UV-Induced Cell Death in Plants
Abstract
:1. Introduction
2. Perception and Signaling of UV Light in Plants
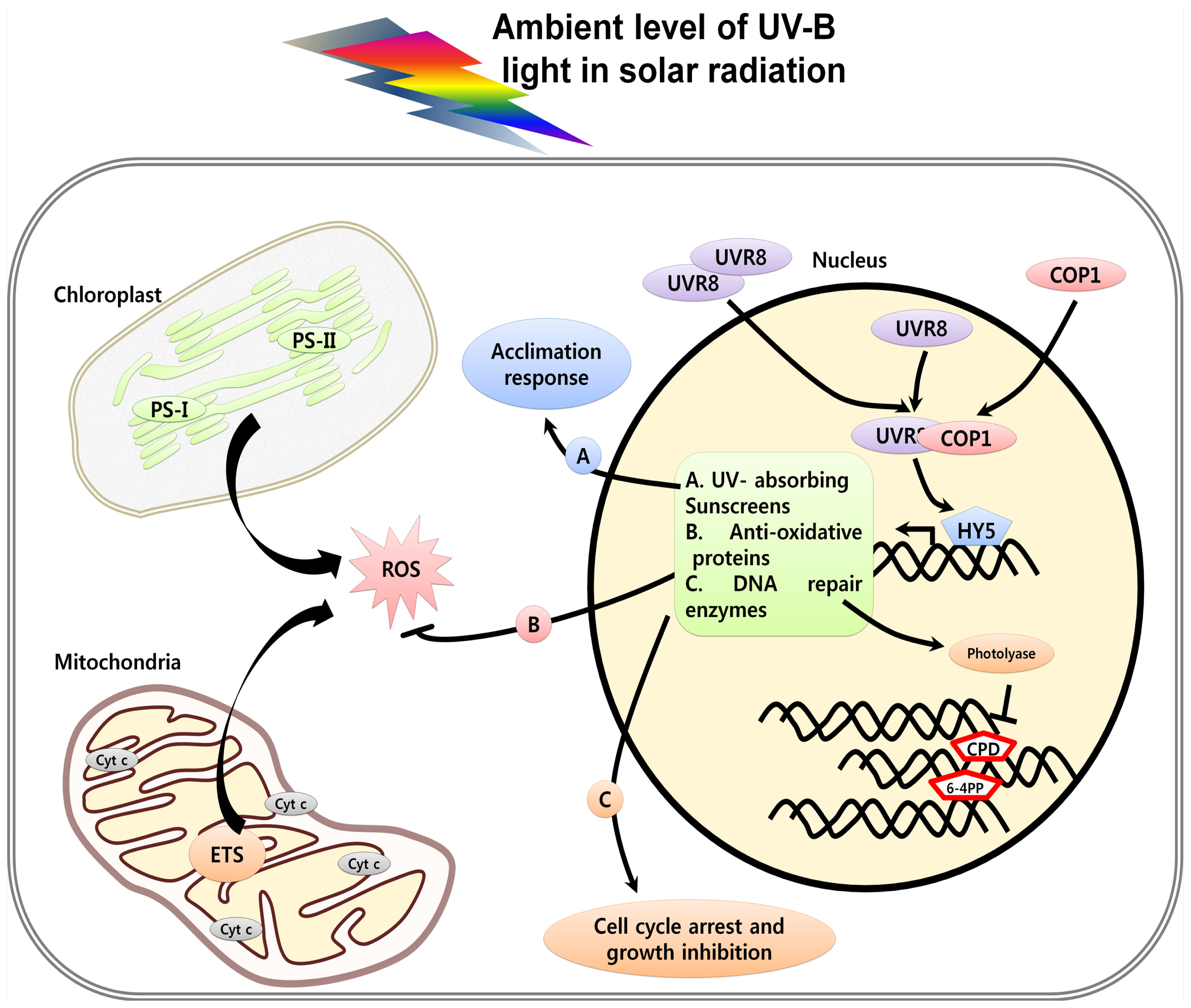
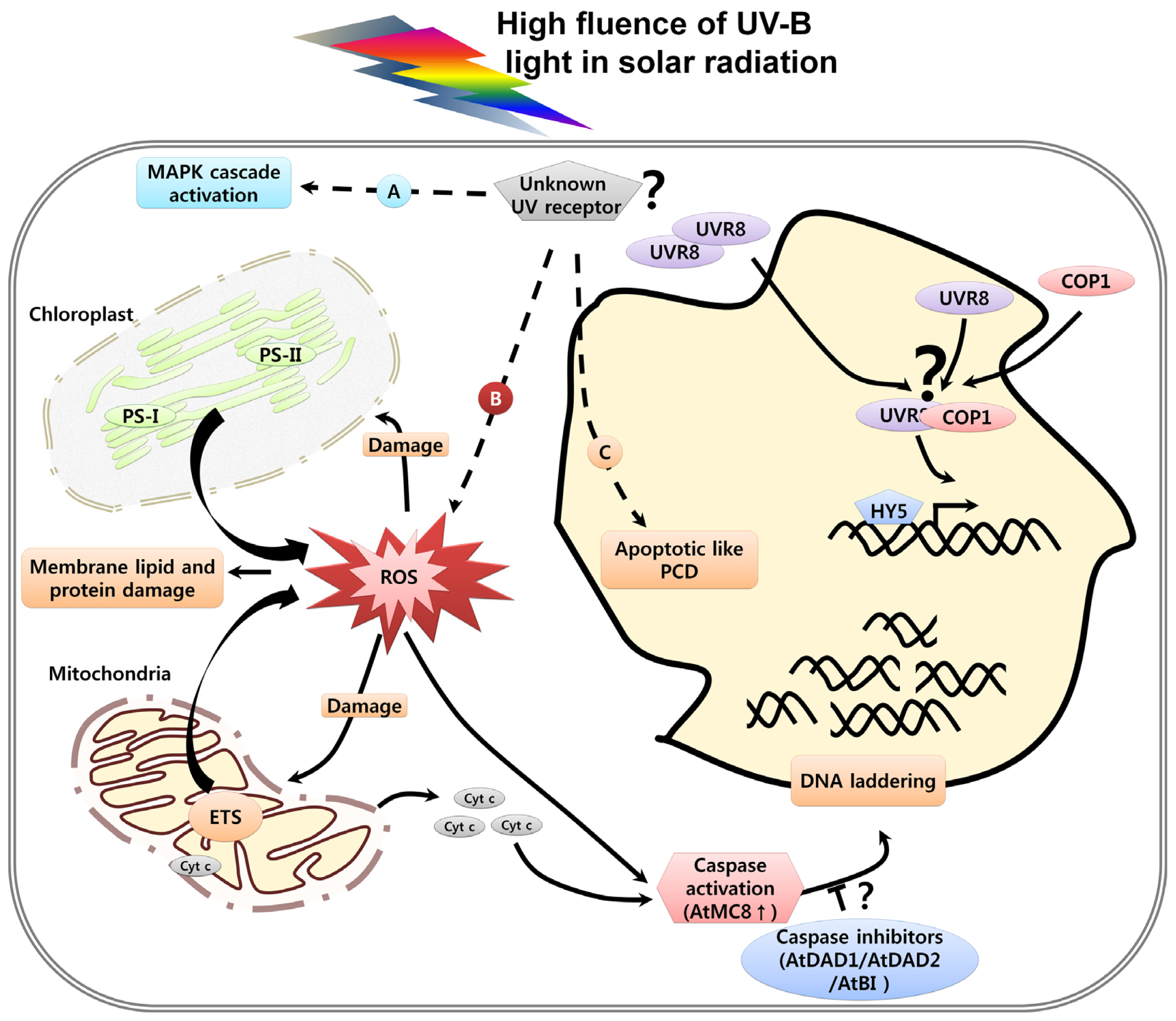
3. UV Induced Damage and Cell Death
3.1. DNA Damage, Cell Cycle Arrest and Cell Death
3.2. Proteins and Lipids Damage
3.3. Organelle Dysfunction in Response to UV and Cell Death
3.4. UV-Induced Caspase Activity and Cell Death
3.5. UV-Induced ROS and Cell Death
4. Conclusions
Acknowledgments
- Conflict of InterestThe authors declare no conflict of interest.
References
- Coohill, T.P. Ultraviolet action spectra (280 nm to 380 nm) and solar effectiveness spectra for higher plants. Photochem. Photobiol 1989, 50, 451–457. [Google Scholar]
- Frederick, J.E. Ultraviolet sunlight reaching the Earth’s surface. A review of recent research. Photochem. Photobiol 1993, 57, 175–178. [Google Scholar]
- Hollosy, F.; Seprodi, J.; Orfi, L.; Eros, D.; Keri, G.; Idei, M. Evaluation of lipophilicity and antitumour activity of parallel carboxamide libraries. J. Chromatogr. B 2002, 780, 355–363. [Google Scholar]
- Frohnmeyer, H.; Staiger, D. Ultraviolet-B radiation-mediated responses in plants. Balancing damage and protection. Plant Physiol 2003, 133, 1420–1428. [Google Scholar]
- McKenzie, R.L.; Bjorn, L.O.; Bais, A., II; yas, M. Changes in biologically active ultraviolet radiation reaching the Earth’s surface. Photochem. Photobiol. Sci 2003, 2, 5–15. [Google Scholar]
- Paul, N.D.; Gwynn, J.D. Ecological roles of solar UV radiation: Towards an integrated approach. Trends Ecol. Evol 2003, 18, 48–55. [Google Scholar]
- Pyle, J.A. Global Ozone Depletion: Observation and Theory. In Plants and UV-B: Responses to Environmental Change; Lumbsden, P.J., Ed.; Cambridge University Press: Cambridge, UK, 1996; pp. 3–12. [Google Scholar]
- McKenzie, R.L.; Aucamp, P.J.; Bais, A.F.; Björn, L.O.; Ilyas, M. Changes in biologically-active ultraviolet radiation reaching the Earth’s surface. Photochem. Photobiol. Sci 2007, 6, 218–231. [Google Scholar]
- Herman, J.R. Global increase in UV irradiance during the past 30 years (1979–2008) estimated from satellite data. J. Geophys. Res 2010, 15, D04203. [Google Scholar]
- Ballare, C.L.; Caldwell, M.M.; Flint, S.D.; Robinson, S.A.; Bornman, J.F. Effects of solar ultraviolet radiation on terrestrial ecosystems. Patterns, mechanisms, and interactions with climate change. Photochem. Photobiol. Sci 2011, 10, 226–241. [Google Scholar]
- Ballare, C.L.; Rousseaux, C.M.; Searles, P.S.; Zaller, J.G.; Giordano, C.V.; Robson, M.T.; Caldwell, M.M.; Sala, O.E.; Scopel, A.L. Impacts of solar ultraviolet-B radiation on terrestrial ecosystems of Tierra del Fuego (Southern Argentina). An overview of recent progress. J. Photochem. Photobiol 2001, 62, 67–77. [Google Scholar]
- Caldwell, M.M.; Ballare, C.L.; Bornman, J.F.; Flint, S.D.; Bjorn, L.O.; Teramura, A.H.; Kulandaivelu, G.; Tevini, M. Terrestrial ecosystems increased solar ultraviolet radiation and interactions with other climatic change factors. Photochem. Photobiol Sci 2003, 2, 29–38. [Google Scholar]
- Caldwell, M.M.; Bornman, J.F.; Ballare, C.L.; Flint, S.D.; Kulandaivelu, G. Terrestrial ecosystems, increased solar ultraviolet radiation, and interactions with other climate change factors. Photochem. Photobiol. Sci 2007, 6, 252–266. [Google Scholar]
- Ulm, R.; Baumann, A.; Oravecz, A.; Mate, Z.; Adam, E.; Oakeley, E.J.; Schafer, E.; Nagy, F. Genome-wide analysis of gene expression reveals function of the bZIP transcription factor HY5 in the UV-B response of Arabidopsis. Proc. Natl. Acad. Sci. USA 2004, 101, 1397–1402. [Google Scholar]
- Suesslin, C.; Frohnmeyer, H. An Arabidopsis mutant defective in UV-B light-mediated responses. Plant J 2003, 33, 1–11. [Google Scholar]
- Danon, A.; Gallois, P. UV-C radiation induces apoptotic-like changes in Arabidopsis thaliana. FEBS Lett 1998, 437, 131–136. [Google Scholar]
- Danon, A.; Rotari, V.I.; Gordon, A.; Mailhac, N.; Gallois, P. Ultraviolet- C overexposure induces programmed cell death in Arabidopsis, which is mediated by caspase-like activities and which can be suppressed by caspase inhibitors, p35 and Defender against apoptotic Death. J. Biol. Chem 2004, 279, 779–787. [Google Scholar]
- Gao, C.; Xing, D.; Li, L.; Zhang, L. Implication of reactive oxygen species and mitochondrial dysfunction in the early stages of plant programmed cell death induced by ultraviolet-C overexposure. Planta 2008, 227, 755–767. [Google Scholar]
- Lytvyn, D.I.; Yemets, A.I.; Blume, Y.B. UV-B overexposure induces programmed cell death in a BY-2 tobacco cell line. Environ. Exp. Bot 2010, 68, 51–57. [Google Scholar]
- Heijde, M.; Ulm, R. UV-B photoreceptor-mediated signaling in plants. Trends Plant Sci 2012, 17, 230–237. [Google Scholar]
- Kami, C.; Lorrain, S. Light-Regulated Plant Growth and Development. In Current Topics in Developmental Biology; Center for Integrative Genomics, University of Lausanne: Lausanne, Switzerland, 2010; Volume 91, pp. 29–65. [Google Scholar]
- Gyula, P.; Scha, E.; Nagy, F. Light perception and signalling in higher plants. Curr. Opin. Plant Biol 2003, 6, 446–452. [Google Scholar]
- Jiao, Y.; Lau, O.S.; Deng, X.W. Light-regulated transcriptional networks in higher plants. Nat. Rev. Genet 2007, 8, 217–230. [Google Scholar]
- Christie, J.M. Phototropin blue light receptors. Annu. Rev. Plant Biol 2007, 58, 21–45. [Google Scholar]
- Demarsy, E.; Fankhauser, C. Higher plants use LOV to perceive blue light. Curr. Opin. Plant Biol 2009, 12, 69–74. [Google Scholar]
- Lin, C.; Shalitin, D. Cryptochome structure and signal transduction. Annu. Rev. Plant Biol 2003, 54, 469–496. [Google Scholar]
- Somers, D.E.; Fujiwara, S. Thinking outside the F-box: Novel ligands for novel receptors. Trends Plant Sci 2009, 14, 206–213. [Google Scholar]
- Franklin, K.A.; Quail, P.H. Phytochrome functions in Arabidopsis development. J. Exp. Bot 2010, 61, 11–24. [Google Scholar]
- Rockwell, N.C.; Su, Y.S.; Lagarias, J.C. Phytochrome structure and signaling mechanisms. Annu. Rev. Plant Biol 2006, 57, 837–858. [Google Scholar]
- Kim, B.C.; Tenessen, D.; Last, R. UV-B-induced photomorphogenesis in Arabidopsis thaliana. Plant J 1998, 15, 667–674. [Google Scholar]
- Boccalandro, H.; Mazza, C.; Mazzella, M.; Casal, J.; Ballare, C. Ultraviolet B radiation enhances a phytochrome-B-mediated photomorphogenic response in Arabidopsis. Plant Physiol 2001, 126, 780–788. [Google Scholar]
- Brosche, M.; Strid, A. Molecular events following perception of ultraviolet-B radiation by plants. Physiol. Plant 2003, 117, 1–10. [Google Scholar]
- Ulm, R.; Nagy, F. Signalling and gene regulation in response to ultraviolet light. Curr. Opin. Plant Biol 2005, 8, 477–482. [Google Scholar]
- Favory, J.J.; Stec, A.; Gruber, H.; Rizzini, L.; Oravecz, A.; Funk, M.; Albert, A.; Cloix, C.; Jenkins, G.I.; Oakeley, E.A.; et al. Interaction of COP1 and UVR8 regulates UVB-induced photomorphogenesis and stress acclimation in Arabidopsis. EMBO J 2009, 28, 591–601. [Google Scholar]
- Kliebenstein, D.J.; Lim, J.E.; Landry, L.G.; Last, R.L. Arabidopsis UVR8 regulates ultraviolet-B signal transduction and tolerance and contains sequence similarity to human regulator of chromatin condensation. Plant Physiol 2002, 130, 234–243. [Google Scholar]
- Brown, B.A.; Cloix, C.; Jiang, G.H.; Kaiserli, E.; Herzyk, P.; Kliebenstein, D.J.; Jenkins, G.I. A UV-B specific signaling component orchestrates plant UV protection. Proc. Natl. Acad. Sci. USA 2005, 102, 18225–18230. [Google Scholar]
- Rizzini, L.; Favory, J.J.; Cloiax, C.; Faggionato, D.; Hara, A.O.; Kaiserli, E.; Baumeister, R.; Schafer, E.; Nagy, F.; Jenkins, G.I.; et al. Perception of UV-B by the Arabidopsis UVR8 protein. Science 2011, 332, 103–106. [Google Scholar]
- Christie, J.M.; Arvai, A.S.; Baxter, K.J.; Heilmann, M.; Pratt, A.J.; O’Hara, A.; Kelly, S.M.; Hothorn, M.; Smith, B.O.; Hitomi, K.; Jenkins, G.I.; Getzoff, E.D. Plant UVR8 photoreceptor senses UV-B by tryptophan-mediated disruption of cross-dimer salt bridges. Science 2012, 335, 1492–1496. [Google Scholar]
- Wu, D.; Hu, Q.; Yan, Z.; Chen, W.; Yan, C.; Huang, X.; Zhang, J.; Yang, P.; Deng, H.; Wang, J.; et al. Structural basis of ultraviolet-B perception by UVR8. Nature 2012, 484, 214–219. [Google Scholar]
- Rice-Evans, C.A.; Miller, N.J.; Papaga, G. Antioxidant properties of phenolic compounds. Trends Plant Sci 1997, 2, 152–159. [Google Scholar]
- Kaiserli, E.; Jenkins, G.I. UV-B promotes rapid nuclear translocation of the Arabidopsis UV-B specific signaling component UVR8 and activates its function in the nucleus. Plant Cell 2007, 19, 2662–2673. [Google Scholar]
- Oravecz, A.; Baumann, A.; Mate, Z.; Brzezinska, M.J.; Oakeley, E.J.; Adam, E.; Schafer, E.; Nagy, F.; Ulm, R. CONSTITUTIVELY PHOTOMORPHOGENIC1 is required for the UV-B response in Arabidopsis. Plant Cell 2006, 18, 1975–1990. [Google Scholar]
- Brown, B.A.; Jenkins, G.I. UV-B signaling pathways with different fluence-rate response profiles are distinguished in mature Arabidopsis leaf tissue by requirement for UVR8, HY5, and HYH. Plant Physiol 2008, 146, 276–588. [Google Scholar]
- Jiang, L.; Wang, Y.; Li, Q.F.; Bjorm, L.O.; He, J.X.; Li, S.S. Arabidopsis STO/BBX24 negatively regulates UV-B signaling by interacting with COP1 and repressing HY5 transcriptional activity. Cell Res 2012, 22, 1046–1057. [Google Scholar]
- Gruber, H.; Heijde, M.; Heller, W.; Albert, A.; Seidlitz, H.K.; Ulm, R. Negative feedback regulation of UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. Proc. Natl. Acad. Sci. USA 2010, 107, 20132–20137. [Google Scholar]
- Gonzalez Besteiro, M.A.; Bartels, S.; Albert, A.; Ulm, R. Arabidopsis MAP kinase phosphatase 1 and its target MAP kinases 3 and 6 antagonistically determine UV-B stress tolerance, independent of the UVR8 photoreceptor pathway. Plant J 2011, 68, 727–737. [Google Scholar]
- Li, J.; Ou-Lee, T.M.; Raba, R.; Amundson, R.G.; Last, R.L. Arabidopsis flavonoid mutants are hypersensitive to UV-B irradiation. Plant Cell 1993, 5, 171–179. [Google Scholar]
- Stracke, R.; Favory, J.J.; Gruber, H.; Bartelniewoehner, L.; Bartels, S.; Binkert, M.; Funk, M.; Weisshaar, B.; Ulm, R. The Arabidopsis bZIP transcription factor HY5 regulates expression of the PFG1/MYB12 gene in response to light and ultraviolet-B radiation. Plant Cell Environ 2010, 33, 88–103. [Google Scholar]
- Xie, Y.; Xu, D.; Cui, W.; Shen, W. Mutation of Arabidopsis HY1 causes UV-C hypersensitivity by impairing carotenoid and flavonoid biosynthesis and the down-regulation of antioxidant defence. J. Exp. Bot 2012, 63, 3869–3883. [Google Scholar]
- Gardner, G.; Lin, C.; Tobin, E.M.; Loehrer, H.; Brinkman, D. Photobiological properties of the inhibition of etiolated Arabidopsis seedling growth by ultraviolet-B irradiation. Plant Cell Environ 2009, 32, 1573–1583. [Google Scholar]
- Degols, G.; Russell, P. Discrete roles of the Spc1 Kinase and the Atf1 transcription factor in the UV response of Schizosacchararomyces peombe. Mol. Cell. Biol 1997, 17, 3356–3363. [Google Scholar]
- Ulm, R. Molecular genetics of genotoxic stress signalling in plants. Topics Curr. Genet 2003, 4, 217–240. [Google Scholar]
- Herrlich, P.; Karin, M.; Weiss, C. Supreme EnLIGHTenment: Damage recognition and signalling in the mammalian response. Mol. Cell 2008, 29, 279–290. [Google Scholar]
- Bartels, S.; Anderson, J.C.; Gonza lez Besteiro, M.A.; Carreri, A.; Hirt, H.; Buchala, A.; Me traux, J.P.; Peck, S.C.; Ulm, R. MAP KINASE PHOSPHATASE1 and PROTEIN TYROSINE PHOSPHATASE1 are repressors of salicylic acid synthesis and SNC1-mediated responses in Arabidopsis. Plant Cell 2009, 21, 2884–2897. [Google Scholar]
- Zhou, C.; Cai, Z.; Guo, Y.; Gan, S. An Arabidopsis mitogen-activated protein kinase cascade MKK9-MPK6 plays a role in leaf senescence. Plant Physiol 2009, 150, 167–177. [Google Scholar]
- Samuel, M.A.; Hall, H.; Krzymowska, M.; Drzewiecka, K.; Hennig, J.; Ellis, B.E. SIPK signalling controls multiple components of harpin-induced cell death in tobacco. Plant J 2005, 42, 406–416. [Google Scholar]
- Adrain, C.; Martin, S.J. The mitochondrial apoptosome: A killer unleashed by the cytochrome seas. Trends Biochem. Sci 2001, 26, 390–397. [Google Scholar]
- Reape, T.J.; McCabe, P.F. Apoptotic-like programmed cell death in plants. New Phytol 2008, 180, 13–26. [Google Scholar]
- Danon, A.; Delorme, V.; Mailhac, N.; Gallois, P. Plant programmed cell death: A common way to die. Plant Physiol. Biochem 2000, 38, 647–655. [Google Scholar]
- Mittler, R.; Lam, E. Sacrifice in the face of foes: Pathogen-induced programmed cell death in plants. Trends Microbiol 1996, 4, 10–15. [Google Scholar]
- Lam, E.; Pontier, D.; del Pozo, O. Die and let live: Programmed cell death in plants. Curr. Opin. Plant Biol 1999, 2, 502–507. [Google Scholar]
- Lam, E.; Pozo, O. Caspase-like protease involvement in the control of plant cell death. Plant Mol. Biol 2000, 44, 417–428. [Google Scholar]
- Wang, H.; Li, J.; Bostock, R.M.; Gilchrist, D.G. Apoptosis: A functional paradigm for programmed cell death induced by a host selective phytotoxin and invoked during development. Plant Cell 1996, 8, 375–391. [Google Scholar]
- McCabe, P.F.; Levine, A.; Meijer, P.J.; Tapon, N.A.; Pennell, R.I. A programmed cell death pathway activated in carrot cells cultured at low cell density. Plant J 1997, 712, 267–280. [Google Scholar]
- McCabe, P.F.; Leaver, C.J. Programmed cell death in cell cultures. Plant Mol. Biol 2000, 44, 359–368. [Google Scholar]
- Kawai, M.; Pan, L.; Reed, J.C.; Uchimiya, H. Evolutionally conserved plant homologue of the Bax inhibitor-1 (BI-1) gene capable of suppressing Bax-induced cell death in yeast. FEBS Lett 1999, 464, 143–147. [Google Scholar]
- He, R.; Drury, G.E.; Rotari, V.I.; Gordon, A.; Willer, M.; Farzaneh, T.; Woltering, E.J.; Gallois, P. Metacaspase-8 modulates programmed cell death induced by ultraviolet light and H2O2 in Arabidopsis. J. Biol. Chem 2008, 283, 774–783. [Google Scholar]
- Jiang, L.; Wang, Y.; Bjorn, L.O.; Li, S. Arabidopsis radical-induced cell death is involved in UV-B signaling. Photochem. Photobiol. Sci 2009, 8, 838–846. [Google Scholar]
- Ries, G.; Heller, W.; Puchta, H.; Sandermann, H.; Seidlitz, H.K.; Hohn, B. Elevated UV-B radiation reduces genome stability in plants. Nature 2000, 406, 98–101. [Google Scholar]
- Hutchinson, F. A review of some topics concerning mutagenesis by ultraviolet light. Photochem. Photobiol 1987, 45, 897–903. [Google Scholar]
- Molinier, J.; Lechner, E.; Dumbliauskas, E.; Genschik, P. Regulation and role of Arabidopsis CUL4-DDB1A-DDB2 in maintaining genome Integrity upon UV stress. PLoS Genet 2008, 4, e1000093. [Google Scholar]
- Liu, Z.; Hossain, G.S.; Islas-Osuna, M.A.; Mitchell, D.L.; Mount, D.W. Repair of UV damage in plants by nucleotide excision repair: Arabidopsis UVH1 DNA repair gene is a homolog of Saccharomyces cerevisiae Rad1. Plant J 2000, 21, 519–552. [Google Scholar]
- Dubest, S.; Gallego, M.E.; White, C.I. Role of the AtRad1p endonuclease in homologous recombination in plants. EMBO Rep 2002, 3, 1049–1054. [Google Scholar]
- Ahmad, M.; Jarillo, J.A.; Klimczak, L.J.; Landry, L.G.; Peng, T.; Last, R.L.; Cashmore, A.R. An enzyme similar to animal type II photolyases mediates Photoreactivation in Arabidopsis. Plant Cell 1997, 9, 199–207. [Google Scholar]
- Nakajima, S.; Sugiyama, M.; Iwai, S.; Hitomi, K.; Otoshi, E.; Kim, S.; Jiang, C.Z.; Todo, T.; Britt, A.B.; Yamamoto, K. Cloning and characterization of a gene (UVR3) required for photorepair of 6–4 photoproducts in Arabidopsis thaliana. Nucleic Acids Res 1998, 26, 638–644. [Google Scholar]
- Zhou, B.B.; Elledge, S.J. The DNA damage response: Putting checkpoints in perspective. Nature 2000, 408, 433–439. [Google Scholar]
- Casati, P.; Walbot, V. Gene expression profiling in response to ultraviolet radiation in maize genotypes with varying flavonoid content. Plant Physiol 2003, 132, 1739–1754. [Google Scholar]
- Jiang, L.; Wang, Y.; Bjorn, L.O.; Li, S. UV-B induced DNA damage mediates expression changes of cell cycle regulatory genes in Arabidopsis root tips. Planta 2011, 233, 831–841. [Google Scholar]
- Preuss, S.B.; Britt, A.B. A DNA damage induced cell cycle checkpoint in Arabidopsis. Genetics 2003, 164, 323–334. [Google Scholar]
- Culligan, K.; Tissier, A.; Britt, A. ATR regulates a G2-Phase cell-cycle checkpoint in Arabidopsis thaliana. Plant Cell 2004, 16, 1091–1104. [Google Scholar]
- Garcia, V.; Bruchet, H.; Camescasse, D.; Granier, F.; Bouchez, D.; Tissier, A. AtATM is essential for meiosis and the somatic response to DNA damage in plants. Plant Cell 2003, 15, 119–132. [Google Scholar]
- Landry, L.G.; Stapleton, A.E.; Lim, J.; Hoffman, P.; Hays, J.B.; Walbot, V.; Last, R.L. An Arabidopsis photolyase mutant is hypersensitive to ultraviolet-B radiation. Proc. Natl. Acad. Sci. USA 1997, 94, 328–332. [Google Scholar]
- Molinier, J.; Ramos, C.; Fritsch, O.; Hohn, B. CENTRIN2 modulates holologous recombination and nucleotide excision repair in Arabidopsis. Plant Cell 2004, 16, 1633–1643. [Google Scholar]
- Furukawa, T.; Curtis, M.J.; Tominey, C.M.; Duong, Y.H.; Wilcox, B.W.L.; Aggoune, D.; Hays, J.B.; Britt, A.B. A shared DNA-damage response pathway for induction of stem-cell death by UVB and by gamma irradiation. DNA Repair 2010, 9, 940–948. [Google Scholar]
- Sakamoto, A.N.; Lan, V.T.; Puripunyavanich, V.; Hase, Y.; Yokota, Y.; Shikazono, N.; Nakagawa, M.; Narumi, I.; Tanaka, A. A UVB-hypersensitive mutant in Arabidopsis thaliana is defective in the DNA damage response. Plant J 2009, 60, 509–517. [Google Scholar]
- Stein, J.C.; Hansen, G. Mannose induces an endonuclease responsible for DNA laddering in plant cells. Plant Physiol 1999, 121, 71–80. [Google Scholar]
- Houot, V.; Etienne, P.; Petitot, A.S.; Barbier, S.; Blein, J.P.; Suty, L. Hydrogen peroxide induces programmed cell death features in cultured tobacco BY-2 cells, in a dose-dependent manner. J. Exp. Bot 2001, 52, 1721–1730. [Google Scholar]
- Gorczyca, W.; Tuziak, T.; Kram, A.; Melamed, M.R.; Darzynkiewicz, Z. Detection of apoptosis-associated DNA strand breaks in fine-needle aspiration biopsies by in situ end labeling of fragmented DNA. Cytometry 1994, 15, 169–175. [Google Scholar]
- Khoroshilova, E.V.; Repeyev, Y.A.; Nikogosyan, D.N. UV photolysis of aromatic amino acids and related dipeptides and tripeptides. Photochem. Photobiol 1990, 7, 159–172. [Google Scholar]
- Caldwell, C.R. Ultraviolet-induced photodegradation of cucumber (Cucumis sativus L.) microsomal and soluble protein tryptophanyl residues in vitro. Plant Physiol 1993, 101, 947–953. [Google Scholar]
- Fiscus, E.L.; Booker, F.L. Is increased UV-B a threat to crop photosynthesis and productivity? Photosynth. Res 1995, 43, 81–92. [Google Scholar]
- Casati, P.; Walbot, V. Crosslinking of ribosomal proteins to RNA in maize ribosomes by UV-B and its effects on translation. Plant Physiol 2004, 136, 3319–3332. [Google Scholar]
- Kramer, G.F.; Norman, H.L.; Krizek, D.T.; Mirecki, R.M. Influence of UV-B radiation on polyamines, lipid peroxidation and membrane lipids in cucumber. Phytochemistry 1991, 30, 2101–2108. [Google Scholar]
- Panagopoulos, I.; Bomman, J.F.; Bjorn, L.O. Effects of ultraviolet radiation and visible light on growth, fluorescence induction, ultra weak luminescence and peroxidase activity in sugar beet plants. Photochem. Photobiol 1990, 8, 73–87. [Google Scholar]
- Mishra, R.K.; Singhal, G.S. Function of photosynthetic apparatus of intact wheat leaves under high light and heat stress and its relationship with peroxidation of thylakoid lipids. Plant Physiol 1992, 98, 1–6. [Google Scholar]
- Murphy, T.M.; Vu, H. Photoactivation of superoxide synthases of the plasma membrane from rose (Rosa damascene Mill.) cells. Photochem. Photobiol 1996, 64, 106–109. [Google Scholar]
- Predieri, S.; Norman, H.A.; Krizek, D.T.; Pillai, P.; Mirecki, R.M.; Zimmerman, R.H. Influence of UV-B radiation on membrane lipid composition and ethylene evolution in (Doyenne D’ Hiver) pear shoots grown in vitro under different photosynthetic photon fluxes. Environ. Exp. Bot 1995, 35, 151–160. [Google Scholar]
- Dat, J.; Vandenabeele, S.; Vranova, E.; van Montagu, M.; Inze, D.; van Breusegem, F. Dual action of the active oxygen species during plant stress responses. Cell Mol. Life Sci 2000, 57, 779–795. [Google Scholar]
- Foyer, C.H.; Noctor, G. Redox homeostasis and antioxidant signaling: A metabolic interface between stress perception and physiological responses. Plant Cell 2005, 17, 1866–1875. [Google Scholar]
- Zapata, J.M.; Guera, A.; Esteban-Carrasco, A.; Martin, M.; Sabater, B. Chloroplasts regulate leaf senescence: Delayed senescence in transgenic ndhF-defective tobacco. Cell Death Differ 2005, 12, 1277–1284. [Google Scholar]
- Chen, S.R.; Dickman, M.B. Bcl-2 family localize to tobacco chloroplasts and inhibit programmed cell death induced by chloroplast- targeted herbicides. J. Exp. Bot 2004, 55, 2617–2623. [Google Scholar]
- Yao, N.; Eisfelder, B.J.; Marvin, J.; Greenberg, J.T. The mitochondrion-an organelle commonly involved in programmed cell death in Arabidopsis thaliana. Plant J 2004, 40, 596–610. [Google Scholar]
- Wang, X. The expanding role of mitochondria in apoptosis. Genes Dev 2001, 15, 2922–2933. [Google Scholar]
- Wolf, B.B.; Green, D.R. Suicidal tendencies: Apoptotic cell death by caspase family proteinases. J. Biol. Chem 1999, 274, 20049–20052. [Google Scholar]
- Mateo, A.; Muhlenbock, P.; Rusterucci, C.; Chang, C.C.; Miszalski, Z.; Karpinska, B.; Parker, J.E.; Mullineaux, P.M.; Karpinski, S. Lesion simulating disease is required for acclimation to conditions that promote excess excitation energy. Plant Physiol 2004, 136, 2818–2830. [Google Scholar]
- Del Pozo, O.; Lam, E. Caspases and programmed cell death in the hypersensitive response of plants to pathogens. Curr. Biol 1998, 8, 1129–1132. [Google Scholar]
- Tian, R.; Zhang, G.Y.; Yan, C.H.; Dai, Y.R. Involvement of poly (ADP-ribose) polymerase and activation of caspase-3-like protease in heat shock-induced apoptosis in tobacco suspension cells. FEBS Lett 2000, 474, 5–11. [Google Scholar]
- Uren, A.G.; Rourke, K.; Aravind, L.; Pisabarro, M.T.; Seshagiri, S.; Koonin, E.V.; Dixit, V.M. Identification of paracaspases and metacaspases: Two ancient families of caspase-like proteins, one of which plays a key role in MALT lymphoma. Mol. Cell 2000, 6, 961–967. [Google Scholar]
- Watanabe, N.; Lam, E. Recent advance in the study of caspase-like proteases and Bax inhibitor-1 in plants: Their possible roles as regulator of programmed cell death. Mol. Plant Pathol 2004, 5, 65–70. [Google Scholar]
- Vercammen, D.; van de Cotte, B.; de Jaeger, G.; Eeckhout, D.; Casteels, P.; Vandepoele, K.; Vandenberghe, I.; van Beeumen, J.; Inze, D.; van Breusegem, F. Type II metacaspases Atmc4 and Atmc9 of Arabidopsis thaliana cleave substrates after arginine and lysine. J. Biol. Chem 2004, 279, 45329–45336. [Google Scholar]
- Watanabe, N.; Lam, E. Two Arabidopsis metacaspases AtMCP1 band AtMCP2b are arginine/lysine-specific cysteine proteases and activate apoptosis-like cell death in yeast. J. Biol. Chem 2005, 280, 14691–14699. [Google Scholar]
- Zhang, L.; Xu, Q.; Xing, D.; Gao, C.; Xiong, H. Real-time detection of caspase-3-like protease activation in vivo using fluorescence resonance energy transfer during plant programmed cell death induced by ultraviolet C overexposure. Plant Physiol 2009, 150, 1773–1783. [Google Scholar]
- Vranova, E.; Inze, D.; van Breusegem, F. Signal transduction during oxidative stress. J. Exp. Bot 2002, 53, 1227–1236. [Google Scholar]
- Apel, K.; Hirt, H. Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol 2004, 55, 373–399. [Google Scholar]
- Breusegem, F.V.; Dat, J.F. Reactive oxygen species in plant cell death. Plant Physiol 2006, 141, 384–390. [Google Scholar]
- Mittler, R.; Vanderauwera, S.; Suzuki, N.; Miller, G.; Tognetti, V.B.; Vandepoele, K.; Gollery, M.; Shulaey, V.; van Breusegem, F. ROS signalling: The new wave? Trends Plant Sci 2011, 16, 300–309. [Google Scholar]
- De Tullio, M.C. Antioxidants and redox regulation: Changing notions in a changing world. Plant Physiol. Biochem 2010, 48, 289–291. [Google Scholar]
- Hideg, E.; Vass, I. UV-B induced free radical production in plant leaves and isolated thylakoid membranes. Plant Sci 1996, 115, 251–260. [Google Scholar]
- Kalbina, I.; Strid, A. The role of NADPH oxidase and MAP kinase phosphatase in UV-B-dependent gene expression in Arabidopsis. Plant Cell Environ 2006, 29, 1783–1793. [Google Scholar]
- Mackerness, S.; John, C.F.; Jordan, B.; Thomas, B. Early signaling components in ultraviolet-B responses: Distinct roles for different reactive oxygen species and nitric oxide. FEBS Lett 2001, 489, 237–242. [Google Scholar]
- Pontier, D.; Balague, C.; Roby, D. The hypersensitive response. A programmed cell death associated with plant resistance. C R Acad. Sci. III 1998, 321, 721–734. [Google Scholar]
- Jabs, T. Reactive oxygen intermediates as mediators of programmed cell death in plants and animals. Biochem. Pharmacol 1999, 57, 231–245. [Google Scholar]
- Vacca, R.A.; Valenti, D.; Bobba, A.; Merafina, R.S.; Passarella, S.; Marra, E. Cytochrome c is released in a reactive oxygen species-dependent manner and is degraded via caspase-like proteases in tobacco Bright-Yellow 2 cells en route to heat shock-induced cell death. Plant Physiol 2006, 141, 208–219. [Google Scholar]
- Overmyer, K.; Brosche, M.; Pellinen, R.; Kuittinen, T.; Tuominen, H.; Ahlfors, R.; Keinanen, M.; Saarma, M.; Scheel, D.; Kangasjarvi, J. Ozone-induced programmed cell death in the Arabidopsis radical-induced cell death1 mutant. Plant Physiol 2005, 137, 1092–1104. [Google Scholar]
- Surplus, S.L.; Jordan, B.R.; Murphy, A.M.; Carr, J.P.; Thomas, B.; Mackerness, S. UV-B induced responses in Arabidopsis thaliana: Role of salicylic acid and ROS in the regulation of transcripts and acidic PR proteins. Plant Cell Environ 1998, 21, 685–694. [Google Scholar]
- Mackerness, S. Plant responses to ultraviolet-B (UV-B: 280–320 nm) stress: What are the key regulators? Plant Growth Regul 2000, 32, 27–39. [Google Scholar]
- Demkura, P.V.; Abdala, G.; Baldwin, I.T.; Ballare, C.L. Jasmonate-dependent and independent pathways mediate specific effects of solar ultraviolet B radiation on leaf phenolics and antiherbivore defense. Plant Physiol 2010, 152, 1084–1095. [Google Scholar]
- Demkura, P.V.; Ballare, C.L. UVR8 mediates UV-B induced Arabidopsis defense responses against Botrytis cinerea by controlling sinapate accumulation. Mol. Plant 2012, 5, 642–652. [Google Scholar]
- Mackerness, S.; Surplus, S.L.; Blake, P.; John, C.F.; Buchanan-Wollaston, V.; Jordan, B.R.; Thomas, B. UV-B induced stress and changes in gene expression in Arabidopsis thalaina: Role of signaling pathways controlled by jasmonic acid, ethylene and reactive oxygen species. Plant Cell Environ 1999, 22, 1413–1424. [Google Scholar]
- Lawton, K.A.; Potter, S.L.; Uknes, S.; Ryals, J. Acquired resistance signal transduction in Arabidopsis is ethylene independent. Plant Cell 1994, 6, 581–588. [Google Scholar]
| Symbol | AGI code | Full name | Phenotype of UV-grown mutant seedlings | Reference |
|---|---|---|---|---|
| UVR8 | AT5G63860 | UVB-RESISTANCE 8 | hypersensitive to UV-B and blocks expression of UV-B induced genes, acts as a UV-B photoreceptor | [35, 37, 41] |
| COP1 | AT2G32950 | CONSTITUTIVE PHOTOMORPHOGENIC 1 | hypersensitive to UV-B and blocks UVR8-dependent expression of UV-B induced genes | [42] |
| HY5 | AT5G11260 | ELONGATED HYPOCOTYL 5 | hypersensitive to UV-B and blocks UVR8-COP1 dependent expression of chalcone synthase and flavonoid synthase pathway | [14] |
| HYH | AT3G17609 | HY5-HOMOLOG | hypersensitive to UV-B and showed overlapping functions with HY5 in affecting UV-B induced gene expression | [43] |
| BBX24 | AT1G06040 | B-BOX DOMAIN PROTEIN 24 | hypersensitive to UV-B and BBX24 interacts with COP1 and antagonizes the transcriptional activity of HY5 in response to UV-B | [44] |
| RUP1 | AT5G52250 | REPRESSOR OF UV-B PHOTOMORPHOGENESIS 1 | no obvious phenotype but acts redundantly to RUP2. The double rup1rup2 mutant is hypersensitive to UV-B | [45] |
| RUP2 | AT5G23730 | REPRESSOR OF UV-B PHOTOMORPHOGENESIS 2 | hypersensitive to UV-B and acts in feedback regulation of UV signaling | [45] |
| MKP1 | AT1G10210 | MITOGEN-ACTIVATED PROTEIN KINASE 1 | mutant is hypersensitive to acute UV-B stress due to mis-regulation of MPK3 and MPK6 | [46] |
| MPK3 | AT3G45640 | MITOGEN-ACTIVATED PROTEIN KINASE 3 | mutants display increased tolerance to UV-B radiation | [46] |
| MPK6 | AT2G43790 | MITOGEN-ACTIVATED PROTEIN KINASE 6 |
| Symbol | AGI code | Full name | Phenotype of UV-grown mutant seedlings | Reference |
|---|---|---|---|---|
| TT4 | AT5G13930 | TRANSPARENT TESTA 4/ CHALCONE SYNTHASE | has reduced flavonoids and normal levels of sinapate esters, is more sensitive to UV-B | [47] |
| TT5 | AT3G55120 | TRANSPARENT TESTA 5/ CHALCONE ISOMERASE | has reduced levels of UV-absorptive leaf flavonoids and the monocyclic sinapic acid ester phenolic compounds, are highly sensitive to UV-B | [47] |
| TT6 | AT3G51240 | TRANSPARENT TESTA 6/ FLAVANONE 3 HYDROXYLASE | similar to tt5, mutants are highly sensitive to UV-B light damage | [47] |
| PFG1 | AT2G47460 | PRODUCTION OF FLAVONOL GLYCOSIDES 1 | acts downstream of HY5 and overexpression is sufficient to increase UV-B tolerance | [48] |
| PFG2 | AT3G62610 | PRODUCTION OF FLAVONOL GLYCOSIDES 2 | hypersensitive to UV-B due to low levels of flavonol compounds n response to UV-B | [48] |
| HO1 | AT2G26670 | HEME OXYGENASE 1 | hypersensitive to UV-C due to down regulation of flavonoid and carotenoid metabolism as well as antioxidant defense mechanisms | [49] |
| ULI3 | AT5G59920 | UV-B LIGHT INSENSITIVE 3 | mutant is hyposensitive to UV-B and affected in its hypocotyls elongation but was also impaired in CHS and PR-1gene expression after irradiation with continuous UV-B | [15] |
| Symbol | AGI code | Full name | Effect on UV-induced cell death | Reference |
|---|---|---|---|---|
| AtDAD1 | AT1G32210 | DEFENDER AGAINST APOPTOTIC DEATH 1 | overexpression can suppress the DNA fragmentation caused by DEVDase activity and retard UV-B induced cell death | [17] |
| AtDAD2 | AT2G35520 | DEFENDER AGAINST CELL DEATH 2 (DAD2) | ||
| AtBI | AT5G47120 | BAX INHIBITOR 1 | anti-apoptotic protein increases cell survivability under abiotic stresses | [66] |
| AtMC8 | AT1G16420 | METACASPASE 8 | induced in response to UV-C radiation and induces cell death since knock out mutant is tolerant | [67] |
| RCD1 | AT1G32230 | RADICAL-INDUCED CELL DEATH1 | mutants are sensitive to ozone and apoplastic superoxides but tolerant to ROS and UV-B stress | [67, 68] |
| Symbol | AGI code | Full name | Phenotype of UV-grown mutant seedlings | Reference |
|---|---|---|---|---|
| UVR2/PHR1 | AT1G12370 | UV REPAIR DEFECTIVE 2/ PHOTOLYASE 1 | mutant is impaired in the CPD photolyase gene PHR1 and thus, hypersensitive to high doses of UV-B | [82] |
| UVR3 | AT3G15620 | UV REPAIR DEFECTIVE 3 | mutant is hypersensitive to high doses of UV-B as it is defective in photoreactivation of 6-4 photoproducts | [75] |
| UVR1/UVH1 | AT3G28030 | UV REPAIR DEFECTIVE 1 | mutants are defective in dark repair or Nucleotide Excision Repair and shows hypersensitive to UV radiation | [72] |
| CEN2 | AT4G37010 | CENTRIN 2 | mutants exhibited a moderate UV-C sensitivity due to defective Homologous Recombination and Nucleotide Excision Repair | [83] |
| ATR | AT5G40820 | ATAXIA TELANGIECTASIA-MUTATED AND RAD3-RELATED | mutants is defective in cell-cycle arrest in response to DNA damage, and this causes elevated cell death under high level of UV-B | [80] |
| ATM | AT3G48190 | ATAXIA-TELANGIECTASIA MUTATED | mutants show elevated cell death under high level UV-B due to DNA damage | [84] |
| SOG1 | AT1G25580 | SUPPRESSOR OF GAMMA RADIATION 1 | mutant show reduced PCD in response to UV-B induced DNA photoproducts | [84] |
| SUV2 | AT5G45610 | SENSITIVE TO UV 2 | mutant is UV-B sensitive which has a weaker expression of CYCB1;1 in response to DNA damage | [85] |
© 2013 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Nawkar, G.M.; Maibam, P.; Park, J.H.; Sahi, V.P.; Lee, S.Y.; Kang, C.H. UV-Induced Cell Death in Plants. Int. J. Mol. Sci. 2013, 14, 1608-1628. https://doi.org/10.3390/ijms14011608
Nawkar GM, Maibam P, Park JH, Sahi VP, Lee SY, Kang CH. UV-Induced Cell Death in Plants. International Journal of Molecular Sciences. 2013; 14(1):1608-1628. https://doi.org/10.3390/ijms14011608
Chicago/Turabian StyleNawkar, Ganesh M., Punyakishore Maibam, Jung Hoon Park, Vaidurya Pratap Sahi, Sang Yeol Lee, and Chang Ho Kang. 2013. "UV-Induced Cell Death in Plants" International Journal of Molecular Sciences 14, no. 1: 1608-1628. https://doi.org/10.3390/ijms14011608




