Structural DNA Nanotechnology: From Design to Applications
Abstract
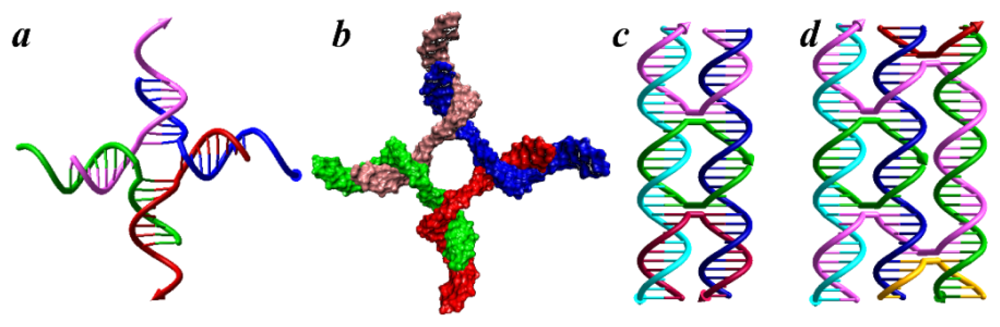
:1. DNA Self-Assembly
2. Tile-based DNA Structures and Applications
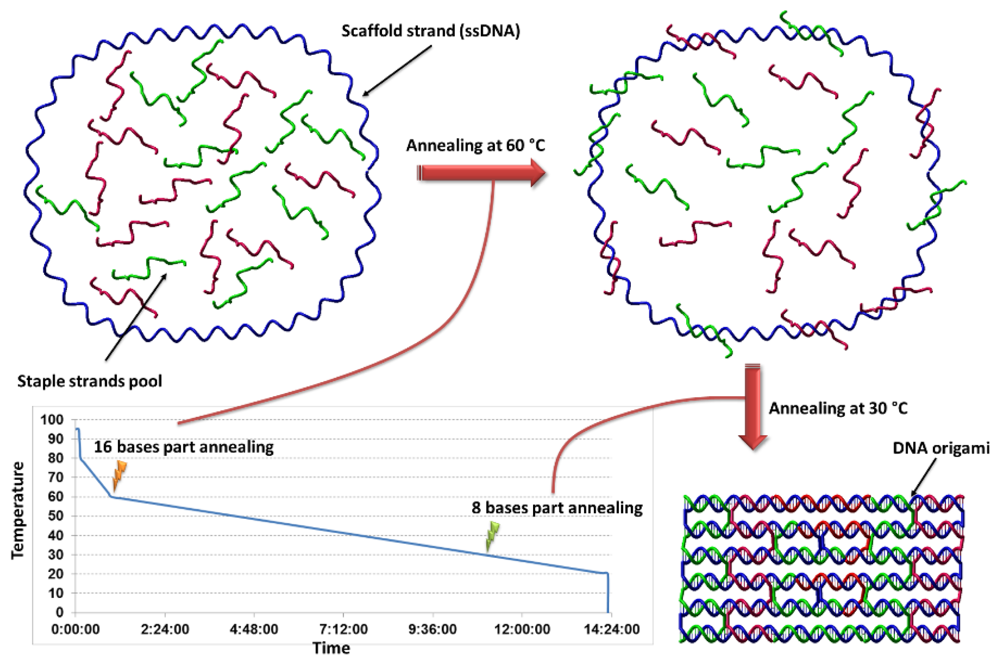
3. Origins of DNA Origami
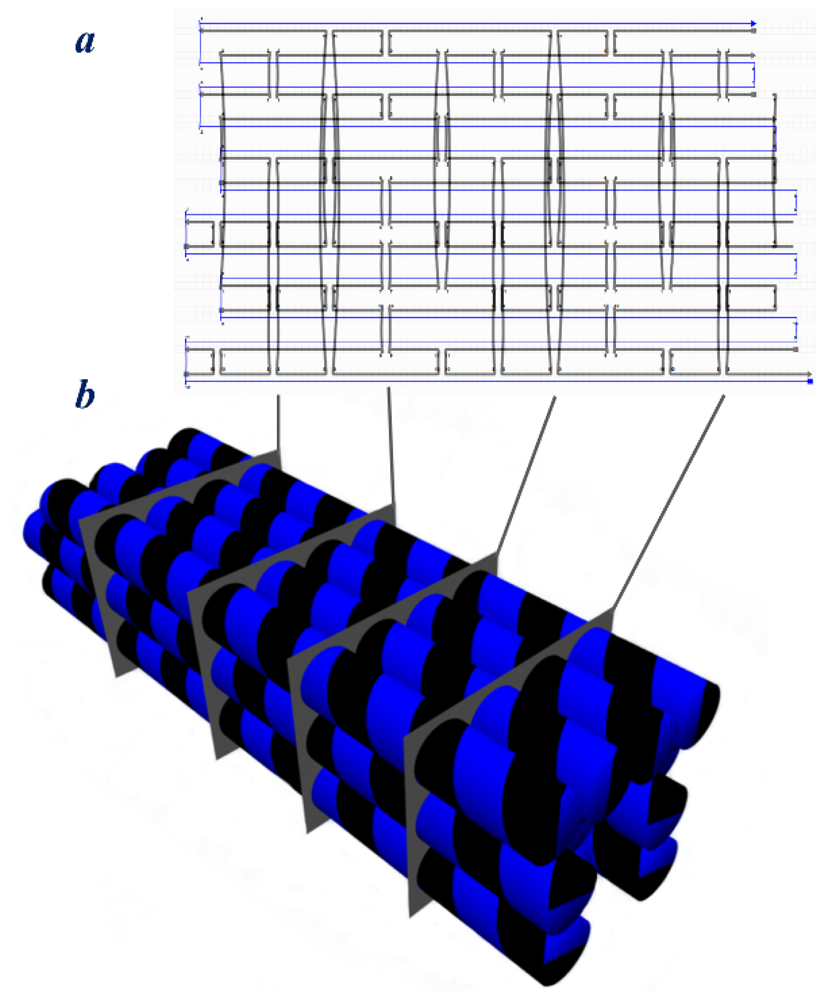
4. 2D DNA Origami
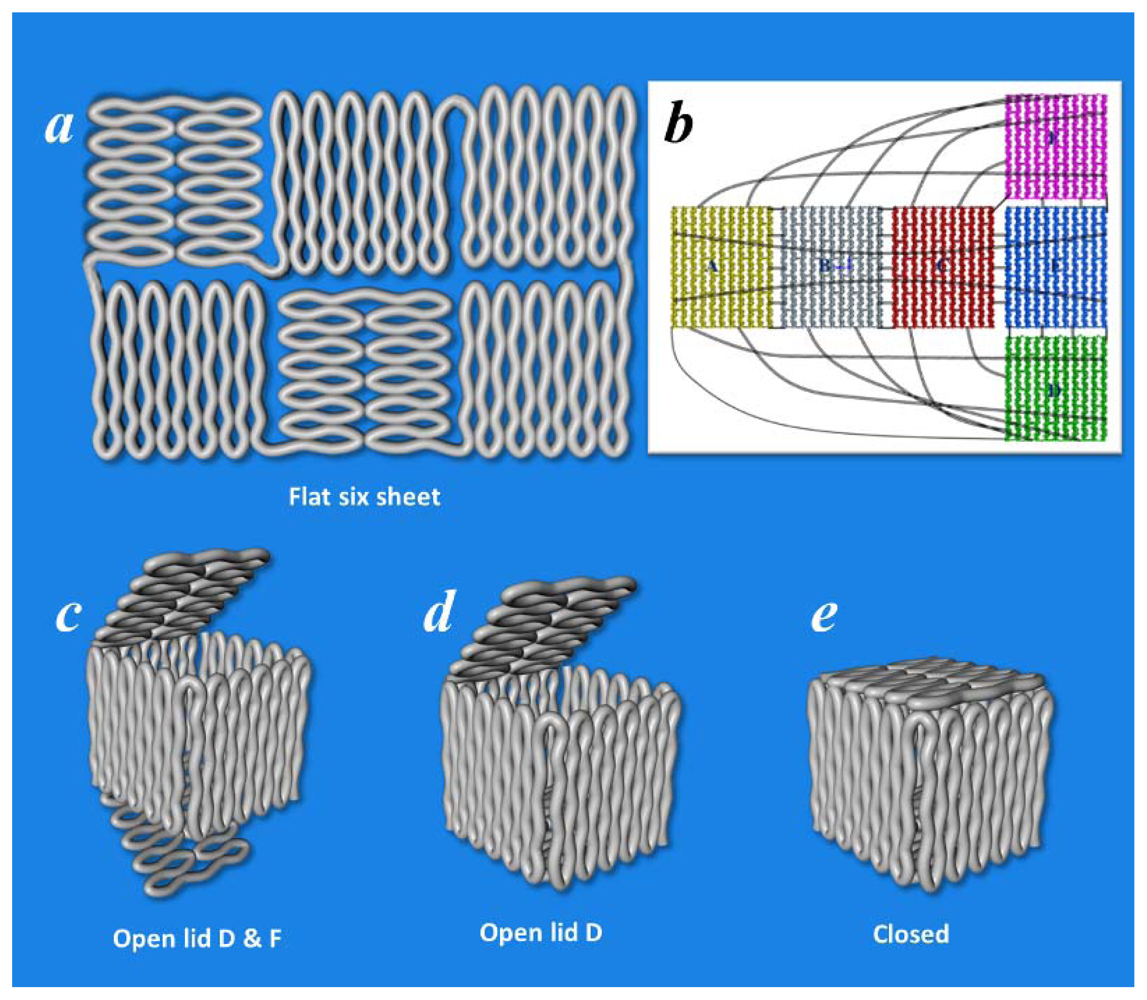
5. 3D DNA Origami
6. Discussion
Acknowledgments
References
- Seeman, N.C. Nanomaterials based on DNA. Annu. Rev. Biochem 2010, 79, 65–87. [Google Scholar]
- Seeman, N.C. Nucleic acid junctions and lattices. J. Theor. Biol 1982, 99, 237–247. [Google Scholar]
- Fu, T.-J.; Tse-Dinh, Y.-C.; Seeman, N.C. Holliday junction crossover topology. J. Mol. Biol 1994, 236, 91–105. [Google Scholar]
- Seeman, N.C.; Kallenbach, N.R. Design of immobile nucleic acid junctions. Biophys. J 1983, 44, 201–209. [Google Scholar]
- Seeman, N.C. Construction of three-dimensional stick figures from branched DNA. DNA Cell Biol 1991, 10, 475–486. [Google Scholar]
- Seeman, N.C. The design and engineering of nucleic acid nanoscale assemblies. Curr. Opin. Struct. Biol 1996, 6, 519–526. [Google Scholar]
- Winfree, E.; Liu, F.; Wenzler, L.A.; Seeman, N.C. Design and self-assembly of two-dimensional DNA crystals. Nature 1998, 394, 539–544. [Google Scholar]
- LaBean, T.H.; Yan, H.; Kopatsch, J.; Liu, F.; Winfree, E.; Reif, J.H.; Seeman, N.C. Construction, analysis, ligation, and self-assembly of DNA triple crossover complexes. J. Am. Chem. Soc 2000, 122, 1848–1860. [Google Scholar]
- Feng, L.; Park, S.H.; Reif, J.H.; Yan, H. A two-state DNA lattice switched by DNA nanoactuator. Angew. Chem. Int. Ed 2003, 42, 4342–4346. [Google Scholar]
- Endo, M.; Sugiyama, H. Chemical approaches to DNA nanotechnology. ChemBioChem 2009, 10, 2420–2443. [Google Scholar]
- Li, X.; Yang, X.; Qi, J.; Seeman, N.C. Antiparallel DNA double crossover molecules as components for nanoconstruction. J. Am. Chem. Soc 1996, 118, 6131–6140. [Google Scholar]
- Malo, J.; Mitchell, J.C.; Turberfield, A.J. A two-dimensional DNA array: The three-layer logpile. J. Am. Chem. Soc 2009, 131, 13574–13575. [Google Scholar]
- Mao, C.; Sun, W.; Shen, Z.; Seeman, N.C. A nanomechanical device based on the B-Z transition of DNA. Nature 1999, 397, 144–146. [Google Scholar]
- Yan, H.; Park, S.H.; Finkelstein, G.; Reif, J.H.; LaBean, T.H. DNA-templated self-assembly of protein arrays and highly conductive nanowires. Science 2003, 301, 1882–1884. [Google Scholar]
- Liu, D.; Wang, M.; Deng, Z.; Walulu, R.; Mao, C. Tensegrity: Construction of rigid DNA triangles with flexible four-arm DNA junctions. J. Am. Chem. Soc 2004, 126, 2324–2325. [Google Scholar]
- Mathieu, F.; Liao, S.; Kopatsch, J.; Wang, T.; Mao, C.; Seeman, N.C. Six-helix bundles designed from DNA. Nano Lett 2005, 5, 661–665. [Google Scholar]
- Liu, H.; He, Y.; Ribbe, A.E.; Mao, C. Two-dimensional (2D) DNA crystals assembled from two DNA strands. Biomacromolecules 2005, 6, 2943–2945. [Google Scholar]
- Zheng, J.; Birktoft, J.J.; Chen, Y.; Wang, T.; Sha, R.; Constantinou, P.E.; Ginell, S.L.; Mao, C.; Seeman, N.C. From molecular to macroscopic via the rational design of a self-assembled 3D DNA crystal. Nature 2009, 461, 74–77. [Google Scholar]
- Yin, P.; Hariadi, R.F.; Sahu, S.; Choi, H.M.T.; Park, S.H.; LaBean, T.H.; Reif, J.H. Programming DNA tube circumferences. Science 2008, 321, 824–826. [Google Scholar]
- Hansen, M.N.; Zhang, A.M.; Rangnekar, A.; Bompiani, K.M.; Carter, J.D.; Gothelf, K.V.; LaBean, T.H. Weave tile architecture construction strategy for DNA nanotechnology. J. Am. Chem. Soc 2010, 132, 14481–14486. [Google Scholar]
- Chen, J.; Seeman, N.C. Synthesis from DNA of a molecule with the connectivity of a cube. Nature 1991, 350, 631–633. [Google Scholar]
- Shih, W.M.; Quispe, J.D.; Joyce, G.F. A 1.7-kilobase single-stranded DNA that folds into a nanoscale octahedron. Nature 2004, 427, 618–621. [Google Scholar]
- Goodman, R.P.; Berry, R.M.; Turberfield, A.J. The single-step synthesis of a DNA tetrahedron. Chem. Commun 2004, 12, 1372–1373. [Google Scholar]
- Goodman, R.P.; Heilemann, M.; Doose, S.; Erben, C.M.; Kapanidis, A.N.; Turberfield, A.J. Reconfigurable, braced, three-dimensional DNA nanostructures. Nat. Nanotechnol 2008, 3, 93–96. [Google Scholar]
- Aldaye, F.A.; Sleiman, H.F. Modular access to structurally switchable 3D discrete DNA assemblies. J. Am. Chem. Soc 2007, 129, 13376–13377. [Google Scholar]
- He, Y.; Su, M.; Fang, P.-A.; Zhang, C.; Ribbe, A.E.; Jiang, W.; Mao, C. On the chirality of self-assembled DNA octahedra. Angew. Chem. Int. Ed 2010, 49, 748–751. [Google Scholar]
- Jungmann, R.; Steinhauer, C.; Scheible, M.; Kuzyk, A.; Tinnefeld, P.; Simmel, F.C. Single-molecule kinetics and super-resolution microscopy by fluorescence imaging of transient binding on DNA origami. Nano Lett 2010, 10, 4756–4761. [Google Scholar]
- Sannohe, Y.; Endo, M.; Katsuda, Y.; Hidaka, K.; Sugiyama, H. Visualization of dynamic conformational switching of the G-quadruplex in a DNA nanostructure. J. Am. Chem. Soc 2010, 132, 16311–16313. [Google Scholar]
- Shin, J.-S.; Pierce, N.A. A synthetic DNA walker for molecular transport. J. Am. Chem. Soc 2004, 126, 10834–10835. [Google Scholar]
- Wang, Z.-G.; Elbaz, J.; Willner, I. DNA machines: Bipedal walker and stepper. Nano Lett 2010, 11, 304–309. [Google Scholar]
- Yin, P.; Yan, H.; Daniell, X.G.; Turberfield, A.J.; Reif, J.H. A unidirectional DNA walker that moves autonomously along a track. Angew. Chem. Int. Ed 2004, 43, 4906–4911. [Google Scholar]
- Yurke, B.; Turberfield, A.J.; Mills, A.P.; Simmel, F.C.; Neumann, J.L. A DNA-fuelled molecular machine made of DNA. Nature 2000, 406, 605–608. [Google Scholar]
- Bath, J.; Green, S.J.; Turberfield, A.J. A free-running DNA motor powered by a nicking enzyme. Angew. Chem. Int. Ed 2005, 44, 4358–4361. [Google Scholar]
- Omabegho, T.; Sha, R.; Seeman, N.C. A bipedal DNA brownian motor with coordinated legs. Science 2009, 324, 67–71. [Google Scholar]
- Sherman, W.B.; Seeman, N.C. A precisely controlled DNA biped walking device. Nano Lett 2004, 4, 1203–1207. [Google Scholar]
- Tian, Y.; He, Y.; Chen, Y.; Yin, P.; Mao, C. A DNAzyme that walks processively and autonomously along a one-dimensional track. Angew. Chem. Int. Ed 2005, 44, 4355–4358. [Google Scholar]
- Rothemund, P.W. Folding DNA to create nanoscale shapes and patterns. Nature 2006, 440, 297–302. [Google Scholar]
- Yan, H.; LaBean, T.H.; Feng, L.; Reif, J.H. Directed nucleation assembly of DNA tile complexes for barcode-patterned lattices. Proc. Natl. Acad. Sci. USA 2003, 100, 8103–8108. [Google Scholar]
- Williamson, J.R. RNA origami. Nat. Struct. Mol. Biol 1994, 1, 270–272. [Google Scholar]
- Qian, L.; Wang, Y.; Zhang, Z.; Zhao, J.; Pan, D.; Zhang, Y.; Liu, Q.; Fan, C.; Hu, J.; He, L. Analogic China map constructed by DNA. Chin. Sci. Bull 2006, 51, 2973–2976. [Google Scholar]
- Andersen, E.S.; Dong, M.; Nielsen, M.M.; Jahn, K.; Lind-Thomsen, A.; Mamdouh, W.; Gothelf, K.V.; Besenbacher, F.; Kjems, J. DNA origami design of dolphin-shaped structures with flexible tails. ACS Nano 2008, 2, 1213–1218. [Google Scholar]
- Douglas, S.M.; Marblestone, A.H.; Teerapittayanon, S.; Vazquez, A.; Church, G.M.; Shih, W.M. Rapid prototyping of 3D DNA-origami shapes with caDNAno. Nucleic Acids Res 2009, 37, 5001–5006. [Google Scholar]
- Castro, C.E.; Kilchherr, F.; Kim, D.N.; Shiao, E.L.; Wauer, T.; Wortmann, P.; Bathe, M.; Dietz, H. A primer to scaffolded DNA origami. Nat. Methods 2011, 8, 221–229. [Google Scholar]
- Högberg, B.; Liedl, T.; Shih, W.M. Folding DNA origami from a double-stranded source of scaffold. J. Am. Chem. Soc 2009, 131, 9154–9155. [Google Scholar]
- Li, Z.; Liu, M.; Wang, L.; Nangreave, J.; Yan, H.; Liu, Y. Molecular behavior of DNA origami in higher-order self-assembly. J. Am. Chem. Soc 2010, 132, 13545–13552. [Google Scholar]
- Endo, M.; Sugita, T.; Rajendran, A.; Katsuda, Y.; Emura, T.; Hidaka, K.; Sugiyama, H. Two-dimensional DNA origami assemblies using a four-way connector. Chem. Commun 2011, 47, 3213–3215. [Google Scholar]
- Rajendran, A.; Endo, M.; Katsuda, Y.; Hidaka, K.; Sugiyama, H. Programmed two-dimensional self-assembly of multiple DNA origami jigsaw pieces. ACS Nano 2010, 5, 665–671. [Google Scholar]
- Kuzuya, A.; Kimura, M.; Numajiri, K.; Koshi, N.; Ohnishi, T.; Okada, F.; Komiyama, M. Precisely programmed and robust 2D streptavidin nanoarrays by using periodical nanometer-scale wells embedded in DNA origami assembly. ChemBioChem 2009, 10, 1811–1815. [Google Scholar]
- Shen, W.; Zhong, H.; Neff, D.; Norton, M.L. NTA directed protein nanopatterning on DNA origami nanoconstructs. J. Am. Chem. Soc 2009, 131, 6660–6661. [Google Scholar]
- Kuzyk, A.; Laitinen, K.T.; Törmä, P. DNA origami as a nanoscale template for protein assembly. Nanotechnology 2009, 20. [Google Scholar] [CrossRef]
- Saccà, B.; Meyer, R.; Erkelenz, M.; Kiko, K.; Arndt, A.; Schroeder, H.; Rabe, K.S.; Niemeyer, C.M. Orthogonal protein decoration of DNA origami. Angew. Chem. Int. Ed 2010, 49, 9378–9383. [Google Scholar]
- Stephanopoulos, N.; Liu, M.; Tong, G.J.; Li, Z.; Liu, Y.; Yan, H.; Francis, M.B. Immobilization and one-dimensional arrangement of virus capsids with nanoscale precision using DNA origami. Nano Lett 2010, 10, 2714–2720. [Google Scholar]
- Rinker, S.; Ke, Y.; Liu, Y.; Chhabra, R.; Yan, H. Self-assembled DNA nanostructures for distance-dependent multivalent ligand-protein binding. Nat. Nanotechnol 2008, 3, 418–422. [Google Scholar]
- Stearns, L.A.; Chhabra, R.; Sharma, J.; Liu, Y.; Petuskey, W.T.; Yan, H.; Chaput, J.C. Template-directed nucleation and growth of inorganic nanoparticles on DNA scaffolds. Angew. Chem. Int. Ed 2009, 48, 8494–8496. [Google Scholar]
- Sharma, J.; Chhabra, R.; Andersen, C.S.; Gothelf, K.V.; Yan, H.; Liu, Y. Toward reliable gold nanoparticle patterning on self-assembled DNA nanoscaffold. J. Am. Chem. Soc 2008, 130, 7820–7821. [Google Scholar]
- Steinhauer, C.; Jungmann, R.; Sobey, T.L.; Simmel, F.C.; Tinnefeld, P. DNA origami as a nanoscopic ruler for super-resolution microscopy. Angew. Chem. Int. Ed 2009, 48, 8870–8873. [Google Scholar]
- Ding, B.; Wu, H.; Xu, W.; Zhao, Z.; Liu, Y.; Yu, H.; Yan, H. Interconnecting gold islands with DNA origami nanotubes. Nano Lett 2010, 10, 5065–5069. [Google Scholar]
- Liu, J.; Geng, Y.; Pound, E.; Gyawali, S.; Ashton, J.R.; Hickey, J.; Woolley, A.T.; Harb, J.N. Metallization of branched DNA origami for nanoelectronic circuit fabrication. ACS Nano 2011, 5, 2240–2247. [Google Scholar]
- Maune, H.T.; Han, S.-P.; Barish, R.D.; Bockrath, M.; Goddard, W.A.; Rothemund, P.W.K.; Winfree, E. Self-assembly of carbon nanotubes into two-dimensional geometries using DNA origami templates. Nat. Nanotechnol 2010, 5, 61–66. [Google Scholar]
- Eskelinen, A.-P.; Kuzyk, A.; Kaltiaisenaho, T.K.; Timmermans, M.Y.; Nasibulin, A.G.; Kauppinen, E.I.; Törmä, P. Assembly of single-walled carbon nanotubes on DNA-origami templates through streptavidin-biotin interaction. Small 2011, 7, 746–750. [Google Scholar]
- Kershner, R.J.; Bozano, L.D.; Micheel, C.M.; Hung, A.M.; Fornof, A.R.; Cha, J.N.; Rettner, C.T.; Bersani, M.; Frommer, J.; Rothemund, P.W.K.; Wallraff, G.M. Placement and orientation of individual DNA shapes on lithographically patterned surfaces. Nat. Nanotechnol 2009, 4, 557–561. [Google Scholar]
- Hung, A.M.; Micheel, C.M.; Bozano, L.D.; Osterbur, L.W.; Wallraff, G.M.; Cha, J.N. Large-area spatially ordered arrays of gold nanoparticles directed by lithographically confined DNA origami. Nat. Nanotechnol 2010, 5, 121–126. [Google Scholar]
- Kuzyk, A.; Yurke, B.; Toppari, J.J.; Linko, V.; Törmä, P. Dielectrophoretic trapping of DNA origami. Small 2008, 4, 447–450. [Google Scholar]
- Kuzyk, A.; Schreiber, R.; Fan, Z.; Pardatscher, G.; Roller, E.M.; Hogele, A.; Simmel, F.C.; Govorov, A.O.; Liedl, T. DNA-based self-assembly of chiral plasmonic nanostructures with tailored optical response. Nature 2012, 483, 311–314. [Google Scholar]
- Subramanian, H.K.K.; Chakraborty, B.; Sha, R.; Seeman, N.C. The label-free unambiguous detection and symbolic display of single nucleotide polymorphisms on DNA origami. Nano Lett 2011, 11, 910–913. [Google Scholar]
- Barish, R.D.; Schulman, R.; Rothemund, P.W.K.; Winfree, E. An information-bearing seed for nucleating algorithmic self-assembly. Proc. Natl. Acad. Sci. USA 2009, 106, 6054–6059. [Google Scholar]
- Fujibayashi, K.; Hariadi, R.; Park, S.H.; Winfree, E.; Murata, S. Toward reliable algorithmic self-assembly of DNA tiles: A fixed-width cellular automaton pattern. Nano Lett 2007, 8, 1791–1797. [Google Scholar]
- Endo, M.; Katsuda, Y.; Hidaka, K.; Sugiyama, H. Regulation of DNA methylation using different tensions of double strands constructed in a defined DNA nanostructure. J. Am. Chem. Soc 2010, 132, 1592–1597. [Google Scholar]
- Subramani, R.; Juul, S.; Rotaru, A.; Andersen, F.F.; Gothelf, K.V.; Mamdouh, W.; Besenbacher, F.; Dong, M.; Knudsen, B.R. A novel secondary DNA binding site in human topoisomerase I unravelled by using a 2D DNA origami platform. ACS Nano 2010, 4, 5969–5977. [Google Scholar]
- Lund, K.; Manzo, A.J.; Dabby, N.; Michelotti, N.; Johnson-Buck, A.; Nangreave, J.; Taylor, S.; Pei, R.; Stojanovic, M.N.; Walter, N.G.; et al. Molecular robots guided by prescriptive landscapes. Nature 2010, 465, 206–210. [Google Scholar]
- Gu, H.; Chao, J.; Xiao, S.-J.; Seeman, N.C. A proximity-based programmable DNA nanoscale assembly line. Nature 2010, 465, 202–205. [Google Scholar]
- Wickham, S.F.; Endo, M.; Katsuda, Y.; Hidaka, K.; Bath, J.; Sugiyama, H.; Turberfield, A.J. Direct observation of stepwise movement of a synthetic molecular transporter. Nat. Nanotechnol 2011, 6, 166–169. [Google Scholar] [Green Version]
- Endo, M.; Hidaka, K.; Kato, T.; Namba, K.; Sugiyama, H. DNA prism structures constructed by folding of multiple rectangular arms. J. Am. Chem. Soc 2009, 131, 15570–15571. [Google Scholar]
- Douglas, S.M.; Dietz, H.; Liedl, T.; Hogberg, B.; Graf, F.; Shih, W.M. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature 2009, 459, 414–418. [Google Scholar]
- Andersen, E.S.; Dong, M.; Nielsen, M.M.; Jahn, K.; Subramani, R.; Mamdouh, W.; Golas, M.M.; Sander, B.; Stark, H.; Oliveira, C.L.P.; et al. Self-assembly of a nanoscale DNA box with a controllable lid. Nature 2009, 459, 73–76. [Google Scholar]
- Kuzuya, A.; Komiyama, M. Design and construction of a box-shaped 3D-DNA origami. Chem. Commun 2009, 28, 4182–4184. [Google Scholar]
- Ke, Y.; Sharma, J.; Liu, M.; Jahn, K.; Liu, Y.; Yan, H. Scaffolded DNA origami of a DNA tetrahedron molecular container. Nano Lett 2009, 9, 2445–2447. [Google Scholar]
- Dietz, H.; Douglas, S.M.; Shih, W.M. Folding DNA into twisted and curved nanoscale shapes. Science 2009, 325, 725–730. [Google Scholar]
- Han, D.; Pal, S.; Liu, Y.; Yan, H. Folding and cutting DNA into reconfigurable topological nanostructures. Nat. Nanotechnol 2010, 5, 712–717. [Google Scholar]
- Douglas, S.M.; Bachelet, I.; Church, G.M. A logic-gated nanorobot for targeted transport of molecular payloads. Science 2012, 335, 831–834. [Google Scholar]
- Kuzuya, A.; Sakai, Y.; Yamazaki, T.; Xu, Y.; Komiyama, M. Nanomechanical DNA origami “single-molecule beacons” directly imaged by atomic force microscopy. Nat. Commun 2011, 2. [Google Scholar] [CrossRef]
- Birac, J.J.; Sherman, W.B.; Kopatsch, J.; Constantinou, P.E.; Seeman, N.C. Architecture with GIDEON, a program for design in structural DNA nanotechnology. J. Mol. Graphics Modell 2006, 25, 470–480. [Google Scholar]
- Nanorex NanoEngineer-1. Available online: http://www.nanoengineer-1.com accessed on 25 March 2004.
- Kim, D.N.; Kilchherr, F.; Dietz, H.; Bathe, M. Quantitative prediction of 3D solution shape and flexibility of nucleic acid nanostructures. Nucleic Acids Res 2012, 40, 2862–2868. [Google Scholar]
© 2012 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Zadegan, R.M.; Norton, M.L. Structural DNA Nanotechnology: From Design to Applications. Int. J. Mol. Sci. 2012, 13, 7149-7162. https://doi.org/10.3390/ijms13067149
Zadegan RM, Norton ML. Structural DNA Nanotechnology: From Design to Applications. International Journal of Molecular Sciences. 2012; 13(6):7149-7162. https://doi.org/10.3390/ijms13067149
Chicago/Turabian StyleZadegan, Reza M., and Michael L. Norton. 2012. "Structural DNA Nanotechnology: From Design to Applications" International Journal of Molecular Sciences 13, no. 6: 7149-7162. https://doi.org/10.3390/ijms13067149
APA StyleZadegan, R. M., & Norton, M. L. (2012). Structural DNA Nanotechnology: From Design to Applications. International Journal of Molecular Sciences, 13(6), 7149-7162. https://doi.org/10.3390/ijms13067149



