Reconstitution of Membrane Proteins into Model Membranes: Seeking Better Ways to Retain Protein Activities
Abstract
:1. Introduction
2. Reconstitution of Membrane Protein into Model Membranes
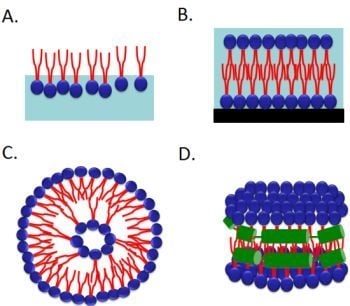
2.1. Langmuir Monolayer at the Air–Water Interface
2.1.1. Transmembrane Protein Structure in Monolayers
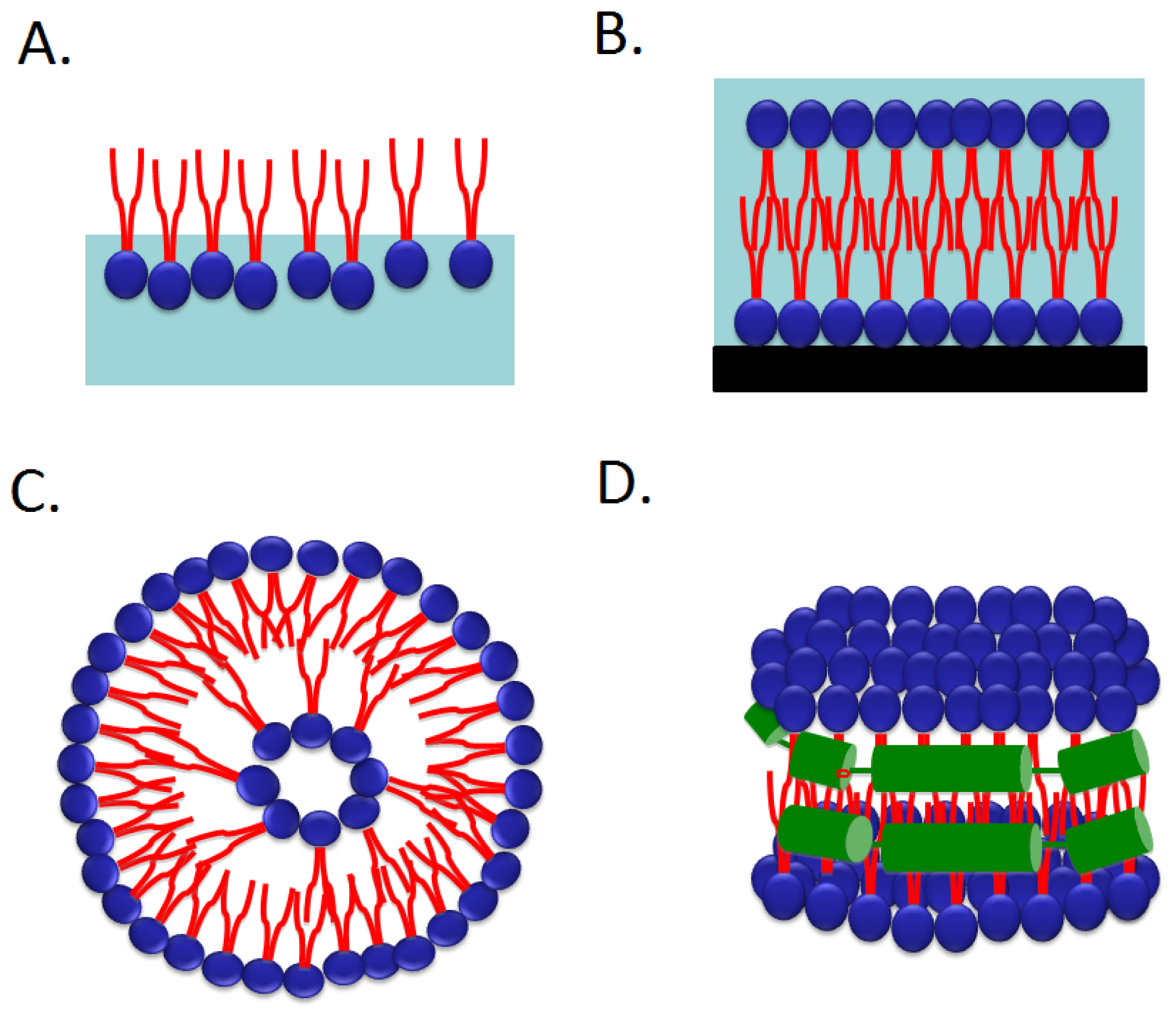
2.1.2. Binding of Peripheral Proteins onto Monolayer
2.2. Supported Planar Lipid Bilayer
2.3. Liposomes
2.3.1. Activity of Membrane-Bound Enzymes
2.3.2. Transporters
2.4. Nanodiscs
2.4.1. G Protein Coupled Receptors [55]
2.4.2. Enzymatic Activities, Cytochrome P450 and Its Ligand Binding
2.4.3. Transporters and Channels
3. Comparisons
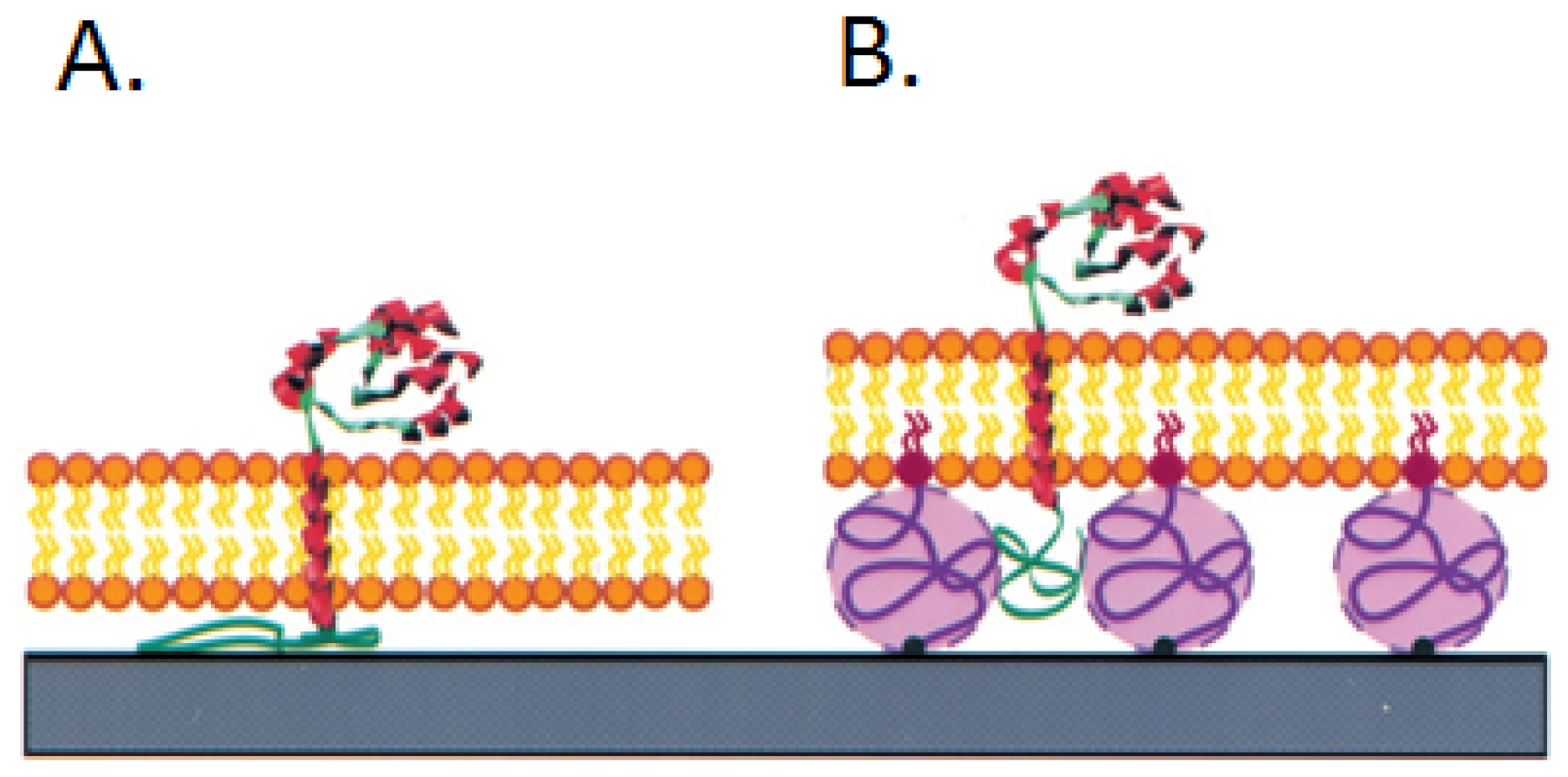
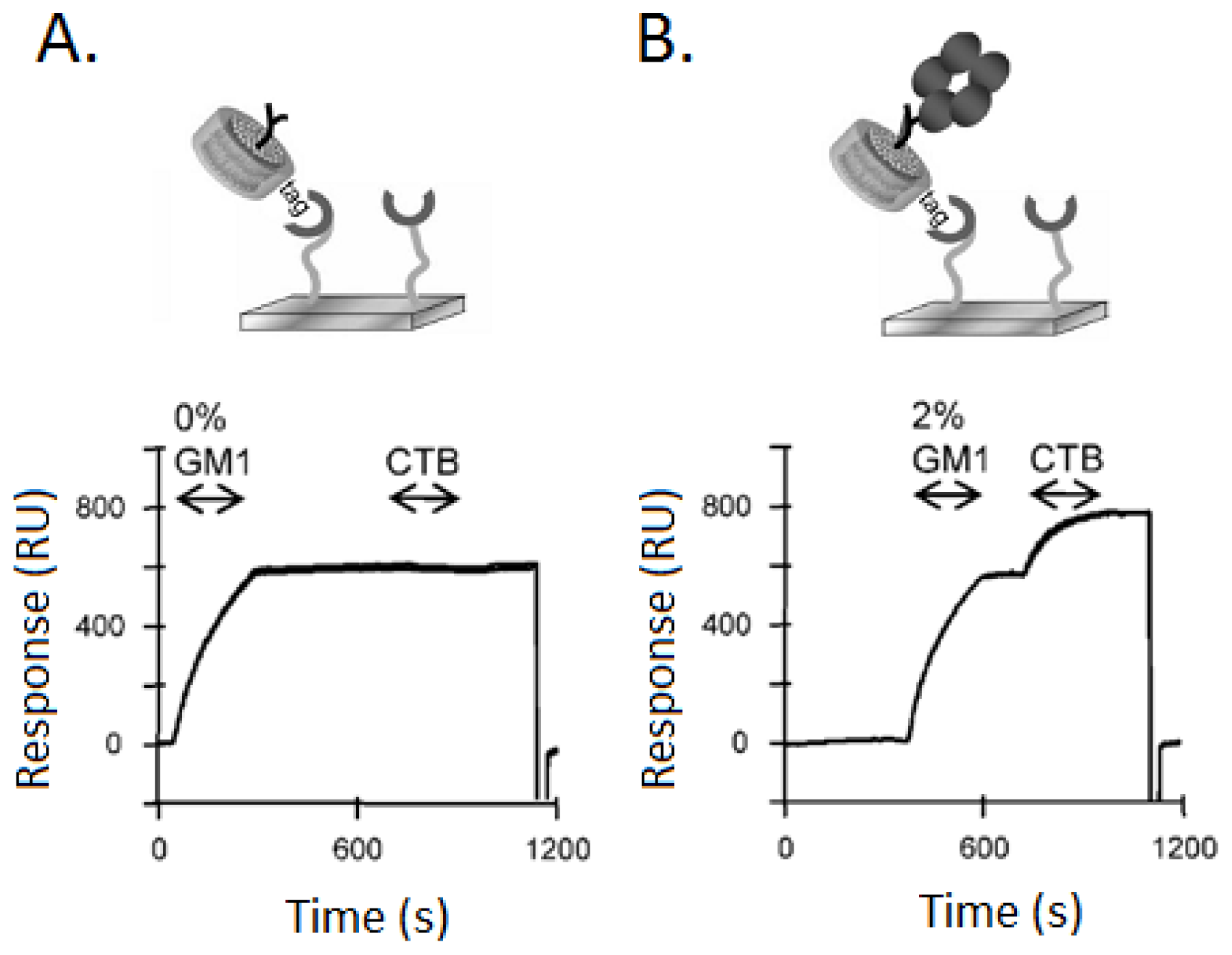
3.1. Ganglioside GM1 Receptors Binding Activity
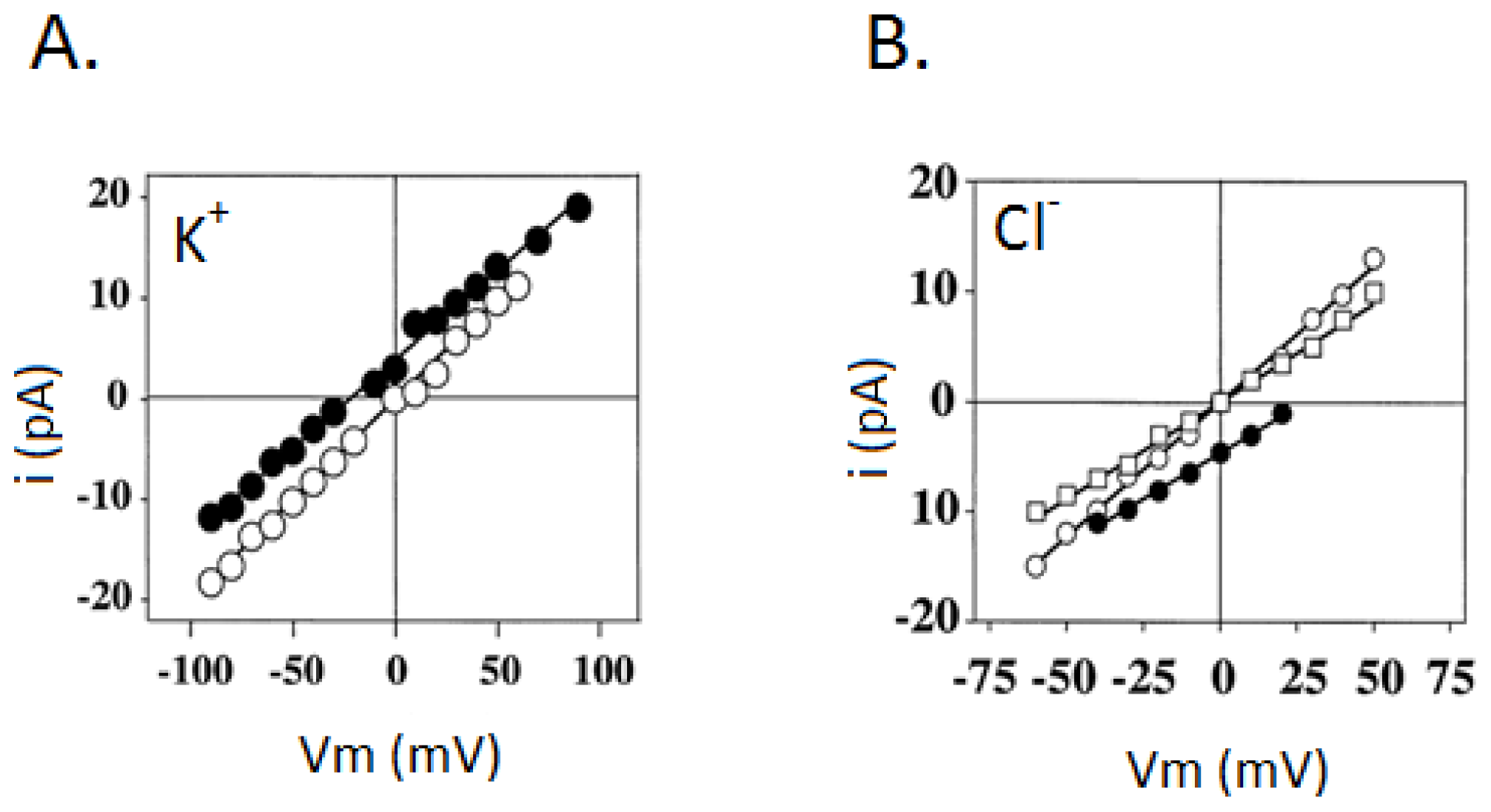
3.2. Liver Nuclear Ionic Channels
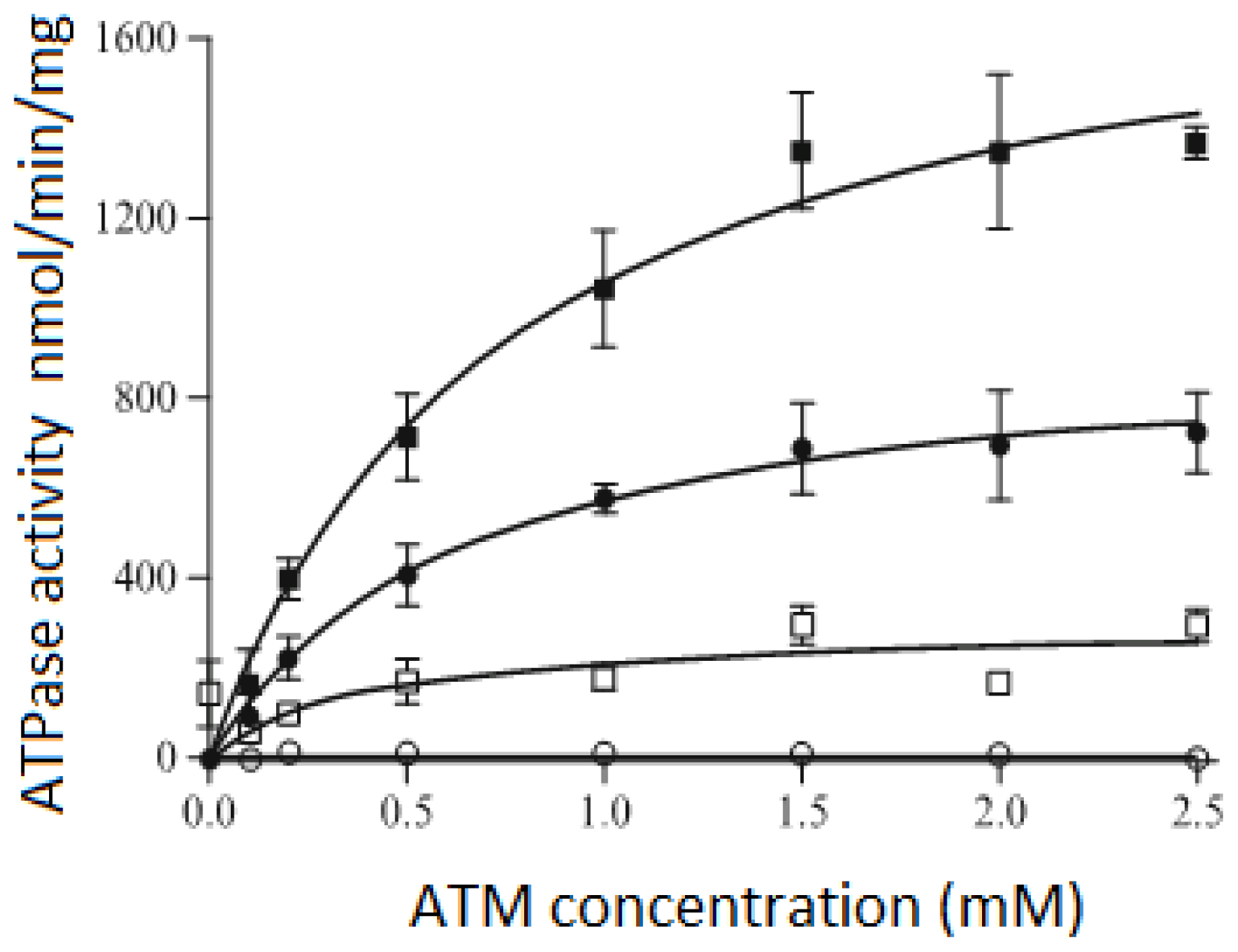
3.3. ATPase Activity of the P-Glycoprotein Transporter
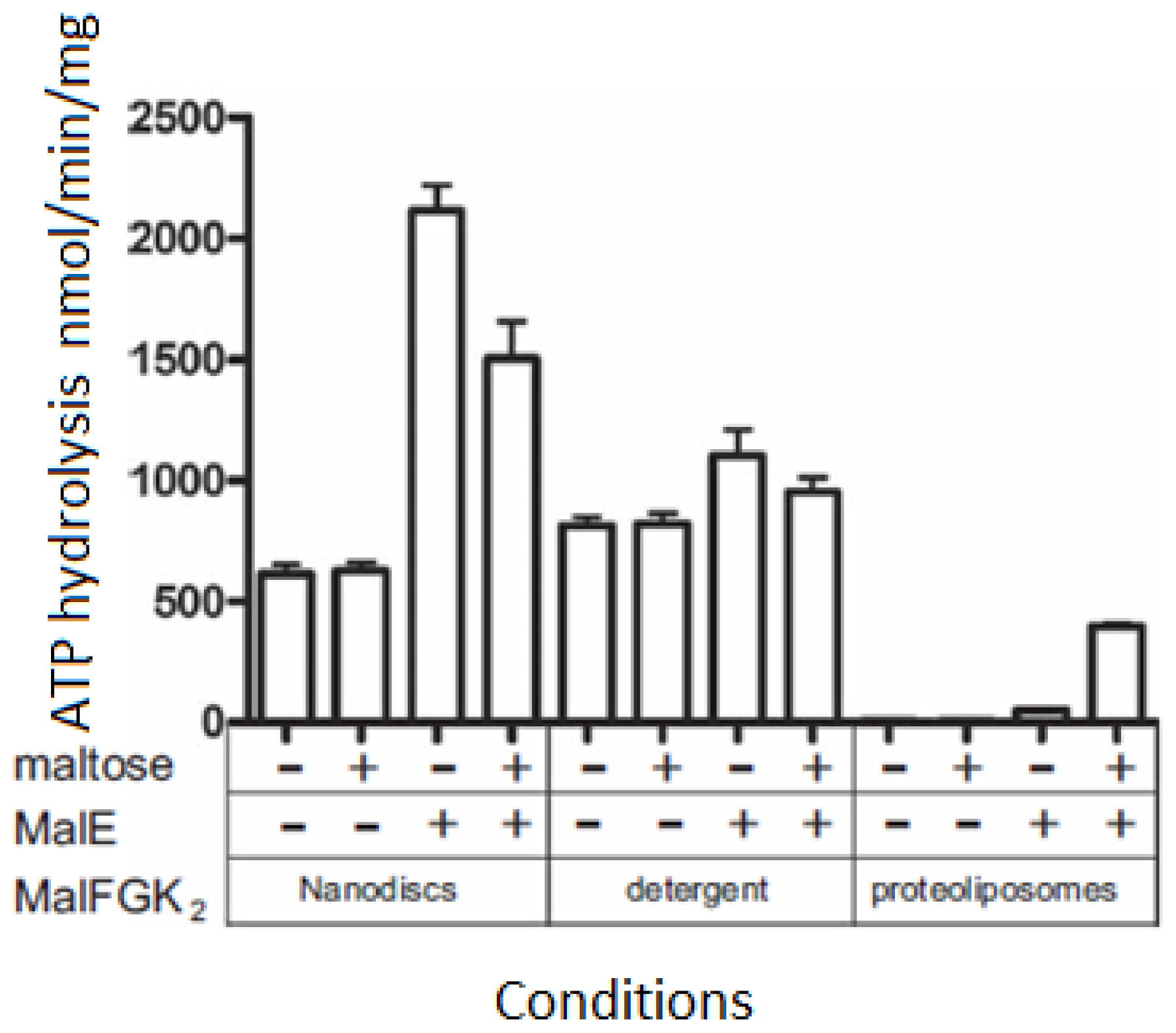
3.4. ATPase Activity of the MalFGK2 Complex in Nanodiscs, Detergents and Proteoliposomes
4. Conclusions
Acknowledgements
References
- Liao, Y.; Yuan, Q.; Torres, J.; Tam, J.P.; Liu, D.X. Biochemical and functional characterization of the membrane association and membrane permeabilizing activity of the severe acute respiratory syndrome coronavirus envelope protein. Virology 2006, 349, 264–275. [Google Scholar]
- Frenkel, E.J.; Roelofsen, B.; Brodbeck, U.; van Deenen, L.L.; Ott, P. Lipid-protein interactions in human erythrocyte-membrane acetylcholinesterase. Modulation of enzyme activity by lipids. Eur. J. Biochem 1980, 109, 377–382. [Google Scholar]
- Aroca, J.D.; Sanchez-Pinera, P.; Corbalan-Garcia, S.; Conesa-Zamora, P.; de Godos, A.; Gomez-Fernandez, J.C. Correlation between the effect of the anti-neoplastic ether lipid 1-O-octadecyl-2-O-methyl-glycero-3-phosphocholine on the membrane and the activity of protein kinase Calpha. Eur. J. Biochem 2001, 268, 6369–6378. [Google Scholar]
- Kucik, D.F.; Elson, E.L.; Sheetz, M.P. Weak dependence of mobility of membrane protein aggregates on aggregate size supports a viscous model of retardation of diffusion. Biophys. J 1999, 76, 314–322. [Google Scholar]
- Kaganer, V.M. Structure and phase transitions in Langmuir monolayers. Rev. Mod. Phys 1999, 71, 779–819. [Google Scholar]
- Brian, A.A.; McConnell, H.M. Allogeneic stimulation of cytotoxic T cells by supported planar membranes. Proc. Natl. Acad. Sci. USA 1984, 81, 6159–6163. [Google Scholar]
- Tokuda, H.; Konisky, J. Effect of colicins Ia and E1 on ion permeability of liposomes. Proc. Natl. Acad. Sci. USA 1979, 76, 6167–6171. [Google Scholar]
- Civjan, N.R.; Bayburt, T.H.; Schuler, M.A.; Sligar, S.G. Direct solubilization of heterologously expressed membrane proteins by incorporation into nanoscale lipid bilayers. Biotechniques 2003, 35, 556–560. [Google Scholar]
- Mangeney, C.; Dupres, V.; Roche, Y.; Felidj, N.; Levi, G.; Aubard, J.; Bernard, S. Surface enhanced Raman scattering of a lipid Langmuir monolayer at the air-water interface. Biopolymers 2004, 74, 136–140. [Google Scholar]
- Brockman, H. Lipid monolayers: Why use half a membrane to characterize protein-membrane interactions? Curr. Opin. Struc. Biol 1999, 9, 438–443. [Google Scholar]
- Nakahara, H.; Lee, S.; Sugihara, G.; Chang, C.H.; Shibata, O. Langmuir monolayer of artificial pulmonary surfactant mixtures with an amphiphilic peptide at the air/water interface: Comparison of new preparations with surfactant. Langmuir 2008, 24, 3370–3379. [Google Scholar]
- Clausell, A.; Busquets, M.A.; Pujol, M.; Alsina, A.; Cajal, Y. Polymyxin B-lipid interactions in Langmuir-Blodgett monolayers of Escherichia coli lipids: A thermodynamic and atomic force microscopy study. Biopolymers 2004, 75, 480–490. [Google Scholar]
- Costa, A.P.; Xu, X.; Burgess, D.J. Langmuir balance investigation of superoxide dismutase interactions with mixed-lipid monolayers. Langmuir 2012, 28, 10050–10056. [Google Scholar]
- Dynarowicz-Latka, P.; Dhanabalan, A.; Oliveira, O.N., Jr. Modern physicochemical research on Langmuir monolayers. Adv. Colloid Interface Sci. 2001, 91, 221–293. [Google Scholar]
- Lavoie, H.; Desbat, B.; Vaknin, D.; Salesse, C. Structure of rhodopsin in monolayers at the air-water interface: A PM-IRRAS and X-ray reflectivity study. Biochemistry 2002, 41, 13424–13434. [Google Scholar]
- Korenbrot, J.I.; Pramik, M.J. Formation, structure, and spectrophotometry of air-water interface films containing rhodopsin. J. Membr. Boil 1977, 37, 235–262. [Google Scholar]
- Korenbrot, J.I.; Hwang, S.B. Proton transport by bacteriorhodopsin in planar membranes assembled from air-water interface films. J. Gen. Physiol 1980, 76, 649–682. [Google Scholar]
- Hwang, S.B.; Korenbrot, J.I.; Stoeckenius, W. Structural and spectroscopic characteristics of bacteriorhodopsin in air-water interface films. J. Membr. Boil 1977, 36, 115–135. [Google Scholar]
- Lavoie, H.; Blaudez, D.; Vaknin, D.; Desbat, B.; Ocko, B.M.; Salesse, C. Spectroscopic and structural properties of valine gramicidin A in monolayers at the air-water interface. Biophys. J 2002, 83, 3558–3569. [Google Scholar]
- Biswas, S.C.; Rananavare, S.B.; Hall, S.B. Effects of gramicidin-A on the adsorption of phospholipids to the air-water interface. Biochim. Biophys. Acta 2005, 1717, 41–49. [Google Scholar]
- Boucher, J.; Trudel, E.; Methot, M.; Desmeules, P.; Salesse, C. Organization, structure and activity of proteins in monolayers. Colloid. Surface. B 2007, 58, 73–90. [Google Scholar]
- Grandbois, M.; Desbat, B.; Salesse, C. Monitoring of phospholipid monolayer hydrolysis by phospholipase A2 by use of polarization-modulated Fourier transform infrared spectroscopy. Biophys. Chem 2000, 88, 127–135. [Google Scholar]
- McConnell, H.M.; Watts, T.H.; Weis, R.M.; Brian, A.A. Supported planar membranes in studies of cell-cell recognition in the immune system. Biochim. Biophys. Acta 1986, 864, 95–106. [Google Scholar]
- Tamm, L.K.; McConnell, H.M. Supported phospholipid bilayers. Biophys. J. 1985, 47, 105–113. [Google Scholar]
- Urisu, T.; Rahman, M.M.; Uno, H.; Tero, R.; Nonogaki, Y. Formation of high-resistance supported lipid bilayer on the surface of a silicon substrate with microelectrodes. Nanomedicine 2005, 1, 317–322. [Google Scholar]
- Furukawa, K.; Sumitomo, K.; Nakashima, H.; Kashimura, Y.; Torimitsu, K. Supported lipid bilayer self-spreading on a nanostructured silicon surface. Langmuir 2007, 23, 367–371. [Google Scholar]
- Dimitrievski, K.; Reimhult, E.; Kasemo, B.; Zhdanov, V.P. Simulations of temperature dependence of the formation of a supported lipid bilayer via vesicle adsorption. Colloid. Surfaces B 2004, 39, 77–86. [Google Scholar]
- Allerbo, O.; Lundstrom, A.; Dimitrievski, K. Simulations of lipid vesicle rupture induced by an adjacent supported lipid bilayer patch. Colloid. Surfaces B 2011, 82, 632–636. [Google Scholar]
- Deverall, M.A.; Gindl, E.; Sinner, E.-K.; Besir, H.; Ruehe, J.; Saxton, M.J.; Naumann, C.A. Membrane Lateral Mobility Obstructed by Polymer-Tethered Lipids Studied at the Single Molecule Level. Biophys. J 2000, 79, 1400–1414. [Google Scholar]
- Wagner, M.L.; Tamm, L.K. Tethered polymer-supported planar lipid bilayers for reconstitution of integral membrane proteins: Silane-polyethyleneglycol-lipid as a cushion and covalent linker. Biophys. J 2005, 88, 1875–1886. [Google Scholar]
- Rehfeldt, F.; Steitz, R.; Armes, S.P.; von Klitzing, R.; Gast, A.P.; Tanaka, M. Reversible activation of diblock copolymer monolayers at the interface by pH modulation, 1: Lateral chain density and conformation. J. Phys. Chem. B 2006, 110, 9171–9176. [Google Scholar]
- Wong, J.Y.; Majewski, J.; Seitz, M.; Park, C.K.; Israelachvili, J.N.; Smith, G.S. Polymer-cushioned bilayers. I. A structural study of various preparation methods using neutron reflectometry. Biophys. J 1999, 77, 1445–1457. [Google Scholar]
- Wong, J.Y.; Park, C.K.; Seitz, M.; Israelachvili, J. Polymer-cushioned bilayers. II. An investigation of interaction forces and fusion using the surface forces apparatus. Biophys. J 1999, 77, 1458–1468. [Google Scholar]
- Renner, L.; Pompe, T.; Lemaitre, R.; Drechsel, D.; Werner, C. Controlled enhancement of transmembrane enzyme activity in polymer cushioned supported bilayer membranes. Soft Matter 2010, 6, 5382–5389. [Google Scholar]
- Iwahashi, Y.; Nakamura, T. Orientation and reactivity of NADH kinase in proteoliposomes. J. Biochem 1989, 105, 922–926. [Google Scholar]
- Brown, M.F. Modulation of rhodopsin function by properties of the membrane bilayer. Chem. Phys. Lipids 1994, 73, 159–180. [Google Scholar]
- Escriba, P.V.; Ozaita, A.; Ribas, C.; Miralles, A.; Fodor, E.; Farkas, T.; Garcia-Sevilla, J.A. Role of lipid polymorphism in G protein-membrane interactions: Nonlamellar-prone phospholipids and peripheral protein binding to membranes. Proc. Natl. Acad. Sci. USA 1997, 94, 11375–11380. [Google Scholar]
- Yang, Q.; Alemany, R.; Casas, J.; Kitajka, K.; Lanier, S.M.; Escriba, P.V. Influence of the membrane lipid structure on signal processing via G protein-coupled receptors. Mol. Pharmacol 2005, 68, 210–217. [Google Scholar]
- Epand, R.F.; Martinou, J.C.; Fornallaz-Mulhauser, M.; Hughes, D.W.; Epand, R.M. The apoptotic protein tBid promotes leakage by altering membrane curvature. J. Biol. Chem 2002, 277, 32632–32639. [Google Scholar]
- Attard, G.S.; Templer, R.H.; Smith, W.S.; Hunt, A.N.; Jackowski, S. Modulation of CTP:phosphocholine cytidylyltransferase by membrane curvature elastic stress. Proc. Natl. Acad. Sci. USA 2000, 97, 9032–9036. [Google Scholar]
- Epand, R.M.; Lester, D.S. The role of membrane biophysical properties in the regulation of protein kinase C activity. Trends Pharmacol. Sci 1990, 11, 317–320. [Google Scholar]
- Epand, R.M. Membrane lipid polymorphism: Relationship to bilayer properties and protein function. Methods Mol. Biol 2007, 400, 15–26. [Google Scholar]
- Hong, H.; Tamm, L.K. Elastic coupling of integral membrane protein stability to lipid bilayer forces. Proc. Natl. Acad. Sci. USA 2004, 101, 4065–4070. [Google Scholar]
- Uratani, Y.; Cramer, W.A. Reconstitution of colicin E1 into dimyristoylphosphatidylcholine membrane vesicles. J. Biol. Chem 1981, 256, 4017–4023. [Google Scholar]
- Phurtrakul, S.; Jones, M.N. The permeability of bilayer lipid membranes on the incorporation of erythrocyte membrane extracts and the identification of the monosaccharide transport proteins. Biochim. Biophys. Acta 1979, 550, 188–200. [Google Scholar]
- Sogin, D.C.; Hinkle, P.C. Binding of cytochalasin B to human erythrocyte glucose transporter. Biochemistry 1980, 19, 5417–5420. [Google Scholar]
- Geibel, S.; Zimmermann, D.; Zifarelli, G.; Becker, A.; Koenderink, J.B.; Hu, Y.K.; Kaplan, J.H.; Friedrich, T.; Bamberg, E. Conformational dynamics of Na+/K+- and H+/K+-ATPase probed by voltage clamp fluorometry. Ann. N. Y. Acad. Sci. 2003, 986, 31–38. [Google Scholar]
- Neher, E.; Sakmann, B.; Steinbach, J.H. The extracellular patch clamp: A method for resolving currents through individual open channels in biological membranes. Eur. J. Phys 1978, 375, 219–228. [Google Scholar]
- Funakishi, K.; Suzuki, H.; Takeuchi, S. Lipid Bilayer Formation by Contacting Monolayers in a Microfluidic Device for Membrane Protein Analysis. Anal. Chem 2006, 78, 8169–8174. [Google Scholar]
- Maglia, G.; Heron, A.J.; Hwang, W.L.; Holden, M.A.; Mikhailova, E.; Li, Q.; Cheley, S.; Bayley, H. Droplet networks with incorporated protein diodes show collective properties. Nat. Nanotech 2009, 4, 437–440. [Google Scholar] [Green Version]
- Hamada, T.; Yoshikawa, K. Cell-Sized Liposomes and Droplets: Real-World Modeling of Living Cells. Materials 2012, 5, 2292–2305. [Google Scholar]
- Yanagisawa, M.; Masayuki, I.; Kato, A.; Yoshikawa, K.; Oiki, S. Oriented Reconstitution of a Membrane Protein in a Giant Unilamellar Vesicle: Experimental Verification with the Potassium Channel KcsA. J. Am. Chem. Soc 2011, 133, 11774–11779. [Google Scholar]
- Shaw, A.W.; McLean, M.A.; Sligar, S.G. Phospholipid phase transitions in homogeneous nanometer scale bilayer discs. FEBS Lett 2004, 556, 260–264. [Google Scholar]
- Bayburt, T.H.; Sligar, S.G. Membrane protein assembly into Nanodiscs. FEBS Lett 2010, 584, 1721–1727. [Google Scholar]
- Leitz, A.J.; Bayburt, T.H.; Barnakov, A.N.; Springer, B.A.; Sligar, S.G. Functional reconstitution of Beta2-adrenergic receptors utilizing self-assembling Nanodisc technology. BioTechniques 2006, 40, 601–602. [Google Scholar]
- Bayburt, T.H.; Leitz, A.J.; Xie, G.; Oprian, D.D.; Sligar, S.G. Transducin activation by nanoscale lipid bilayers containing one and two rhodopsins. J. Biol. Chem 2007, 282, 14875–14881. [Google Scholar]
- Boldog, T.; Li, M.; Hazelbauer, G.L. Using Nanodiscs to create water-soluble transmembrane chemoreceptors inserted in lipid bilayers. Method. Enzymol 2007, 423, 317–335. [Google Scholar]
- Fotiadis, D.; Liang, Y.; Filipek, S.; Saperstein, D.A.; Engel, A.; Palczewski, K. Atomic-force microscopy: Rhodopsin dimers in native disc membranes. Nature 2003, 421, 127–128. [Google Scholar]
- Bayburt, T.H.; Sligar, S.G. Single-molecule height measurements on microsomal cytochrome P450 in nanometer-scale phospholipid bilayer disks. Proc. Natl. Acad. Sci. USA 2002, 99, 6725–6730. [Google Scholar]
- Duan, H.; Civjan, N.R.; Sligar, S.G.; Schuler, M.A. Co-incorporation of heterologously expressed Arabidopsis cytochrome P450 and P450 reductase into soluble nanoscale lipid bilayers. Arch. Biochem. Biophys 2004, 424, 141–153. [Google Scholar]
- Gantt, S.L.; Denisov, I.G.; Grinkova, Y.V.; Sligar, S.G. The critical iron-oxygen intermediate in human aromatase. Biochem. Biophys. Res. Commun 2009, 387, 169–173. [Google Scholar]
- Grinkova, Y.V.; Denisov, I.G.; Waterman, M.R.; Arase, M.; Kagawa, N.; Sligar, S.G. The ferrous-oxy complex of human aromatase. Biochem. Biophys. Res. Commun 2008, 372, 379–382. [Google Scholar]
- Davydov, D.R.; Fernando, H.; Baas, B.J.; Sligar, S.G.; Halpert, J.R. Kinetics of dithionite-dependent reduction of cytochrome P450 3A4: Heterogeneity of the enzyme caused by its oligomerization. Biochemistry 2005, 44, 13902–13913. [Google Scholar]
- Nath, A.; Grinkova, Y.V.; Sligar, S.; Atkins, W.M. Ligand Binding to Cytochrome P450 3A4 in Phospholipid Bilayer Nanodiscs THE EFFECT OF MODEL MEMBRANES. J. Biol. Chem 2007, 282, 28309–28320. [Google Scholar]
- Lill, R.; Dowhan, W.; Wickner, W. The ATPase activity of secA is regulated by acidic phospholipids, secY, and the leader and mature domains of precursor proteins. Cell 1990, 60, 271–280. [Google Scholar]
- Alami, M.; Dalal, K.; Lelj-Garolla, B.; Sligar, S.G.; Duong, F. Nanodiscs unravel the interaction between the SecYEG channel and its cytosolic partner SecA. EMBO J 2007, 26, 1995–2004. [Google Scholar]
- Dalal, K.; Chan, C.S.; Sligar, S.G.; Duong, F. Two copies of the SecY channel and acidic lipids are necessary to activate the SecA translocation ATPase. Proc. Natl. Acad. Sci. USA 2012, 109, 4104–4109. [Google Scholar]
- Frauenfeld, J.; Gumbart, J.; Sluis, E.O.; Funes, S.; Gartmann, M.; Beatrix, B.; Mielke, T.; Berninghausen, O.; Becker, T.; Schulten, K.; Beckmann, R. Cryo-EM structure of the ribosome-SecYE complex in the membrane environment. Nat. Struct. Mol. Biol 2011, 18, 614–621. [Google Scholar]
- Wu, Z.C.; de Keyzer, J.; Kedrov, A.; Driessen, A.J. Competitive binding of the SecA ATPase and ribosomes to the SecYEG translocon. J. Biol. Chem 2012, 287, 7885–7895. [Google Scholar]
- Merritt, E.A.; Sarfaty, S.; van den Akker, F.; L’Hoir, C.; Martial, J.A.; Hol, W.G. Crystal structure of cholera toxin B-pentamer bound to receptor GM1 pentasaccharide. Protein Sci 1994, 3, 166–175. [Google Scholar]
- Marushchak, D.; Gretskaya, N.; Mikhalyov, I.; Johansson, L.B. Self-aggregation—an intrinsic property of G(M1) in lipid bilayers. Mol. Membr. Biol 2007, 24, 102–112. [Google Scholar]
- Bricarello, D.A.; Mills, E.J.; Petrlova, J.; Voss, J.C.; Parikh, A.N. Ganglioside embedded in reconstituted lipoprotein binds cholera toxin with elevated affinity. J. Lipid Res 2010, 51, 2731–2738. [Google Scholar]
- Borch, J.; Torta, F.; Sligar, S.G.; Roepstorff, P. Nanodiscs for immobilization of lipid bilayers and membrane receptors: Kinetic analysis of cholera toxin binding to a glycolipid receptor. Anal. Chem 2008, 80, 6245–6252. [Google Scholar]
- Guihard, G.; Proteau, S.; Payet, M.D.; Escande, D.; Rousseau, E. Patch-clamp study of liver nuclear ionic channels reconstituted into giant proteoliposomes. FEBS Lett 2000, 476, 234–239. [Google Scholar]
- Heikal, A.; Box, K.; Rothnie, A.; Storm, J.; Callaghan, R.; Allen, M. The stabilisation of purified, reconstituted P-glycoprotein by freeze drying with disaccharides. Cryobiology 2009, 58, 37–44. [Google Scholar]
- Ritchie, T.K.; Grinkova, Y.V.; Bayburt, T.H.; Denisov, I.G.; Zolnerciks, J.K.; Atkins, W.M.; Sligar, S.G. Chapter 11—Reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Method. Enzymol 2009, 464, 211–231. [Google Scholar]
- Holland, I.B. ABC transporters, mechanisms and biology: An overview. Essays Biochem 2011, 50, 1–17. [Google Scholar]
- Bao, H.; Duong, F. Discovery of an auto-regulation mechanism for the maltose ABC transporter MalFGK2. PLoS One 2012, 7. [Google Scholar] [CrossRef]

© 2013 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Shen, H.-H.; Lithgow, T.; Martin, L. Reconstitution of Membrane Proteins into Model Membranes: Seeking Better Ways to Retain Protein Activities. Int. J. Mol. Sci. 2013, 14, 1589-1607. https://doi.org/10.3390/ijms14011589
Shen H-H, Lithgow T, Martin L. Reconstitution of Membrane Proteins into Model Membranes: Seeking Better Ways to Retain Protein Activities. International Journal of Molecular Sciences. 2013; 14(1):1589-1607. https://doi.org/10.3390/ijms14011589
Chicago/Turabian StyleShen, Hsin-Hui, Trevor Lithgow, and Lisa Martin. 2013. "Reconstitution of Membrane Proteins into Model Membranes: Seeking Better Ways to Retain Protein Activities" International Journal of Molecular Sciences 14, no. 1: 1589-1607. https://doi.org/10.3390/ijms14011589
APA StyleShen, H. -H., Lithgow, T., & Martin, L. (2013). Reconstitution of Membrane Proteins into Model Membranes: Seeking Better Ways to Retain Protein Activities. International Journal of Molecular Sciences, 14(1), 1589-1607. https://doi.org/10.3390/ijms14011589