Nuclear Factor-κB (NF-κB) Regulates the Expression of Human Testis-Enriched Leucine-Rich Repeats and WD Repeat Domain Containing 1 (LRWD1) Gene
Abstract
:1. Introduction
2. Results and Discussion
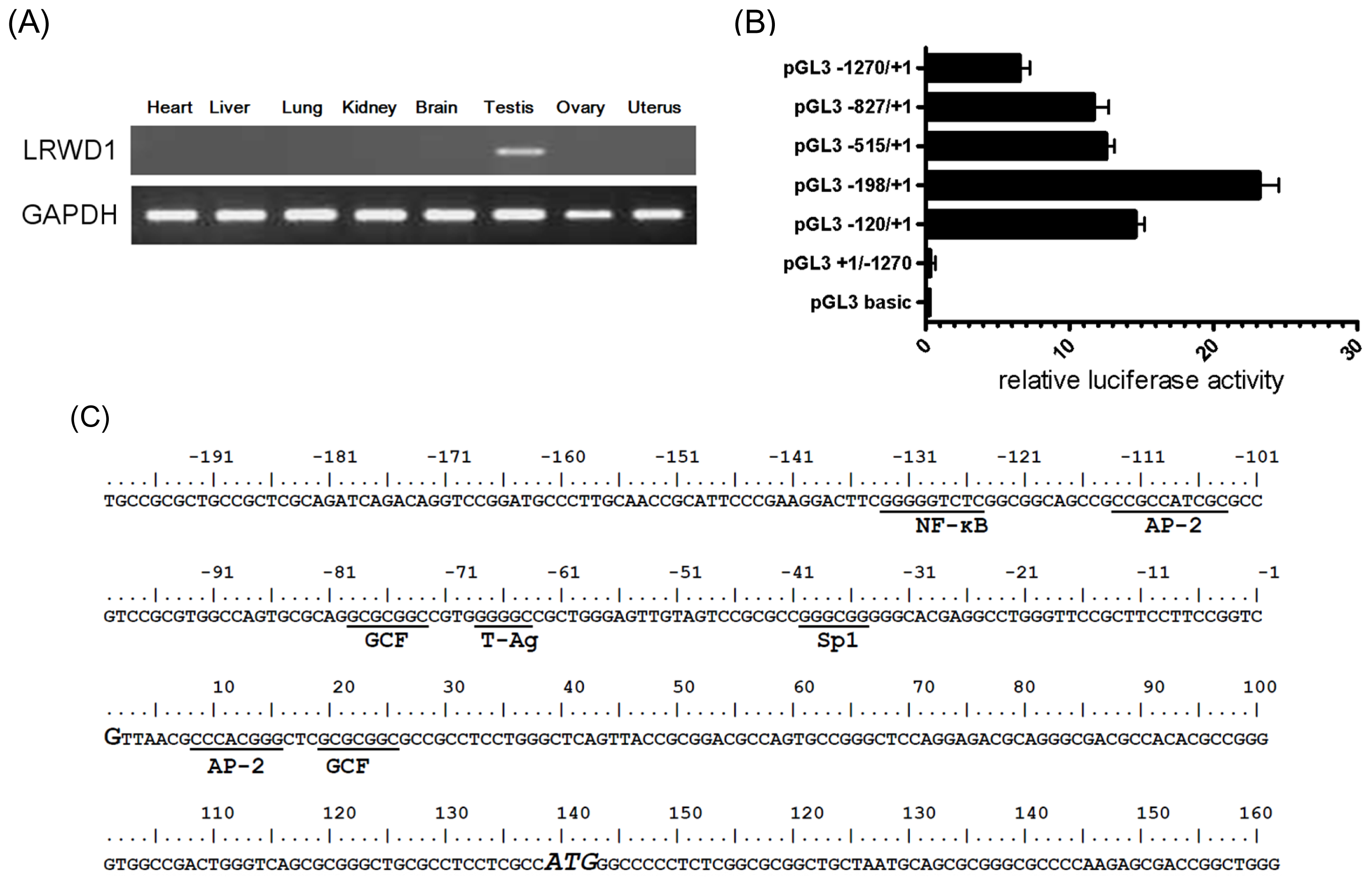
2.1. Promoter Activity in the 5′-Flanking Region of the Testis-Enriched LRWD1 Gene
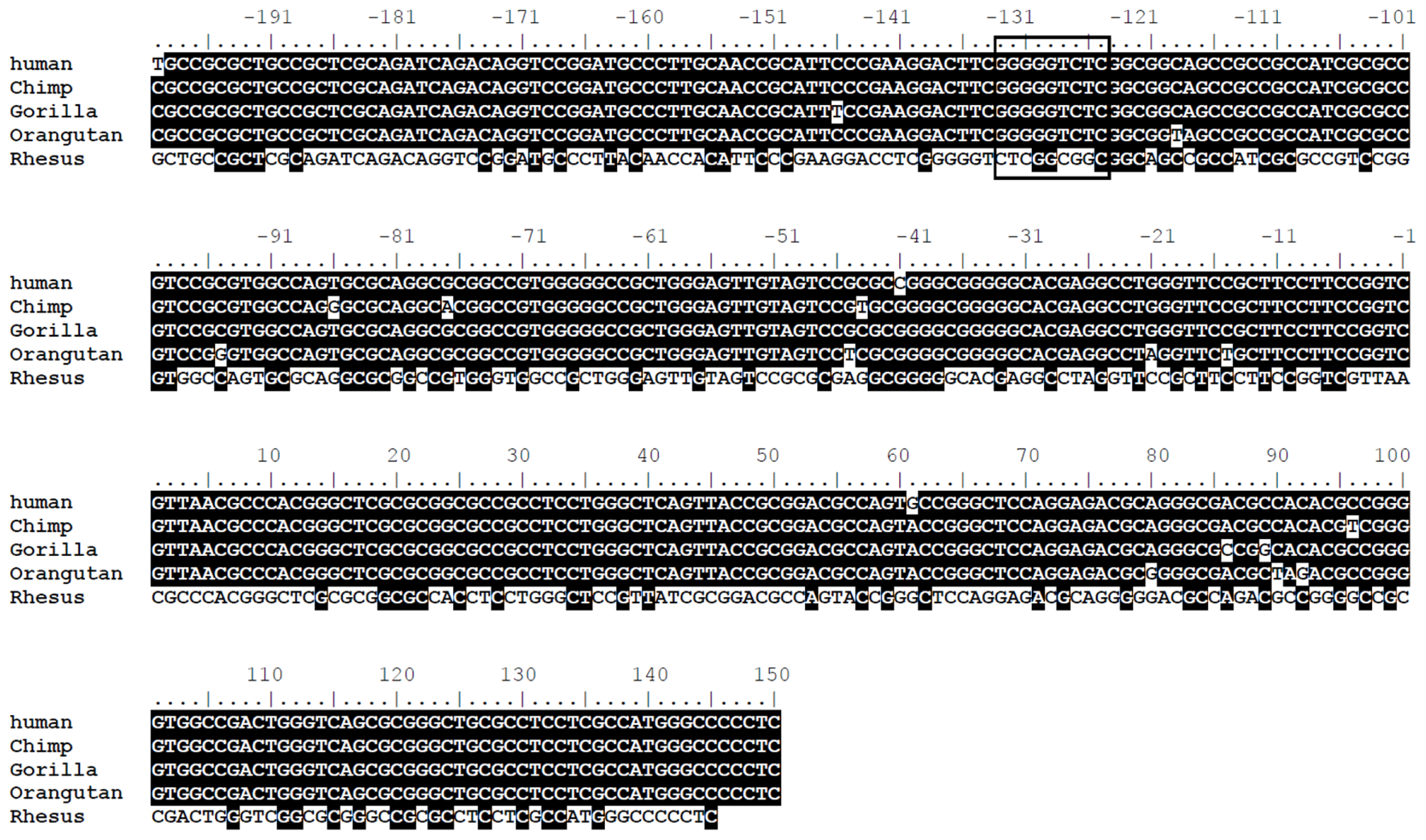
2.2. The LRWD1 Promoter Region Includes an Evolutionarily Conserved κB Site
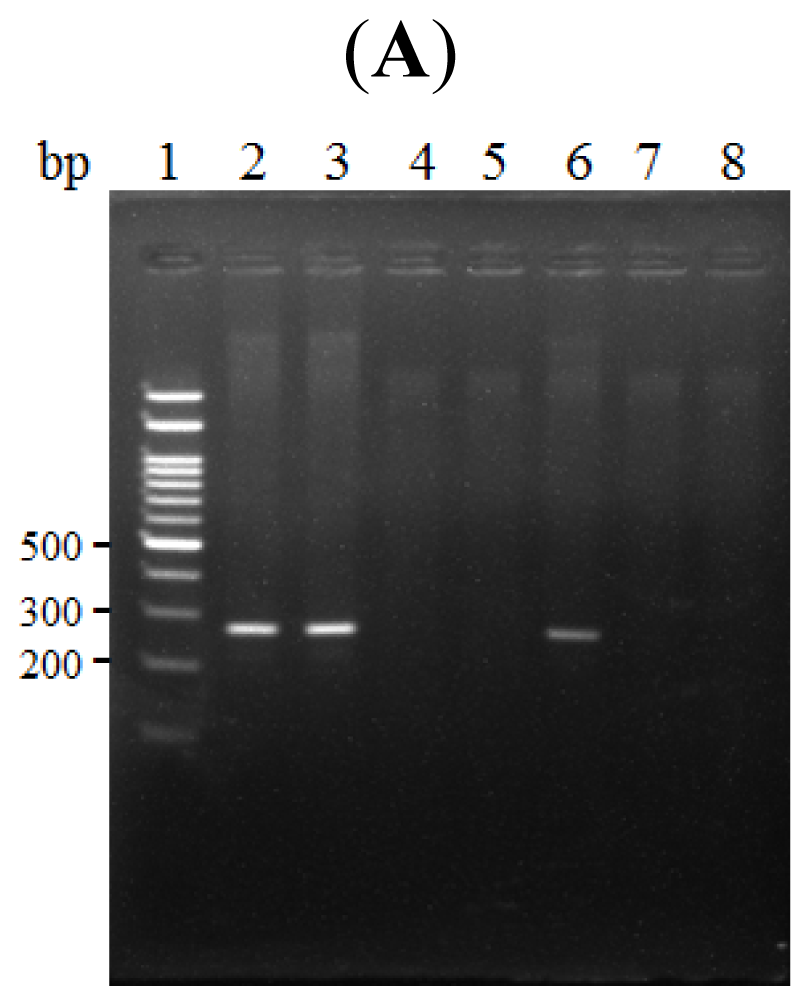
2.3. The κB Site within the LRWD1 Promoter Binds NF-κB in Testicular Germ Cell Nuclei
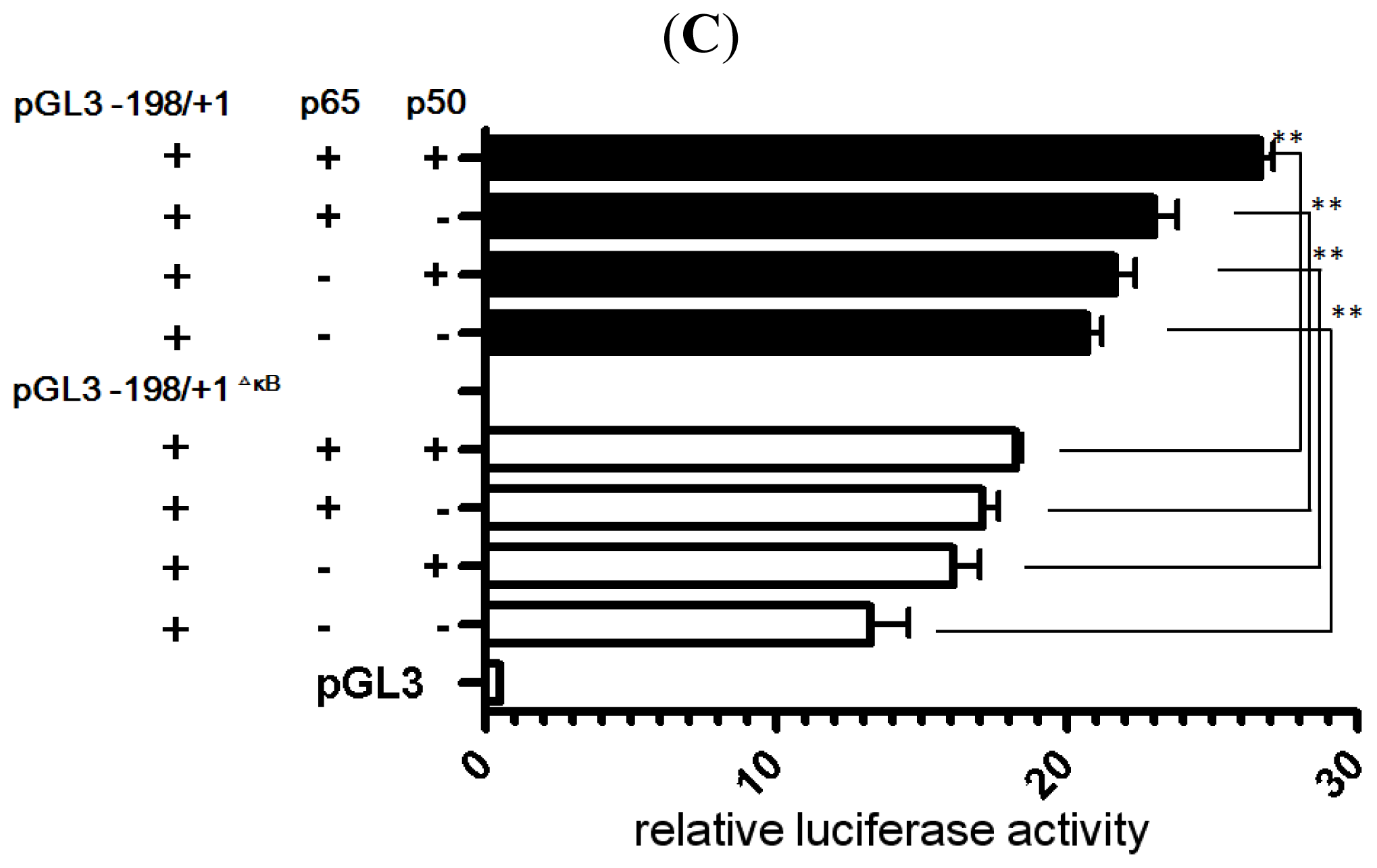
2.4. NF-κB Binds to the LRWD1 Promoter and Regulates Its Activity
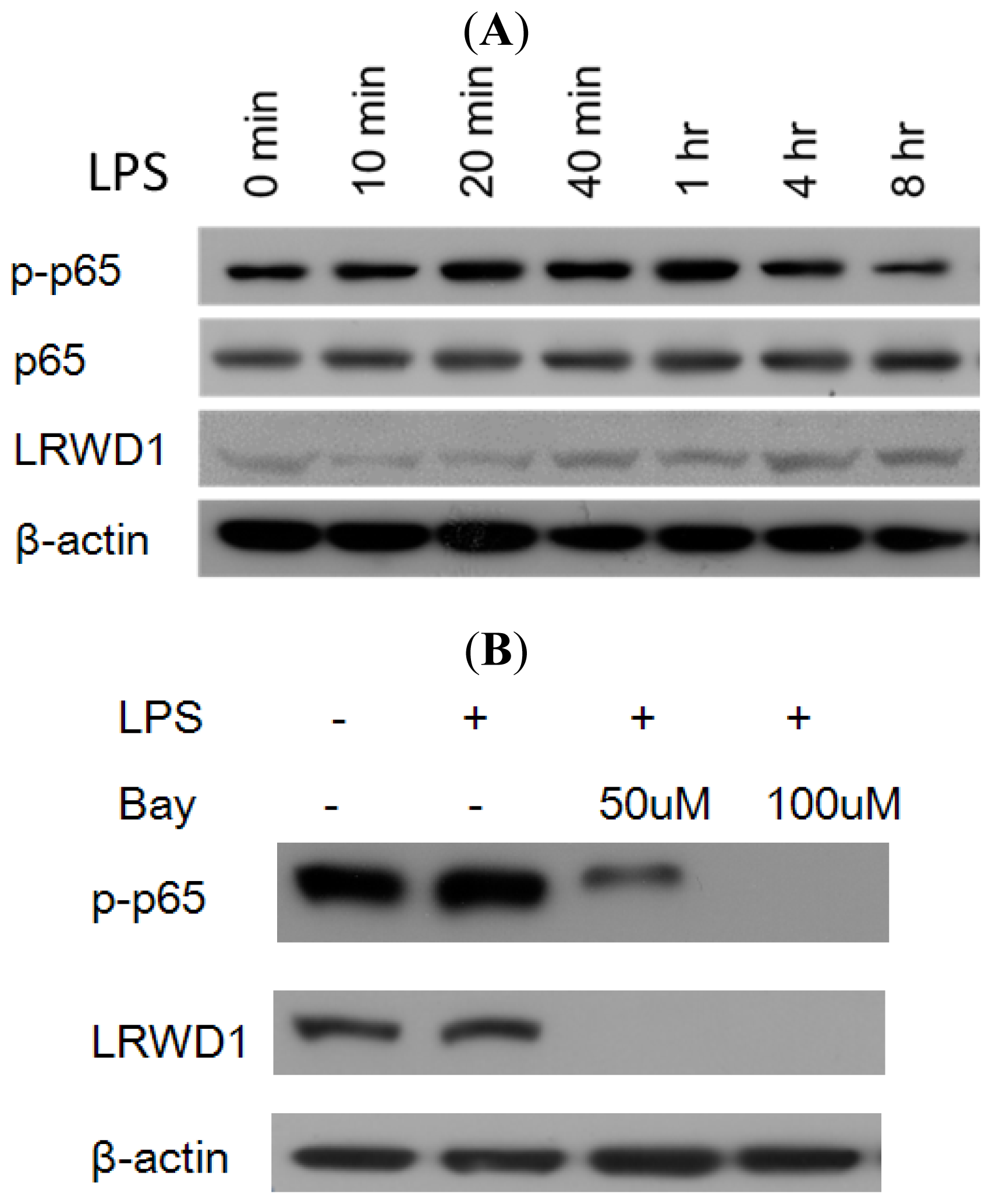
2.5. Activation of NF-κB with LPS Increases LRWD1 Expression
2.6. NF-κB Stimulates LRWD1 Promoter Activity
3. Experimental Section
3.1. LRWD1 Expression Assay by RT-PCR
3.2. Cell Culture and Isolation of Genomic DNA
3.3. Sources of Plasmids and Plasmid Construction
3.4. Transient Transfection and Luciferase Reporter Gene Assay
3.5. Western Blot Analysis
3.6. Chromatin Immunoprecipitation (ChIP)
3.7. Gel Electrophoretic Mobility Shift Assay (EMSA)
3.8. Statistical Analysis and Bioinformatics Software
4. Discussion and Conclusions
Acknowledgments
- Conflict of InterestThe authors declare that they have no conflict of interest.
References
- Lin, Y.H.; Lin, Y.M.; Teng, Y.N.; Hsieh, T.Y.T.; Lin, Y.S.; Kuo, P.L. Identification of ten novel genes involved in human spermatogenesis by microarray analysis of testicular tissue. Fertil. Steril 2006, 86, 1650–1658. [Google Scholar]
- Teng, Y.N.; Liao, M.H.; Lin, Y.B.; Kuo, P.L.; Kuo, T.Y. Expression of lrwd1 in mouse testis and its centrosomal localization. Int. J. Androl 2010, 33, 832–840. [Google Scholar]
- Shen, Z.; Sathyan, K.M.; Geng, Y.; Zheng, R.; Chakraborty, A.; Freeman, B.; Wang, F.; Prasanth, K.V.; Prasanth, S.G. A WD-repeat protein stabilizes ORC binding to chromatin. Mol. Cell 2010, 40, 99–111. [Google Scholar]
- Chakraborty, A.; Shen, Z.; Prasanth, S.G. “ORCanization” on heterochromatin: Linking DNA replication initiation to chromatin organization”. Epigenetics 2011, 6, 665–670. [Google Scholar]
- Bartke, T.; Vermeulen, M.; Xhemalce, B.; Robson, S.C.; Mann, M.; Kouzarides, T. Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell 2010, 143, 470–484. [Google Scholar]
- Baldwin, A.S., Jr. The NF-κB and IκB proteins: New discoveries and insights. Annu. Rev. Immunol. 1996, 14, 649–681. [Google Scholar]
- Ginn-Pease, M.E.; Whisler, R.L. Redox signals and NF-[kappa] B activation in T cells. Free Radic. Biol. Med 1998, 25, 346–361. [Google Scholar]
- Gilmore, T.D. The Rel/NF-kappaB signal transduction pathway: Introduction. Oncogene 1999, 18, 6842–6844. [Google Scholar]
- Baeuerle, P.A.; Baltimore, D. A 65-kappaD subunit of active NF-kappaB is required for inhibition of NF-kappaB by I kappaB. Genes Dev 1989, 3, 1689–1698. [Google Scholar]
- Pahl, H.L. Activators and target genes of Rel/NF-kappaB transcription factors. Oncogene 1999, 18, 6853–6866. [Google Scholar]
- Beg, A.A.; Sha, W.C.; Bronson, R.T.; Ghosh, S.; Baltimore, D. Embryonic lethality and liver degeneration in mice lacking the RelA component of NF-kappa B. Nature 1995, 376, 167–170. [Google Scholar]
- Doi, T.S.; Takahashi, T.; Taguchi, O.; Azuma, T.; Obata, Y. NF-kappa B RelA-deficient lymphocytes: Normal development of T cells and B cells, impaired production of IgA and IgG1 and reduced proliferative responses. J. Exp. Med 1997, 185, 953–961. [Google Scholar]
- Barkett, M.; Gilmore, T.D. Control of apoptosis by Rel/NF-kappaB transcription factors. Oncogene 1999, 18, 6910–6924. [Google Scholar]
- Delfino, F.; Walker, W.H. Hormonal regulation of the NF-[kappa] B signaling pathway. Mol. Cell. Endocrinol 1999, 157, 1–9. [Google Scholar]
- Pentikainen, V.; Suomalainen, L.; Erkkila, K.; Martelin, E.; Parvinen, M.; Pentikainen, M.O.; Dunkel, L. Nuclear factor-kappa B activation in human testicular apoptosis. Am. J. Pathol 2002, 160, 205–218. [Google Scholar]
- Lilienbaum, A.; Sage, J.; Mémet, S.; Rassoulzadegan, M.; Cuzin, F.; Israël, A. NF-κB is developmentally regulated during spermatogenesis in mice. Dev. Dyn 2000, 219, 333–340. [Google Scholar]
- Delfino, F.; Walker, W.H. Stage-specific nuclear expression of NF-κB in mammalian testis. Mol. Endocrinol 1998, 12, 1696–1707. [Google Scholar]
- Erkkila, K.; Henriksen, K.; Hirvonen, V.; Rannikko, S.; Salo, J.; Parvinen, M.; Dunkel, L. Testosterone regulates apoptosis in adult human seminiferous tubules in vitro. J. Clin. Endocrinol. MeTable 1997, 82, 2314–2321. [Google Scholar]
- Schulze-Osthoff, K.; Los, M.; Baeuerle, P.A. Redox signalling by transcription factors NF-kappa B and AP-1 in lymphocytes. Biochem. Pharmacol 1995, 50, 735–741. [Google Scholar]
- Shalini, S.; Bansal, M.P. Co-operative effect of glutathione depletion and selenium induced oxidative stress on API and NFkB expression in testicular cells in vitro: Insights to regulation of spermatogenesis. Biol. Res 2007, 40, 307–317. [Google Scholar]
- Prestridge, D.S. Predicting Pol II promoter sequences using transcription factor binding sites. J. Mol. Biol 1995, 249, 923–932. [Google Scholar]
- Dynan, W.S.; Sazer, S.; Tjian, R.; Schimke, R.T. Transcription factor Sp1 recognizes a DNA sequence in the mouse dihydrofolate reductase promoter. Nature 1986, 319, 246–248. [Google Scholar]
- Pugh, B.F.; Tjian, R. Mechanism of transcriptional activation by Sp1: Evidence for coactivators. Cell 1990, 61, 1187–1197. [Google Scholar]
- Promoter Prediction Server, version 2.0. Available online: http://www.cbs.dtu.dk/services/Promoter/ accessed on 28 December 2012.
- GraphPad Prism, version 5.02; GraphPad Software Inc.: San Diego, CA, USA, 2008.
- TFSEARCH: Searching Transcription Factor Binding Sites, version 1.3. Available online: http://www.cbrc.jp/research/db/TFSEARCH.html accessed on 1 May 2012.
- Briggs, M.R.; Kadonaga, J.T.; Bell, S.P.; Tjian, R. Purification and biochemical characterization of the promoter-specific transcription factor, Sp1. Science 1986, 234, 47–52. [Google Scholar]
- Hilger-Eversheim, K.; Moser, M.; Schorle, H.; Buettner, R. Regulatory roles of AP-2 transcription factors in vertebrate development, apoptosis and cell-cycle control. Gene 2000, 260, 1–12. [Google Scholar]
- Azizkhan, J.C.; Jensen, D.E.; Pierce, A.J.; Wade, M. Transcription from TATA-less promoters: Dihydrofolate reductase as a model. Crit. Rev. Eukaryot. Gene Expr 1993, 3, 229–254. [Google Scholar]
- Santee, S.M.; Owen-Schaub, L.B. Human tumor necrosis factor receptor p75/80 (CD120b) gene structure and promoter characterization. J. Biol. Chem 1996, 271, 21151–21159. [Google Scholar]
- Iguchi, N.; Tanaka, H.; Yamada, S.; Nishimura, H.; Nishimune, Y. Control of mouse hils1 gene expression during spermatogenesis: Identification of regulatory element by transgenic mouse. Biol. Reprod 2004, 70, 1239–1245. [Google Scholar]
- Lee, Y.J.; Ahn, J.K.; Chung, J.H. Expression of murine poly (A) binding protein II gene in testis. IUBMB Life 2000, 49, 57–61. [Google Scholar]
- Barberis, A.; Superti-Furga, G.; Busslinger, M. Mutually exclusive interaction of the CCAAT-binding factor and of a displacement protein with overlapping sequences of a histone gene promoter. Cell 1987, 50, 347–359. [Google Scholar]
- Boer, P.H.; Adra, C.N.; Lau, Y.F.; McBurney, M.W. The testis-specific phosphoglycerate kinase gene pgk-2 is a recruited retroposon. Mol. Cell. Biol 1987, 7, 3107–3112. [Google Scholar]
- Robinson, M.O.; McCarrey, J.R.; Simon, M.I. Transcriptional regulatory regions of testis-specific PGK2 defined in transgenic mice. Proc. Natl. Acad. Sci. USA 1989, 86, 8437–8441. [Google Scholar]
- Ambhaikar, M.; Goldberg, E. DNA-protein interactions in the CCAAT box region of the murine lactate dehydrogenase C promoter. Mol. Reprod. Dev 1999, 52, 360–365. [Google Scholar]
- Iannello, R.; Dahl, H. Transcriptional expression of a testis-specific variant of the mouse pyruvate dehydrogenase E1 alpha subunit. Biol. Reprod 1992, 47, 48–58. [Google Scholar]
- Bonny, C.; Cooker, L.A.; Goldberg, E. Deoxyribonucleic acid-protein interactions and expression of the human testis-specific lactate dehydrogenase promoter: Transcription factor Sp1 plays a major role. Biol. Reprod 1998, 58, 754–759. [Google Scholar]
- Zhang, L.P.; Stroud, J.; Eddy, C.A.; Walter, C.A.; McCarrey, J.R. Multiple elements influence transcriptional regulation from the human testis-specific PGK2 promoter in transgenic mice. Biol. Reprod 1999, 60, 1329–1337. [Google Scholar]
- Bunick, D.; Johnson, P.A.; Johnson, T.R.; Hecht, N.B. Transcription of the testis-specific mouse protamine 2 gene in a homologous in vitro transcription system. Proc. Natl. Acad. Sci. USA 1990, 87, 891–895. [Google Scholar]
- Persengiev, S.P.; Raval, P.J.; Rabinovitch, S.; Millette, C.F.; Kilpatrick, D.L. Transcription factor Sp1 is expressed by three different developmentally regulated messenger ribonucleic acids in mouse spermatogenic cells. Endocrinology 1996, 137, 638–646. [Google Scholar]
- Saffer, J.D.; Jackson, S.P.; Annarella, M.B. Developmental expression of Sp1 in the mouse. Mol. Cell. Biol 1991, 11, 2189–2199. [Google Scholar]

© 2013 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Teng, Y.-N.; Chuang, P.-J.; Liu, Y.-W. Nuclear Factor-κB (NF-κB) Regulates the Expression of Human Testis-Enriched Leucine-Rich Repeats and WD Repeat Domain Containing 1 (LRWD1) Gene. Int. J. Mol. Sci. 2013, 14, 625-639. https://doi.org/10.3390/ijms14010625
Teng Y-N, Chuang P-J, Liu Y-W. Nuclear Factor-κB (NF-κB) Regulates the Expression of Human Testis-Enriched Leucine-Rich Repeats and WD Repeat Domain Containing 1 (LRWD1) Gene. International Journal of Molecular Sciences. 2013; 14(1):625-639. https://doi.org/10.3390/ijms14010625
Chicago/Turabian StyleTeng, Yen-Ni, Po-Jung Chuang, and Yo-Wen Liu. 2013. "Nuclear Factor-κB (NF-κB) Regulates the Expression of Human Testis-Enriched Leucine-Rich Repeats and WD Repeat Domain Containing 1 (LRWD1) Gene" International Journal of Molecular Sciences 14, no. 1: 625-639. https://doi.org/10.3390/ijms14010625



