmelina arborea Roxb. New For 2009, 38, 99–115. [Google Scholar]
- López-Aljorna, A.; Bueno, M.Ã.; Aguinagalde, I.; Martín, J.P. Fingerprinting and genetic variability in cork oak (Quercus suber L.) elite trees using ISSR and SSR markers. Ann. For. Sci 2007, 64, 773–779. [Google Scholar]
- Feyissa, T.; Nybom, H.; Bartish, I.V.; Welander, M. Analysis of genetic diversity in the endangered tropical tree species Hagenia abyssinica using ISSR markers. Genet. Resour. Crop Evol 2007, 54, 947–958. [Google Scholar]
- Angelone, S.; Hilfiker, K.; Holderegger, R.; Bergamini, A.; Hoebee, S.E. Regional population dynamics define the local genetic structure in Sorbus torminalis. Mol. Ecol 2007, 16, 1291–1301. [Google Scholar]
- Çengel, B.; Tayanç, Y.; Kandemir, G.; Velioglu, E.; Alan, M.; Kaya, Z. Magnitude and efficiency of genetic diversity captured from seed stands of Pinus nigra (Arnold) subsp. pallasiana in established seed orchards and plantations. New For 2011. [Google Scholar] [CrossRef]
- Wang, M.B.; Hao, Z.Z. Rangewide genetic diversity in natural populations of Chinese pine (Pinus tabulaeformis). Biochem. Genet 2010, 48, 590–602. [Google Scholar]
- Feng, F.J.; Han, S.J.; Wang, H.M. Genetic diversity and genetic differentiation of natural Pinus koraiensis populations. J. For. Res 2006, 17, 21–24. [Google Scholar]
- Yang, C.P.; Wei, L.; Jiang, J.; Liu, G.F.; Zhao, G.Y. Analysis of genetic diversity for nineteen populations of Pinus sibirica Du Tour with technique of ISSR. J. Northeast For. Univ 2005, 33, 1–3. [Google Scholar]
- Liu, G.F.; Dong, J.X.; Jiang, Y.; Lu, Y.F.; Jiang, J.; Zhao, G.Y. Analysis of genetic relationship in 12 species of Section Strobus with ISSR markers. J. For. Res 2005, 16, 213–215. [Google Scholar]
- Labra, M.; Grassi, F.; Sgorbati, S.; Ferrari, C. Distribution of genetic variability in southern populations of Scots pine (Pinus sylvestris) from the Alps to the Apennines. Flora 2006, 201, 468–476. [Google Scholar]
- Zhang, Z.Y.; Chen, Y.Y.; Li, D.Z. Detection of low genetic variation in a critically endangered Chinese pine, Pinus squamata, using RAPD and ISSR markers. Biochem. Genet 2005, 43, 239–249. [Google Scholar]
- Tiscar-oliver, P.A.; Lucas-Borja, M.E.; Candel-Perez, D. Vreal_IUFRO changes in the structure and composition of two Pinus nigra subsp. salzmannii forest over a century of different silvicultural treatments. For. Syst 2011, 20, 525–535. [Google Scholar]
- Nei, M. Analysis of gene diversity subdivided populations. Proc. Natl. Acad. Sci. USA 1973, 70, 3321–3323. [Google Scholar]
- Rodríguez-Sánchez, F.; Pérez-Barrales, R.; Ojeda, F.; Vargas, P.; Arroy, J. The strait of Gibraltar as a melting pot for plant biodiversity. Quat. Sci. Rev 2008, 27, 2100–2117. [Google Scholar]
- Lonergan, L.; White, N. Origin of the Betic-Rif mountain belt. Tectonics 1997, 16, 504–522. [Google Scholar]
- Duggen, S.; Hoernle, K.; van den Bogaard, P.; Rüpke, L.; Morgan, J. Deep roots of the Messinian salinity crisis. Nature 2003, 422, 602–606. [Google Scholar]
- Krijgsman, W.; Hilgen, F.; Raffi, I.; Sierro, F.; Wilson, D. Chronology, causes and progression of the Messinian salinity crisis. Nature 1999, 400, 652–655. [Google Scholar]
- Hsü, K.; Montadert, L.; Bernoulli, D.; Cita, M.; Erickson, A.; Garrison, R.; Kidd, R.; Melieres, F.; Müller, C.; Wright, R. History of the Mediterranean salinity crisis. Nature 1977, 267, 399–403. [Google Scholar]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure from multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar]
- Hamrick, J.; Godt, M. Allozyme Diversity in Plant Species. In Differentiation Patterns in Higher Plants; Urbanski, K., Ed.; Academic Press: New York NY, USA, 1989; pp. 53–67. [Google Scholar]
- Aguirre-Planter, E.; Furnier, G.R.; Eguiarte, L.E. Low levels of genetic variation within and high levels of genetic differentiation among populations of species of Abies from southern Mexico and Guatemala. Am. J. Bot 2000, 87, 362–371. [Google Scholar]
- Ingvarsson, P.R.K. Nucleotide polymorphism and linkage disequilibrium within and among natural populations of European Aspen (Populus tremula L., Salicaceae). Genetics 2005, 169, 945–953. [Google Scholar]
- de Beaulieu, J.L.; Miras, Y.; Andrieu-Ponel, V.; Guiter, F. Vegetation dynamics in north-western Mediterranean regions: Instability of the Mediterranean bioclimate. Plant Biosyst 2005, 139, 114–126. [Google Scholar]
- Martrat, B.; Grimalt, J.O.; Shackleton, N.J.; de Abreu, L.; Hutterli, M.A.; Stocker, T.F. Four climate cycles of recurring deep and surface water destabilizations on the Iberian margin. Science 2007, 317, 502–507. [Google Scholar]
- Médail, F.; Diadema, K. Glacial refugia influence plant diversity patterns in the Mediterranean Basin. J. Biogeogr 2009, 36, 1333–1345. [Google Scholar]
- Doyle, J.; Doyle, J. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull 1987, 19, 11–15. [Google Scholar]
- Popgene, version 1.32; the user-friendly shareware for population genetic analysis; Molecular Biology and Biotechnology Center, University of Alberta: Edmonton, Canada, 1997. Available online: http://www.ualberta.ca/~fyeh accessed on 23 November 2011.
- Peakall, R.O.D.; Smouse, P.E. GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar]
- Mantel, N. The detection of disease clustering and a generalized regression approach. Cancer Res 1967, 27, 209–220. [Google Scholar]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar]
- Earl, D.A.; vonHoldt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour 2012, 4, 359–361. [Google Scholar]
- Evanno, G.; Reganut, E.; Goudet, J. Detecting the number of clusters of individuals using STRUCTURE: A simulation study. Mol. Ecol 2005, 14, 2611–2620. [Google Scholar]



| Primer Name | Sequence (5′-3′) | Tm (°C) |
|---|---|---|
| ISCS14 | AGTGAGTGAGTGAGTGAGTGA | 52 |
| ISCS17 | DBDBCACCACCACCACCAC | 62 |
| ISCS19 | HVHGTGGTGGTGGTGGTG | 62 |
| ISCS20 | DHBCGACGACGACGACGA | 62 |
| ISCS21 | BDBACAACAACAACAACA | 52 |
| ISCS34 | TGTGTGTGTGTGTGTGRC | 52 |
| ISCS41 | CTCCTCCTCCTCCTCCTC | 62 |
| ISCS69 | CACACACACACACACAA | 52 |
| Population | PPL (%) | na | ne | HE | I |
|---|---|---|---|---|---|
| TrCU | 70.83 | 1.542 | 1.372 | 0.218 | 0.331 |
| ArCU | 79.17 | 1.625 | 1.410 | 0.242 | 0.366 |
| PaCU | 29.17 | 0.875 | 1.222 | 0.123 | 0.178 |
| TaMO | 54.17 | 1.208 | 1.368 | 0.207 | 0.303 |
| MA | 62.50 | 1.375 | 1.273 | 0.170 | 0.268 |
| PaCA | 29.17 | 0.875 | 1.239 | 0.130 | 0.185 |
| NaCA | 45.83 | 1.167 | 1.326 | 0.180 | 0.262 |
| YeCA | 37.50 | 1.083 | 1.228 | 0.133 | 0.199 |
| Mean | 51.04 | 1.219 | 1.305 | 0.175 | 0.262 |
| TrCU | ArCU | PaCU | TaMO | MA | PaCA | NaCA | YeCA | |
|---|---|---|---|---|---|---|---|---|
| TrCU | - | 0.037 | 0.079 | 0.249 | 0.267 | 0.223 | 0.252 | 0.244 |
| ArCU | 0.040 | - | 0.057 | 0.115 | 0.113 | 0.158 | 0.196 | 0.187 |
| PaCU | 0.087 | 0.065 | - | 0.168 | 0.171 | 0.176 | 0.230 | 0.268 |
| TaMO | 0.258 | 0.124 | 0.181 | - | 0.019 | 0.111 | 0.121 | 0.116 |
| MA | 0.274 | 0.120 | 0.183 | 0.031 | - | 0.121 | 0.161 | 0.177 |
| PaCA | 0.233 | 0.168 | 0.191 | 0.126 | 0.134 | - | 0.118 | 0.143 |
| NaCA | 0.264 | 0.208 | 0.247 | 0.138 | 0.176 | 0.136 | - | 0.046 |
| YeCA | 0.254 | 0.198 | 0.283 | 0.131 | 0.191 | 0.160 | 0.064 | - |
| Country, Locality | Site | Population Code | Elev. (m) | Longitude | Latitude | n | Mean Annual Temperature (°C) a | Mean Annual Precipitation (mm) a |
|---|---|---|---|---|---|---|---|---|
| Spain, Cuenca (CU) | Tragacete | TrCU | 1641 | 1°47′22.68″W | 40°20′32.94″N | 50 | 7.54 | 1247 |
| Spain, Cuenca (CU) | Arcas | ArCU | 1099 | 2°04′04.38″W | 39°54′27.00″N | 50 | 10.94 | 735 |
| Spain, Cuenca (CU) | Palancares | PaCU | 1185 | 1°57′38.97″W | 40°00′28.86″N | 10 | 10.38 | 818 |
| Morocco, Talassemtane (MO) | Talassemtane | TaMO | 1710 | 5°08′26.63″W | 35°08′30.33″N | 10 | 9.86 | 1830 |
| Spain, Sierra Mágina (MA) | Sierra Mágina | MA | 1820 | 3°26′50.74″W | 37°42′44.96″N | 10 | 11.51 | 1128 |
| Spain, Cazorla (CA) | Palancares | PaCA | 1084 | 2°51′59.90″W | 38°06′8.25″N | 10 | 12.64 | 939 |
| Spain, Cazorla (CA) | Navaciazo | NaCA | 1400 | 2°51′43.84″W | 37°53′45.75″N | 10 | 10.45 | 1219 |
| Spain, Cazorla (CA) | Yelmo | YeCA | 1565 | 2°39′19.51″W | 38°14′56.97″N | 10 | 9.76 | 975 |
© 2012 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Rubio-Moraga, A.; Candel-Perez, D.; Lucas-Borja, M.E.; Tiscar, P.A.; Viñegla, B.; Linares, J.C.; Gómez-Gómez, L.; Ahrazem, O. Genetic Diversity of Pinus nigra Arn. Populations in Southern Spain and Northern Morocco Revealed By Inter-Simple Sequence Repeat Profiles. Int. J. Mol. Sci. 2012, 13, 5645-5658. https://doi.org/10.3390/ijms13055645
Rubio-Moraga A, Candel-Perez D, Lucas-Borja ME, Tiscar PA, Viñegla B, Linares JC, Gómez-Gómez L, Ahrazem O. Genetic Diversity of Pinus nigra Arn. Populations in Southern Spain and Northern Morocco Revealed By Inter-Simple Sequence Repeat Profiles. International Journal of Molecular Sciences. 2012; 13(5):5645-5658. https://doi.org/10.3390/ijms13055645
Chicago/Turabian StyleRubio-Moraga, Angela, David Candel-Perez, Manuel E. Lucas-Borja, Pedro A. Tiscar, Benjamin Viñegla, Juan C. Linares, Lourdes Gómez-Gómez, and Oussama Ahrazem. 2012. "Genetic Diversity of Pinus nigra Arn. Populations in Southern Spain and Northern Morocco Revealed By Inter-Simple Sequence Repeat Profiles" International Journal of Molecular Sciences 13, no. 5: 5645-5658. https://doi.org/10.3390/ijms13055645









