ADAR Enzyme and miRNA Story: A Nucleotide that Can Make the Difference
Abstract
:1. Introduction
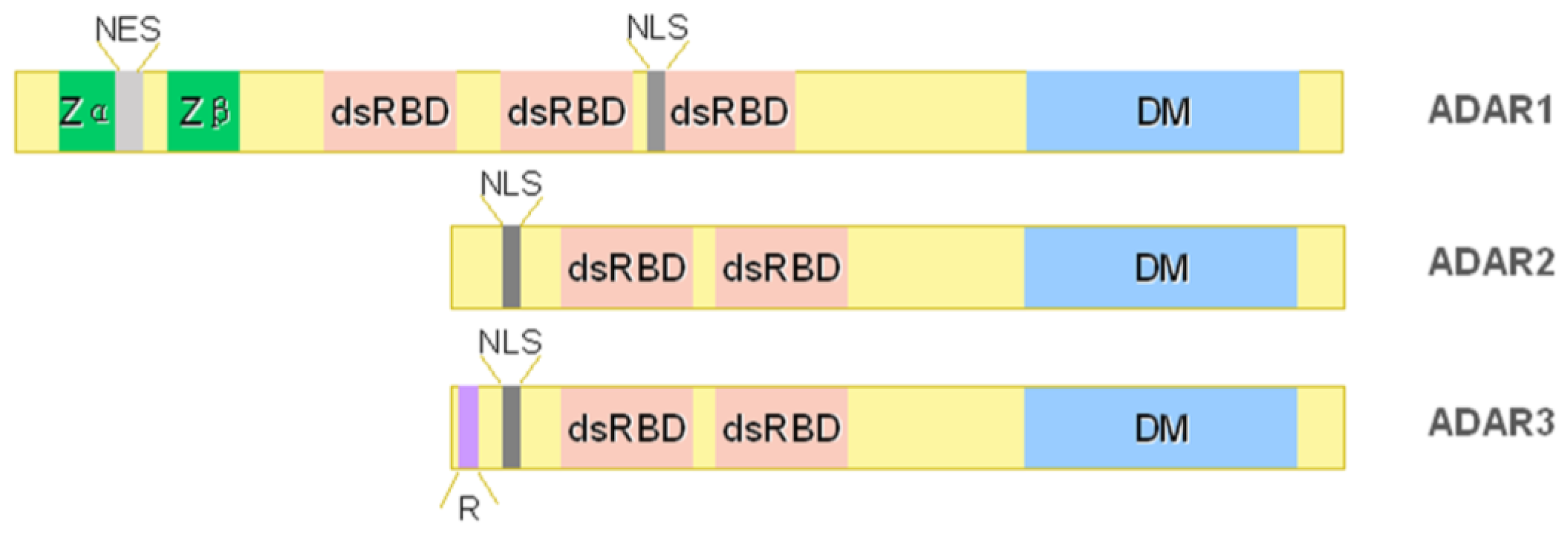
2. ADAR Family
2.1. ADAR1
2.2. ADAR2
2.3. ADAR3
3. ADAR Substrates
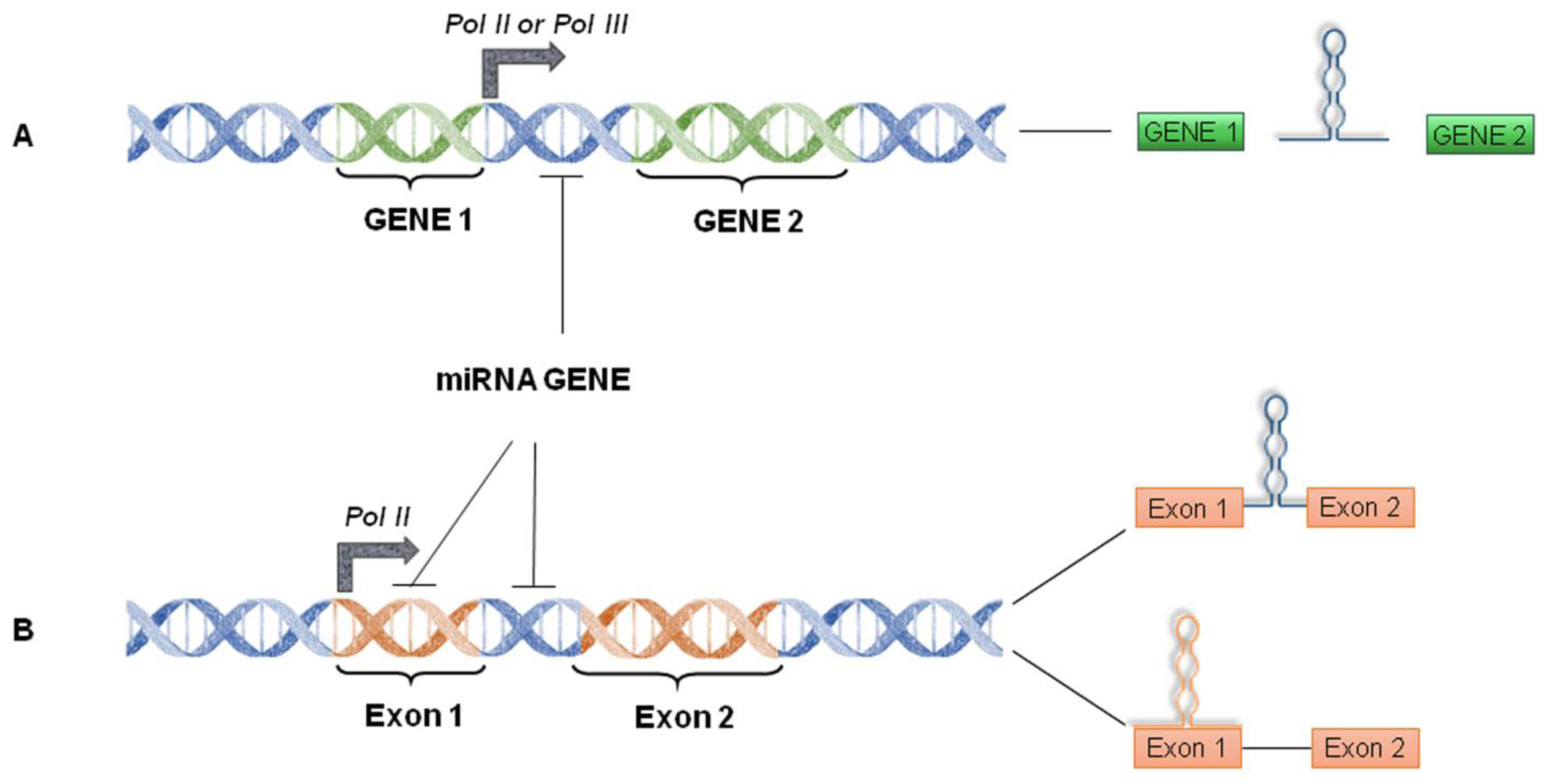
4. miRNA World Machinery Overview
4.1. miRNA Biogenesis and Processing into the Nucleus
4.2. miRNA Processing into the Cytoplasm
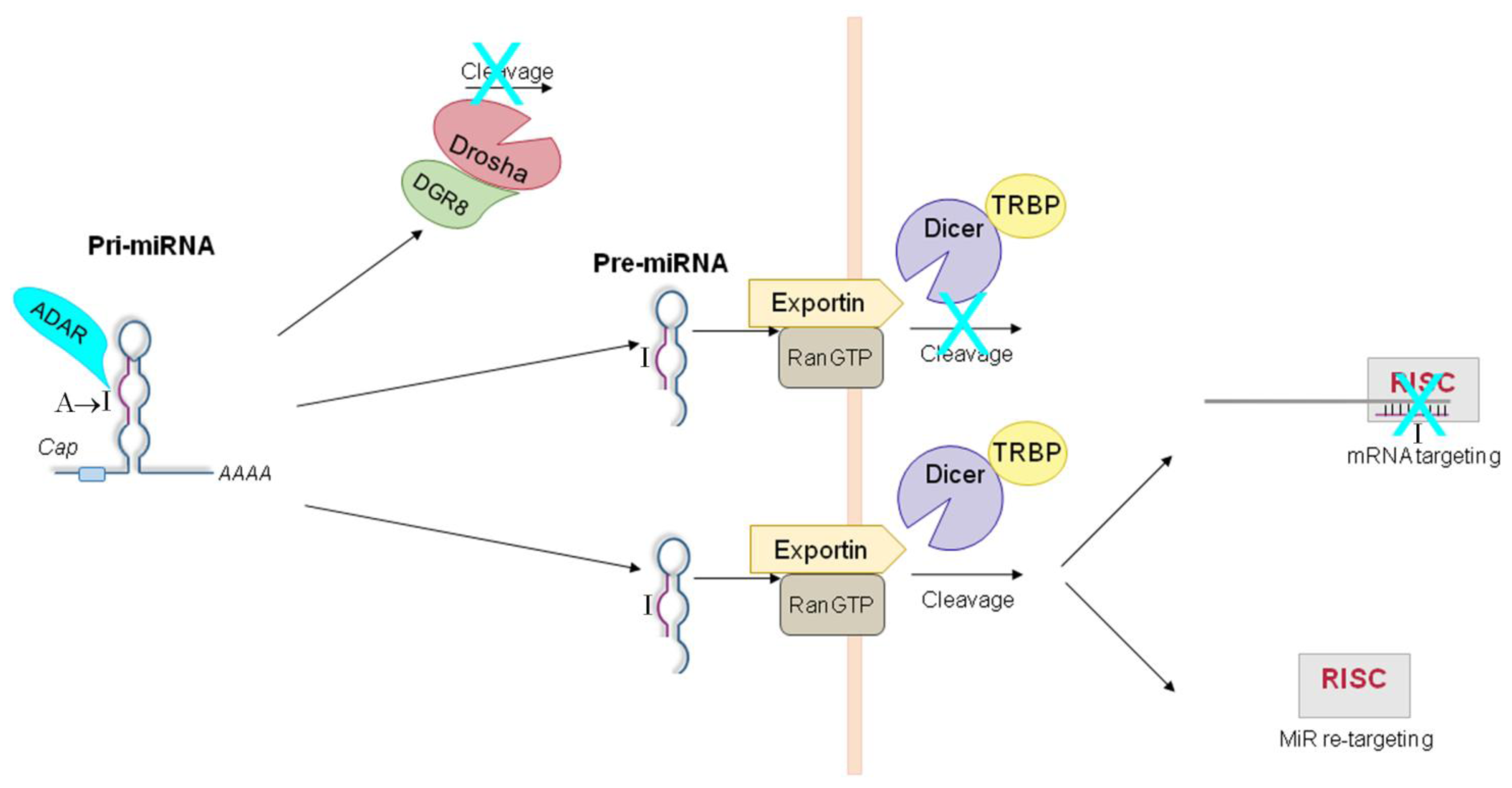
5. ADAR-Dependent Effects on miRNA Pathway
5.1. ADAR-Dependent Effects on Pri-miRs
5.2. ADAR-Dependent Effects on Pre-miRs
5.3. ADAR-Dependent Effects on RISC-Loading
5.4. ADAR-Dependent Effects on Retargeting
5.5. ADAR-Dependent Effects on Target 3′ UTRs
5.6. ADAR-Mediated Editing-Independent Effects on miRNAs
6. Large-Scale Surveys
7. Stimulative Role of ADAR1
8. Conclusions

Acknowledgments
Conflicts of Interest
References
- Lee, J.T. Epigenetic regulation by long noncoding RNAs. Science 2012, 338, 1435–1439. [Google Scholar]
- Baltimore, D. Our genome unveiled. Nature 2001, 409, 814–816. [Google Scholar]
- Gott, J.M.; Emeson, R.B. Functions and mechanisms of RNA editing. Annu. Rev. Genet 2000, 34, 499–531. [Google Scholar]
- Gallo, A.; Locatelli, F. ADARs: Allies or enemies? The importance of A-to-I RNA editing in human disease: From cancer to HIV-1. Biol. Rev. Camb. Philos. Soc 2011, 87, 95–110. [Google Scholar]
- Keegan, L.P.; Gallo, A.; O’Connell, M.A. The many roles of an RNA editor. Nat. Rev. Genet 2001, 2, 869–878. [Google Scholar]
- Nishikura, K. Functions and regulation of RNA editing by ADAR deaminases. Annu. Rev. Biochem 2010, 79, 321–349. [Google Scholar]
- Blow, M.; Futreal, P.A.; Wooster, R.; Stratton, M.R. A survey of RNA editing in human brain. Genome Res 2004, 14, 2379–2387. [Google Scholar]
- Kim, D.D.; Kim, T.T.; Walsh, T.; Kobayashi, Y.; Matise, T.C.; Buyske, S.; Gabriel, A. Widespread RNA editing of embedded alu elements in the human transcriptome. Genome Res 2004, 14, 1719–1725. [Google Scholar]
- Levanon, E.Y.; Eisenberg, E.; Yelin, R.; Nemzer, S.; Hallegger, M.; Shemesh, R.; Fligelman, Z.Y.; Shoshan, A.; Pollock, S.R.; Sztybel, D.; et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat. Biotechnol 2004, 22, 1001–1005. [Google Scholar]
- Athanasiadis, A.; Rich, A.; Maas, S. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol 2004, 2, 2144–2158. [Google Scholar]
- Morse, D.P.; Aruscavage, P.J.; Bass, B.L. RNA hairpins in noncoding regions of human brain and Caenorhabditis elegans mRNA are edited by adenosine deaminases that act on RNA. Proc. Natl. Acad. Sci. USA 2002, 99, 7906–7911. [Google Scholar]
- Maas, S.; Rich, A.; Nishikura, K. A-to-I RNA editing: Recent news and residual mysteries. J. Biol. Chem 2003, 278, 1391–1394. [Google Scholar]
- Nishikura, K. Editor meets silencer: Crosstalk between RNA editing and RNA interference. Nat. Rev. Mol. Cell Biol 2006, 7, 919–931. [Google Scholar]
- Macbeth, M.R.; Schubert, H.L.; Vandemark, A.P.; Lingam, A.T.; Hill, C.P.; Bass, B.L. Inositol hexakisphosphate is bound in the ADAR2 core and required for RNA editing. Science 2005, 309, 1534–1539. [Google Scholar]
- Kuttan, A.; Bass, B.L. Mechanistic insights into editing-site specificity of ADARs. Proc. Natl. Acad. Sci. USA 2012, 109, E3295–E3304. [Google Scholar]
- Ryter, J.M.; Schultz, S.C. Molecular basis of double-stranded RNA-protein interactions: Structure of a dsRNA-binding domain complexed with dsRNA. EMBO J 1998, 17, 7505–7513. [Google Scholar]
- Herbert, A.; Alfken, J.; Kim, Y.G.; Mian, I.S.; Nishikura, K.; Rich, A. A Z-DNA binding domain present in the human editing enzyme, double-stranded RNA adenosine deaminase. Proc. Natl. Acad. Sci. USA 1997, 94, 8421–8426. [Google Scholar]
- Brown, B.A., 2nd; Lowenhaupt, K.; Wilbert, C.M.; Hanlon, E.B.; Rich, A. The zalpha domain of the editing enzyme dsRNA adenosine deaminase binds left-handed Z-RNA as well as Z-DNA. Proc. Natl. Acad. Sci. USA 2000, 97, 13532–13536. [Google Scholar]
- Weier, H.U.; George, C.X.; Greulich, K.M.; Samuel, C.E. The interferon-inducible, double-stranded RNA-specific adenosine deaminase gene (DSRAD) maps to human chromosome 1q21.1-21.2. Genomics 1995, 30, 372–375. [Google Scholar]
- Patterson, J.B.; Samuel, C.E. Expression and regulation by interferon of a double-stranded-RNAspecific adenosine deaminase from human cells: Evidence for two forms of the deaminase. Mol. Cell. Biol 1995, 15, 5376–5388. [Google Scholar]
- Patterson, J.B.; Thomis, D.C.; Hans, S.L.; Samuel, C.E. Mechanism of interferon action: Double-stranded RNA-specific adenosine deaminase from human cells is inducible by alpha and gamma interferons. Virology 1995, 210, 508–511. [Google Scholar]
- George, C.X.; Das, S.; Samuel, C.E. Organization of the mouse RNA-specific adenosine deaminase Adar1 gene 5′-region and demonstration of STAT1-independent, STAT2-dependent transcriptional activation by interferon. Virology 2008, 380, 338–343. [Google Scholar]
- Poulsen, H.; Nilsson, J.; Damgaard, C.K.; Egebjerg, J.; Kjems, J. CRM1 mediates the export of ADAR1 through a nuclear export signal within the Z-DNA binding domain. Mol. Cell. Biol 2001, 21, 7862–7871. [Google Scholar]
- Fritz, J.; Strehblow, A.; Taschner, A.; Schopoff, S.; Pasierbek, P.; Jantsch, M.F. RNA-regulated interaction of transportin-1 and exportin-5 with the double-stranded RNA-binding domain regulates nucleocytoplasmic shuttling of ADAR1. Mol. Cell. Biol 2009, 29, 1487–1497. [Google Scholar]
- Desterro, J.M.; Keegan, L.P.; Jaffray, E.; Hay, R.T.; O’Connell, M.A.; Carmo-Fonseca, M. SUMO-1 modification alters ADAR1 editing activity. Mol. Biol. Cell 2005, 16, 5115–5126. [Google Scholar]
- Gallo, A.; Keegan, L.P.; Ring, G.M.; O’Connell, M.A. An ADAR that edits transcripts encoding ion channel subunits functions as a dimer. EMBO J 2003, 22, 3421–3430. [Google Scholar]
- Valente, L.; Nishikura, K. RNA binding-independent dimerization of adenosine deaminases acting on RNA and dominant negative effects of nonfunctional subunits on dimer functions. J. Biol. Chem 2007, 282, 16054–16061. [Google Scholar]
- Cho, D.S.; Yang, W.; Lee, J.T.; Shiekhattar, R.; Murray, J.M.; Nishikura, K. Requirement of dimerization for RNA editing activity of adenosine deaminases acting on RNA. J. Biol. Chem 2003, 278, 17093–17102. [Google Scholar]
- Chilibeck, K.A.; Wu, T.; Liang, C.; Schellenberg, M.J.; Gesner, E.M.; Lynch, J.M.; MacMillan, A.M. FRET analysis of in vivo dimerization by RNA-editing enzymes. J. Biol. Chem 2006, 281, 16530–16535. [Google Scholar]
- Nie, Y.; Ding, L.; Kao, P.N.; Braun, R.; Yang, J.H. ADAR1 interacts with NF90 through double-stranded RNA and regulates NF90-mediated gene expression independently of RNA editing. Mol. Cell. Biol 2005, 25, 6956–6963. [Google Scholar]
- Clerzius, G.; Gelinas, J.F.; Daher, A.; Bonnet, M.; Meurs, E.F.; Gatignol, A. ADAR1 interacts with PKR during human immunodeficiency virus infection of lymphocytes and contributes to viral replication. J. Virol 2009, 83, 10119–10128. [Google Scholar]
- Lei, M.; Liu, Y.; Samuel, C.E. Adenovirus VAI RNA antagonizes the RNA-editing activity of the ADAR adenosine deaminase. Virology 1998, 245, 188–196. [Google Scholar]
- Liu, Y.; Wolff, K.C.; Jacobs, B.L.; Samuel, C.E. Vaccinia virus E3L interferon resistance protein inhibits the interferon-induced adenosine deaminase A-to-I editing activity. Virology 2001, 289, 378–387. [Google Scholar]
- Mittaz, L.; Scott, H.S.; Rossier, C.; Seeburg, P.H.; Higuchi, M.; Antonarakis, S.E. Cloning of a human RNA editing deaminase (ADARB1) of glutamate receptors that maps to chromosome 21q22.3. Genomics 1997, 41, 210–217. [Google Scholar]
- Maas, S.; Gommans, W.M. Novel exon of mammalian ADAR2 extends open reading frame. PLoS One 2009, 4, e4225. [Google Scholar]
- Peng, P.L.; Zhong, X.; Tu, W.; Soundarapandian, M.M.; Molner, P.; Zhu, D.; Lau, L.; Liu, S.; Liu, F.; Lu, Y. ADAR2-dependent RNA editing of AMPA receptor subunit GluR2 determines vulnerability of neurons in forebrain ischemia. Neuron 2006, 49, 719–733. [Google Scholar]
- Yang, L.; Huang, P.; Li, F.; Zhao, L.; Zhang, Y.; Li, S.; Gan, Z.; Lin, A.; Li, W.; Liu, Y. c-Jun amino-terminal kinase-1 mediates glucose-responsive upregulation of the RNA editing enzyme ADAR2 in pancreatic beta-cells. PLoS One 2012, 7, e48611. [Google Scholar]
- Desterro, J.M.; Keegan, L.P.; Lafarga, M.; Berciano, M.T.; O’Connell, M.; Carmo-Fonseca, M. Dynamic association of RNA-editing enzymes with the nucleolus. J. Cell Sci 2003, 116, 1805–1818. [Google Scholar]
- Maas, S.; Gommans, W.M. Identification of a selective nuclear import signal in adenosine deaminases acting on RNA. Nucleic Acids Res 2009, 37, 5822–5829. [Google Scholar]
- Jaikaran, D.C.; Collins, C.H.; MacMillan, A.M. Adenosine to inosine editing by ADAR2 requires formation of a ternary complex on the GluR-B R/G site. J. Biol. Chem 2002, 277, 37624–37629. [Google Scholar]
- Poulsen, H.; Jorgensen, R.; Heding, A.; Nielsen, F.C.; Bonven, B.; Egebjerg, J. Dimerization of ADAR2 is mediated by the double-stranded RNA binding domain. RNA 2006, 12, 1350–1360. [Google Scholar]
- Mittaz, L.; Antonarakis, S.E.; Higuchi, M.; Scott, H.S. Localization of a novel human RNA-editing deaminase (hRED2 or ADARB2) to chromosome 10p15. Hum. Genet 1997, 100, 398–400. [Google Scholar]
- Chen, C.X.; Cho, D.S.; Wang, Q.; Lai, F.; Carter, K.C.; Nishikura, K. A third member of the RNA-specific adenosine deaminase gene family, ADAR3, contains both single- and double-stranded RNA binding domains. RNA 2000, 6, 755–767. [Google Scholar]
- Melcher, T.; Maas, S.; Herb, A.; Sprengel, R.; Higuchi, M.; Seeburg, P.H. RED2, a brain-specific member of the RNA-specific adenosine deaminase family. J. Biol. Chem 1996, 271, 31795–31798. [Google Scholar]
- Cenci, C.; Barzotti, R.; Galeano, F.; Corbelli, S.; Rota, R.; Massimi, L.; Di Rocco, C.; O’Connell, M.A.; Gallo, A. Down-regulation of RNA editing in pediatric astrocytomas: ADAR2 editing activity inhibits cell migration and proliferation. J. Biol. Chem 2008, 283, 7251–7160. [Google Scholar]
- Bass, B.L. RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem 2002, 71, 817–846. [Google Scholar]
- Kiran, A.; Baranov, P.V. DARNED: A DAtabase of RNa EDiting in humans. Bioinformatics 2010, 26, 1772–1776. [Google Scholar]
- Galeano, F.; Leroy, A.; Rossetti, C.; Gromova, I.; Gautier, P.; Keegan, L.P.; Massimi, L.; Di Rocco, C.; O’Connell, M.A.; Gallo, A. Human BLCAP transcript: New editing events in normal and cancerous tissues. Int. J. Cancer 2010, 127, 127–137. [Google Scholar]
- Wahlstedt, H.; Daniel, C.; Enstero, M.; Ohman, M. Large-scale mRNA sequencing determines global regulation of RNA editing during brain development. Genome Res 2009, 19, 978–986. [Google Scholar]
- Lehmann, K.A.; Bass, B.L. Double-stranded RNA adenosine deaminases ADAR1 and ADAR2 have overlapping specificities. Biochemistry 2000, 39, 12875–12884. [Google Scholar]
- Nishikura, K.; Yoo, C.; Kim, U.; Murray, J.M.; Estes, P.A.; Cash, F.E.; Liebhaber, S.A. Substrate specificity of the dsRNA unwinding/modifying activity. EMBO J 1991, 10, 3523–3532. [Google Scholar]
- Lehmann, K.A.; Bass, B.L. The importance of internal loops within RNA substrates of ADAR1. J. Mol. Biol 1999, 291, 1–13. [Google Scholar]
- Vitali, P.; Basyuk, E.; Le Meur, E.; Bertrand, E.; Muscatelli, F.; Cavaille, J.; Huttenhofer, A. ADAR2-mediated editing of RNA substrates in the nucleolus is inhibited by C/D small nucleolar RNAs. J. Cell Biol 2005, 169, 745–753. [Google Scholar]
- Lev-Maor, G.; Sorek, R.; Levanon, E.Y.; Paz, N.; Eisenberg, E.; Ast, G. RNA-editing-mediated exon evolution. Genome Biol 2007, 8, R29. [Google Scholar]
- Lev-Maor, G.; Sorek, R.; Shomron, N.; Ast, G. The birth of an alternatively spliced exon: 3′ splice-site selection in Alu exons. Science 2003, 300, 1288–1291. [Google Scholar]
- Lev-Maor, G.; Ram, O.; Kim, E.; Sela, N.; Goren, A.; Levanon, E.Y.; Ast, G. Intronic Alus influence alternative splicing. PLoS Genet 2008, 4, e1000204. [Google Scholar]
- Sakurai, M.; Yano, T.; Kawabata, H.; Ueda, H.; Suzuki, T. Inosine cyanoethylation identifies A-to-I RNA editing sites in the human transcriptome. Nat. Chem. Biol 2010, 6, 733–740. [Google Scholar]
- Zhang, Z.; Carmichael, G.G. The fate of dsRNA in the nucleus: A p54(nrb)-containing complex mediates the nuclear retention of promiscuously A-to-I edited RNAs. Cell 2001, 106, 465–475. [Google Scholar]
- Scadden, A.D. The RISC subunit Tudor-SN binds to hyper-edited double-stranded RNA and promotes its cleavage. Nat. Struct. Mol. Biol 2005, 12, 489–496. [Google Scholar]
- He, L.; He, X.; Lim, L.P.; de Stanchina, E.; Xuan, Z.; Liang, Y.; Xue, W.; Zender, L.; Magnus, J.; Ridzon, D.; et al. A microRNA component of the p53 tumour suppressor network. Nature 2007, 447, 1130–1134. [Google Scholar]
- Lagos-Quintana, M.; Rauhut, R.; Lendeckel, W.; Tuschl, T. Identification of novel genes coding for small expressed RNAs. Science 2001, 294, 853–858. [Google Scholar]
- Lau, N.C.; Lim, L.P.; Weinstein, E.G.; Bartel, D.P. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 2001, 294, 858–862. [Google Scholar]
- Lee, R.C.; Ambros, V. An extensive class of small RNAs in Caenorhabditis elegans. Science 2001, 294, 862–864. [Google Scholar]
- Rodriguez, A.; Griffiths-Jones, S.; Ashurst, J.L.; Bradley, A. Identification of mammalian microRNA host genes and transcription units. Genome Res 2004, 14, 1902–1910. [Google Scholar]
- Morlando, M.; Ballarino, M.; Gromak, N.; Pagano, F.; Bozzoni, I.; Proudfoot, N.J. Primary microRNA transcripts are processed co-transcriptionally. Nat. Struct. Mol. Biol 2008, 15, 902–909. [Google Scholar]
- Kim, V.N.; Han, J.; Siomi, M.C. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell. Biol 2009, 10, 126–139. [Google Scholar]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005, 120, 15–20. [Google Scholar]
- Filipowicz, W.; Bhattacharyya, S.N.; Sonenberg, N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat. Rev. Genet 2008, 9, 102–114. [Google Scholar]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar]
- Chen, X.; Liang, H.; Zhang, C.Y.; Zen, K. miRNA regulates noncoding RNA: A noncanonical function model. Trends Biochem. Sci 2011, 37, 457–459. [Google Scholar]
- Fabbri, M.; Paone, A.; Calore, F.; Galli, R.; Gaudio, E.; Santhanam, R.; Lovat, F.; Fadda, P.; Mao, C.; Nuovo, G.J.; et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc. Natl. Acad. Sci. USA 2012, 109, E2110–E2116. [Google Scholar]
- Ota, H.; Sakurai, M.; Gupta, R.; Valente, L.; Wulff, B.E.; Ariyoshi, K.; Iizasa, H.; Davuluri, R.V.; Nishikura, K. ADAR1 forms a complex with Dicer to promote microRNA processing and RNA-induced gene silencing. Cell 2013, 153, 575–589. [Google Scholar]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Radmark, O.; Kim, S.; Kim, V.N. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar]
- Han, J.; Lee, Y.; Yeom, K.H.; Nam, J.W.; Heo, I.; Rhee, J.K.; Sohn, S.Y.; Cho, Y.; Zhang, B.T.; Kim, V.N. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell 2006, 125, 887–901. [Google Scholar]
- Winter, J.; Jung, S.; Keller, S.; Gregory, R.I.; Diederichs, S. Many roads to maturity: MicroRNA biogenesis pathways and their regulation. Nat. Cell Biol 2009, 11, 228–234. [Google Scholar]
- Tang, R.; Li, L.; Zhu, D.; Hou, D.; Cao, T.; Gu, H.; Zhang, J.; Chen, J.; Zhang, C.Y.; Zen, K. Mouse miRNA-709 directly regulates miRNA-15a/16-1 biogenesis at the posttranscriptional level in the nucleus: Evidence for a microRNA hierarchy system. Cell Res 2012, 22, 504–515. [Google Scholar]
- Yi, R.; Qin, Y.; Macara, I.G.; Cullen, B.R. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev 2003, 17, 3011–3016. [Google Scholar]
- Lund, E.; Guttinger, S.; Calado, A.; Dahlberg, J.E.; Kutay, U. Nuclear export of microRNA precursors. Science 2004, 303, 95–98. [Google Scholar]
- Weinmann, L.; Hock, J.; Ivacevic, T.; Ohrt, T.; Mutze, J.; Schwille, P.; Kremmer, E.; Benes, V.; Urlaub, H.; Meister, G. Importin 8 is a gene silencing factor that targets argonaute proteins to distinct mRNAs. Cell 2009, 136, 496–507. [Google Scholar]
- Gregory, R.I.; Chendrimada, T.P.; Cooch, N.; Shiekhattar, R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell 2005, 123, 631–640. [Google Scholar]
- Chendrimada, T.P.; Gregory, R.I.; Kumaraswamy, E.; Norman, J.; Cooch, N.; Nishikura, K.; Shiekhattar, R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 2005, 436, 740–744. [Google Scholar]
- Lee, Y.; Hur, I.; Park, S.Y.; Kim, Y.K.; Suh, M.R.; Kim, V.N. The role of PACT in the RNA silencing pathway. EMBO J 2006, 25, 522–532. [Google Scholar]
- MacRae, I.J.; Ma, E.; Zhou, M.; Robinson, C.V.; Doudna, J.A. In vitro reconstitution of the human RISC-loading complex. Proc. Natl. Acad. Sci. USA 2008, 105, 512–517. [Google Scholar]
- Diederichs, S.; Haber, D.A. Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell 2007, 131, 1097–1108. [Google Scholar]
- Nilsen, T.W. Mechanisms of microRNA-mediated gene regulation in animal cells. Trends Genet 2007, 23, 243–249. [Google Scholar]
- Luciano, D.J.; Mirsky, H.; Vendetti, N.J.; Maas, S. RNA editing of a miRNA precursor. RNA 2004, 10, 1174–1177. [Google Scholar]
- Takeda, A.; Iwasaki, S.; Watanabe, T.; Utsumi, M.; Watanabe, Y. The mechanism selecting the guide strand from small RNA duplexes is different among argonaute proteins. Plant Cell Physiol 2008, 49, 493–500. [Google Scholar]
- Valencia-Sanchez, M.A.; Liu, J.; Hannon, G.J.; Parker, R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev 2006, 20, 515–524. [Google Scholar]
- Carthew, R.W.; Sontheimer, E.J. Origins and Mechanisms of miRNAs and siRNAs. Cell 2009, 136, 642–655. [Google Scholar]
- Blow, M.J.; Grocock, R.J.; van Dongen, S.; Enright, A.J.; Dicks, E.; Futreal, P.A.; Wooster, R.; Stratton, M.R. RNA editing of human microRNAs. Genome Biol 2006, 7, R27. [Google Scholar]
- Kawahara, Y.; Zinshteyn, B.; Sethupathy, P.; Iizasa, H.; Hatzigeorgiou, A.G.; Nishikura, K. Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science 2007, 315, 1137–1140. [Google Scholar]
- Kawahara, Y.; Megraw, M.; Kreider, E.; Iizasa, H.; Valente, L.; Hatzigeorgiou, A.G.; Nishikura, K. Frequency and fate of microRNA editing in human brain. Nucleic Acids Res 2008, 36, 5270–5280. [Google Scholar]
- Vesely, C.; Tauber, S.; Sedlazeck, F.J.; von Haeseler, A.; Jantsch, M.F. Adenosine deaminases that act on RNA induce reproducible changes in abundance and sequence of embryonic miRNAs. Genome Res 2012, 22, 1468–1476. [Google Scholar]
- Alon, S.; Mor, E.; Vigneault, F.; Church, G.M.; Locatelli, F.; Galeano, F.; Gallo, A.; Shomron, N.; Eisenberg, E. Systematic identification of edited microRNAs in the human brain. Genome Res 2012, 22, 1533–1540. [Google Scholar] [Green Version]
- Warf, M.B.; Shepherd, B.A.; Johnson, W.E.; Bass, B.L. Effects of ADARs on small RNA processing pathways in C. elegans. Genome Res 2012, 22, 1488–1498. [Google Scholar]
- Ekdahl, Y.; Farahani, H.S.; Behm, M.; Lagergren, J.; Ohman, M. A-to-I editing of microRNAs in the mammalian brain increases during development. Genome Res 2012, 22, 1477–1487. [Google Scholar]
- Heale, B.S.; Keegan, L.P.; McGurk, L.; Michlewski, G.; Brindle, J.; Stanton, C.M.; Caceres, J.F.; O’Connell, M.A. Editing independent effects of ADARs on the miRNA/siRNA pathways. EMBO J 2009, 28, 3145–3156. [Google Scholar]
- Yang, W.; Chendrimada, T.P.; Wang, Q.; Higuchi, M.; Seeburg, P.H.; Shiekhattar, R.; Nishikura, K. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat. Struct. Mol. Biol 2006, 13, 13–21. [Google Scholar]
- Caudy, A.A.; Ketting, R.F.; Hammond, S.M.; Denli, A.M.; Bathoorn, A.M.; Tops, B.B.; Silva, J.M.; Myers, M.M.; Hannon, G.J.; Plasterk, R.H. A micrococcal nuclease homologue in RNAi effector complexes. Nature 2003, 425, 411–414. [Google Scholar]
- Weissbach, R.; Scadden, A.D. Tudor-SN and ADAR1 are components of cytoplasmic stress granules. RNA 2012, 18, 462–471. [Google Scholar]
- Kawahara, Y.; Zinshteyn, B.; Chendrimada, T.P.; Shiekhattar, R.; Nishikura, K. RNA editing of the microRNA-151 precursor blocks cleavage by the Dicer-TRBP complex. EMBO Rep 2007, 8, 763–769. [Google Scholar]
- Cullen, B.R. Viral and cellular messenger RNA targets of viral microRNAs. Nature 2009, 457, 421–425. [Google Scholar]
- Skalsky, R.L.; Cullen, B.R. Viruses, microRNAs, and host interactions. Annu. Rev. Microbiol 2010, 64, 123–141. [Google Scholar]
- Iizasa, H.; Wulff, B.E.; Alla, N.R.; Maragkakis, M.; Megraw, M.; Hatzigeorgiou, A.; Iwakiri, D.; Takada, K.; Wiedmer, A.; Showe, L.; Lieberman, P.; Nishikura, K. Editing of Epstein-Barr virus-encoded BART6 microRNAs controls their dicer targeting and consequently affects viral latency. J. Biol. Chem 2010, 285, 33358–33370. [Google Scholar]
- Lei, T.; Yuen, K.S.; Tsao, S.W.; Chen, H.; Kok, K.H.; Jin, D.Y. Perturbation of biogenesis and targeting of Epstein-Barr virus-encoded miR-BART3 microRNA by A-to-I editing. J. Gen. Virol 2013. [Google Scholar] [CrossRef]
- Gandy, S.Z.; Linnstaedt, S.D.; Muralidhar, S.; Cashman, K.A.; Rosenthal, L.J.; Casey, J.L. RNA editing of the human herpesvirus 8 kaposin transcript eliminates its transforming activity and is induced during lytic replication. J. Virol 2007, 81, 13544–13551. [Google Scholar]
- Pfeffer, S.; Sewer, A.; Lagos-Quintana, M.; Sheridan, R.; Sander, C.; Grasser, F.A.; van Dyk, L.F.; Ho, C.K.; Shuman, S.; Chien, M.; et al. Identification of microRNAs of the herpesvirus family. Nat. Methods 2005, 2, 269–276. [Google Scholar]
- Choudhury, Y.; Tay, F.C.; Lam, D.H.; Sandanaraj, E.; Tang, C.; Ang, B.T.; Wang, S. Attenuated adenosine-to-inosine editing of microRNA-376a* promotes invasiveness of glioblastoma cells. J. Clin. Investig 2012, 122, 4059–4076. [Google Scholar]
- Maas, S.; Patt, S.; Schrey, M.; Rich, A. Underediting of glutamate receptor GluR-B mRNA in malignant gliomas. Proc. Natl. Acad. Sci. USA 2001, 98, 14687–14692. [Google Scholar]
- Galeano, F.; Rossetti, C.; Tomaselli, S.; Cifaldi, L.; Lezzerini, M.; Pezzullo, M.; Boldrini, R.; Massimi, L.; Di Rocco, C.M.; Locatelli, F.; Gallo, A. ADAR2-editing activity inhibits glioblastoma growth through the modulation of the CDC14B/Skp2/p21/p27 axis. Oncogene 2012, 32, 998–1009. [Google Scholar]
- Liang, H.; Landweber, L.F. Hypothesis: RNA editing of microRNA target sites in humans? RNA 2007, 13, 463–467. [Google Scholar]
- Peng, Z.; Cheng, Y.; Tan, B.C.; Kang, L.; Tian, Z.; Zhu, Y.; Zhang, W.; Liang, Y.; Hu, X.; Tan, X.; et al. Comprehensive analysis of RNA-Seq data reveals extensive RNA editing in a human transcriptome. Nat. Biotechnol 2012, 30, 253–260. [Google Scholar]
- Gu, T.; Buaas, F.W.; Simons, A.K.; Ackert-Bicknell, C.L.; Braun, R.E.; Hibbs, M.A. Canonical A-to-I and C-to-U RNA editing is enriched at 3′UTRs and microRNA target sites in multiple mouse tissues. PLoS One 2012, 7, e33720. [Google Scholar]
- Wang, Q.; Hui, H.; Guo, Z.; Zhang, W.; Hu, Y.; He, T.; Tai, Y.; Peng, P.; Wang, L. ADAR1 regulates ARHGAP26 gene expression through RNA editing by disrupting miR-30b-3p and miR-573 binding. RNA 2013, 19, 1525–1536. [Google Scholar]
- De Hoon, M.J.; Taft, R.J.; Hashimoto, T.; Kanamori-Katayama, M.; Kawaji, H.; Kawano, M.; Kishima, M.; Lassmann, T.; Faulkner, G.J.; Mattick, J.S.; et al. Cross-mapping and the identification of editing sites in mature microRNAs in high-throughput sequencing libraries. Genome Res 2010, 20, 257–264. [Google Scholar]
- Chiang, H.R.; Schoenfeld, L.W.; Ruby, J.G.; Auyeung, V.C.; Spies, N.; Baek, D.; Johnston, W.K.; Russ, C.; Luo, S.; Babiarz, J.E.; et al. Mammalian microRNAs: Experimental evaluation of novel and previously annotated genes. Genes Dev 2010, 24, 992–1009. [Google Scholar]
- Galeano, F.; Tomaselli, S.; Locatelli, F.; Gallo, A. A-to-I RNA editing: The ADAR side of human cancer. Semin. Cell Dev. Biol 2012, 23, 244–250. [Google Scholar]
© 2013 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Tomaselli, S.; Bonamassa, B.; Alisi, A.; Nobili, V.; Locatelli, F.; Gallo, A. ADAR Enzyme and miRNA Story: A Nucleotide that Can Make the Difference. Int. J. Mol. Sci. 2013, 14, 22796-22816. https://doi.org/10.3390/ijms141122796
Tomaselli S, Bonamassa B, Alisi A, Nobili V, Locatelli F, Gallo A. ADAR Enzyme and miRNA Story: A Nucleotide that Can Make the Difference. International Journal of Molecular Sciences. 2013; 14(11):22796-22816. https://doi.org/10.3390/ijms141122796
Chicago/Turabian StyleTomaselli, Sara, Barbara Bonamassa, Anna Alisi, Valerio Nobili, Franco Locatelli, and Angela Gallo. 2013. "ADAR Enzyme and miRNA Story: A Nucleotide that Can Make the Difference" International Journal of Molecular Sciences 14, no. 11: 22796-22816. https://doi.org/10.3390/ijms141122796





