Characterization of the Expression Profile and Genetic Polymorphism of the Cellular Retinol-Binding Protein (CRBP IV) Gene in Erlang Mountainous Chickens
Abstract
:1. Introduction
2. Results
2.1. Molecular Cloning and Sequence of the Chicken CRBP IV Gene
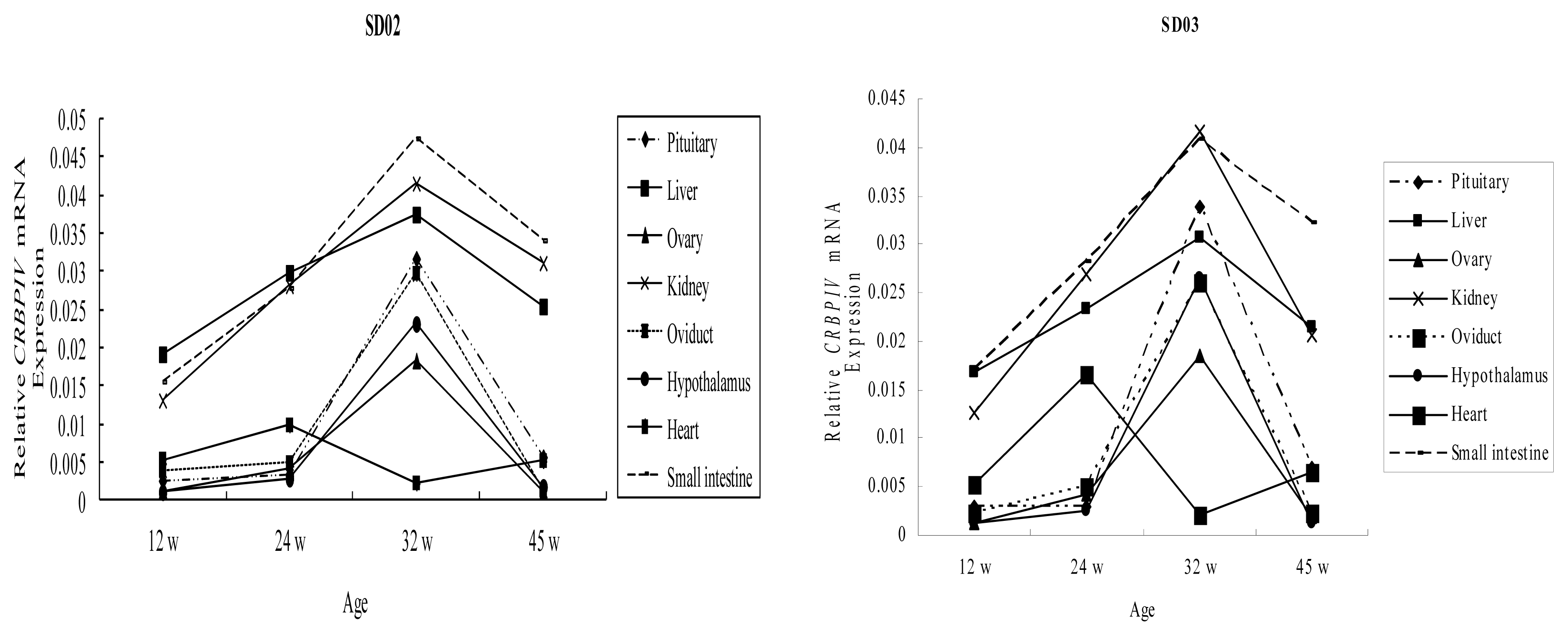
2.2. Ontogenic Expression of CRBP IV Gene in Chickens
2.3. Comparison of CRBP IV mRNA Expression among Different Tissues in Chickens
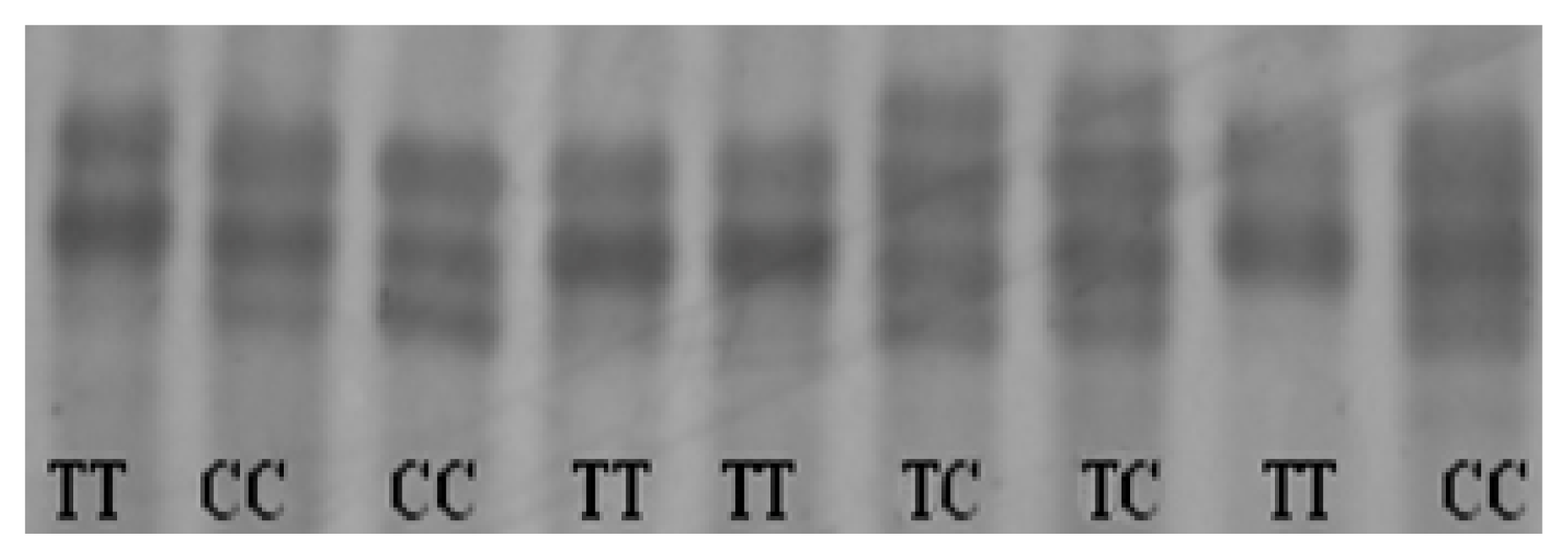
2.4. Identification of Genetic Variants in Chicken CRBP IV Gene
2.5. Genotype Frequencies and Association Analysis with Egg Production Traits
3. Discussion
4. Experimental Section
4.1. Chicken Populations and Trait Measurement
4.2. Total DNA Extraction, RNA Isolation and Reverse Transcription
4.3. Cloning and Sequencing of PCR Product
4.4. Real-Time PCR, SNPs Identification and Association Analysis
4.5. Data Analysis
5. Conclusion
Acknowledgments
References
- Zile, M.H. Function of vitamin A in vertebrate embryonic development. J. Nutr 2001, 131, 705–708. [Google Scholar]
- Duester, G. Retinoic acid synthesis and signaling during early organogenesis. Cell 2008, 134, 921–931. [Google Scholar]
- Ertesvag, A.; Naderi, S.; Blomhoff, H.K. Regulation of B cell proliferation and differentiation by retinoic acid. Semin. Immunol 2009, 21, 36–41. [Google Scholar]
- Maden, M. Vitamin A and the developing embryo. Postgrad. Med. J 2001, 77, 489–491. [Google Scholar]
- Levi, L.; Levavi-Sivan, B.; Lubzens, E. Expression of genes associated with retinoid metabolism in the trout Ovary follicle. Biol. Reprod 2008, 79, 570–577. [Google Scholar]
- Sporn, M.B.; Roberts, A.B.; Goodman, D.W.S. The Retinoids: Biology, Chemistry, and Medicine; Raven Press: New York, New York, NY, USA, 1994. [Google Scholar]
- Harrison, E.H.; Hussain, M.M. Mechanisms involved in the intestinal digestion and absorption of dietary vitamin A. J. Nutr 2001, 131, 1405–1408. [Google Scholar]
- Newcomer, M.E. Retinoid-binding proteins: structural determinants important for function. FASEB. J 1995, 9, 229–239. [Google Scholar]
- Mezaki, Y.; Morii, M.; Yoshikawa, K.; Yamaguchi, N.; Miura, M.; Imai, K.; Yoshino, H.; Senoo, H. Characterization of a cellular retinol-binding protein from lamprey, Lethenteron japonicum. Comp. Biochem. Phys. B 2011, 161, 233–239. [Google Scholar]
- Napoli, J. Biosynthesis and metabolism of retinoic acid: Roles of CRBP and CRABP in retinoic acid: roles of CRBP and CRABP in retinoic acid homeostasis. J. Nutr 1993, 123, 362. [Google Scholar]
- Conforti, L.; Tarlton, A.; Mack, T.G.A.; Mi, W.; Buckmaster, E.A.; Wagner, D.; Perry, V.H.; Coleman, M.P. A Ufd2/D4Cole1e chimeric protein and overexpression of Rbp7 in the slow Wallerian degeneration (WldS) mouse. Proc. Natl. Acad. Sci 2000, 97, 11377–11382. [Google Scholar]
- Yin, H.D.; Zhang, Z.C.; Lan, X.; Zhao, X.L.; Wang, Y.; Zhu, Q. Association of MyF5, MyF6 and MyOG gene polymorphisms with carcass traits in Chinese Meat Type Quality chicken populations. J. Anim. Vet. Adv 2011, 10, 704–708. [Google Scholar]
- Lake, P. Recent progress in poultry reproduction. World Poultry Sci. J 1989, 45, 53–59. [Google Scholar]
- Park, S.; Birkhold, S.; Kubena, L.; Nisbet, D.; Ricke, S. Review on the role of dietary zinc in poultry nutrition, immunity, and reproduction. Biol. Trace. Elem. Res 2004, 101, 147–163. [Google Scholar]
- Wilson, H. Effects of maternal nutrition on hatchability. Poult. Sci 1997, 76, 134–143. [Google Scholar]
- Zile, M.H.; Cullum, M.E. The function of vitamin A: current concepts. Proc. Soc. Exp. Biol. Med 1983, 172, 139–152. [Google Scholar]
- Ong, D.E. Cellular transport and metabolism of vitamin A: Roles of the cellular retinoid-binding proteins. Nutr. Rev 1994, 52, S24–S31. [Google Scholar]
- Gustafson, A.L.; Eriksson, U.; Dencker, L. CRBP I and CRABP I localisation during olfactory nerve development. Dev. Brain. Res 1999, 114, 121–126. [Google Scholar]
- Folli, C.; Calderone, V.; Ottonello, S.; Bolchi, A.; Zanotti, G.; Stoppini, M.; Berni, R. Identification, retinoid binding, and x-ray analysis of a human retinol-binding protein. Proc. Natl. Acad. Sci. USA 2001, 98, 3710–3715. [Google Scholar]
- Vogel, S.; Mendelsohn, C.L.; Mertz, J.R.; Piantedosi, R.; Waldburger, C.; Gottesman, M.E.; Blaner, W.S. Characterization of a new member of the fatty acid-binding protein family that binds all-trans-retinol. J. Biol. Chem 2001, 276, 1353–1360. [Google Scholar]
- Squires, M.W.; Naber, E.C. Vitamin profiles of eggs as indicators of nutritional status in the laying hen: vitamin A study. Poult. Sci 1993, 72, 154–164. [Google Scholar]
- Folli, C.; Calderone, V.; Ramazzina, I.; Zanotti, G.; Berni, R. Ligand binding and structural analysis of a human putative cellular retinol-binding protein. J. Biol. Chem 2002, 277, 41970–41977. [Google Scholar]
- Gong, W.; Tang, Z.; Han, J.; Yang, S.; Wang, H.; Li, Y.; Li, K. Mapping, tissue distribution and polymorphism of porcine retinol binding protein genes (RBP5 and RBP7). Asian Austral. J. Anim 2008, 21, 1544–1550. [Google Scholar]
- Goodman, D.W.S. Vitamin A metabolism. Proc. Soc. Exp. Biol. Med 1980, 39, 2716–2722. [Google Scholar]
- Schmitz, H.H.; Poor, C.L.; Wellman, R.; Erdman, J.W., Jr. Concentrations of selected carotenoids and vitamin A in human liver, kidney and lung tissue. J. Nutr. 1991, 121, 1613–1621. [Google Scholar]
- Zetterstrom, R.; Simon, A.; Giacobini, M.; Eriksson, U.; Olson, L. Localization of cellular retinoid-binding proteins suggests specific roles for retinoids in the adult central nervous system. Neuro. Sci 1994, 62, 899–918. [Google Scholar]
- Morley, J.; Melmed, S.; Reed, A.; Kasson, B.; Levin, S.; Pekary, A.; Hershman, J. Effect of vitamin A on the hypothalamo-pituitary-thyroid axis. Am. J. Physiol-Endoc. M 1980, 238, E174–E179. [Google Scholar]
- Cui, J.; Du, H.; Liang, Y.; Deng, X.; Li, N.; Zhang, X. Association of polymorphisms in the promoter region of chicken prolactin with egg production. Poult. Sci 2006, 85, 26–31. [Google Scholar]
- Zhang, L.; Li, D.; Liu, Y.; Wang, Y.; Zhao, X.; Zhu, Q. The genetic effect of prolactin receptor gene on egg production traits in chickens. Genet. Mol. Res 2012, 11, 4307–4315. [Google Scholar]
- Fatemi, S.; Mehrabani-Yeganeh, H.; Nejati-Javaremi, A.; Niknafs, S. Association of neuropeptide Y and gonadotrophin-releasing hormone receptor gene SNPs with breeding value for growth and egg production traits in Mazandaran native chickens. Genet. Mol. Res 2012, 11, 2539–2547. [Google Scholar]
- Kansaku, N.; Nakada, A.; Okabayashi, H.; Guemene, D.; KuhnleinN, U.; Zadworny, D.; Shimada, K. DNA polymorphism in the chicken growth hormone gene: Association with egg production. Anim. Sci. J 2003, 74, 243–244. [Google Scholar]
- Ledur, M.; Fairfull, R.; McMillan, I.; Asseltine, L. Genetic effects of aging on egg production traits in the first laying cycle of White Leghorn strains and strain crosses. Poult. Sci 2000, 79, 296–304. [Google Scholar]
- McFadden, J.T.; Cooper, E.L.; Andersen, J.K. Some Effects of Environment on Egg Production in Brown Trout (Salmo trutta). Limnol. Oceanogr 1965, 10, 88–95. [Google Scholar]
- Parkhurst, C.R.; Mountney, G.J. Poultry Meat and Egg Production; Van Nostrand Reinhold Company: New York, NY, USA, 1988. [Google Scholar]
- Yin, H.D.; Wang, Y.; Zhao, X.L.; Chen, S.Y.; Zhang, Z.C.; Zhu, Q. Effect Analysis of Heart Fatty Acid-Binding Protein Gene Assisted Selection on Intramuscular Fat Content in Chicken. J. Anim. Vet. Adv 2012, 11, 1595–1600. [Google Scholar]
- Stich, B.; Melchinger, A.E.; Frisch, M.; Maurer, H.P.; Heckenberger, M.; Reif, J.C. Linkage disequilibrium in European elite maize germplasm investigated with SSRs. Theor. Appl. Genet 2005, 111, 723–730. [Google Scholar]
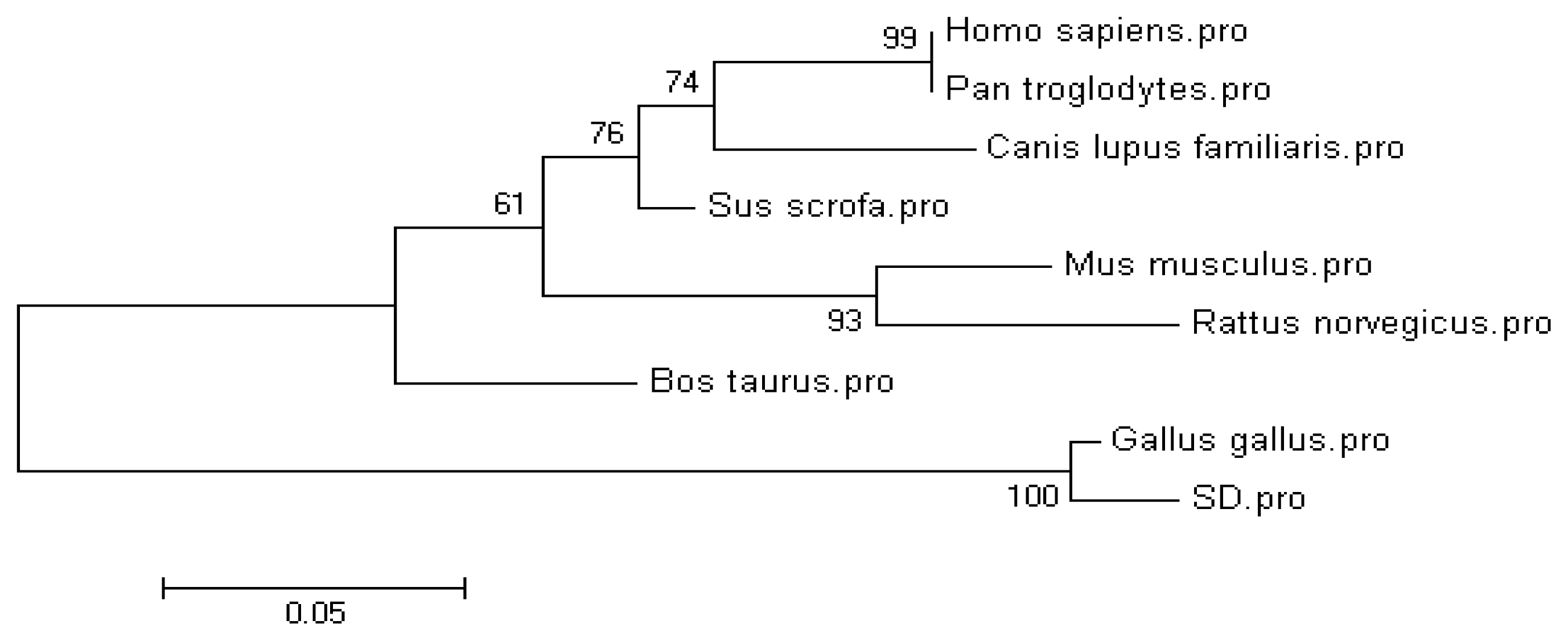

| Species | GenBank accession number | Amino acid Identity 1 |
|---|---|---|
| Gallus gallus | XP_417606.2 | 98.4% (125/127) |
| Sus scrofa | NP_001138694.1 | 74.8% (95/127) |
| Mus musculus | NP_071303.1 | 72.4% (92/127) |
| Rattus norveicus | NP_001102163 | 71.7% (91/127) |
| Homo sapiens | NP_443192.1 | 70.9% (90/127) |
| Pan troglodytes | XP_001161299.1 | 70.9% (90/127) |
| Bos taurus | XP_871364.3 | 66.9% (85/127) |
| Canis lupus familiaris | XP_536740.1 | 57.5% (73/127) |
| Lines | N. | Alleles frequency | Genotypes frequency | p values of chi-square 1 | PIC2 | |||
|---|---|---|---|---|---|---|---|---|
| T | C | TT | TC | CC | ||||
| SD02 | 188 | 0.641 | 0.359 | 0.399 | 0.484 | 0.117 | 0.471 | 0.354 |
| SD03 | 161 | 0.630 | 0.370 | 0.373 | 0.515 | 0.112 | 0.182 | 0.357 |
| Total | 349 | 0.636 | 0.364 | 0.386 | 0.499 | 0.115 | 0.091 | 0.356 |
| Items | N | Traits | ||||||
|---|---|---|---|---|---|---|---|---|
| AFE | BWFE | FEW | TE300 | HCLD | ALI | |||
| Genotypes | CC | 40 | 169.3 ± 10.0 b | 1981.5 ± 296.5 | 40.4 ± 4.9 | 85.6 ± 20.4 a,A | 9.7 ± 3.8 a | 1.8 ± 1.2 |
| TC | 174 | 175.1 ± 11.2 a | 1892.1 ± 244.5 | 39.9 ± 5.8 | 81.1 ± 16.9 b,A,B | 7.8 ± 3.6 b | 1.6 ± 0.7 | |
| TT | 135 | 176.5 ± 11.5 a | 1898.0 ± 247.9 | 40.5 ± 6.8 | 77.9 ± 18.2 c,B | 7.2 ± 3.2 b | 1.7 ± 0.8 | |
| Lines | SD02 | 188 | 169.8 ± 10.2 b | 1903.0 ± 241.2 | 40.2 ± 6.6 | 83.3 ± 18.4 | 7.7 ± 4.0 | 1.6 ± 0.9 |
| SD03 | 161 | 176.8 ± 10.9 a | 1919.5 ± 250.2 | 41.3 ± 5.3 | 81.3 ± 17.3 | 7.8 ± 4.5 | 1.5 ± 0.8 | |
| Primers | Primer Sequence (5′→3′) | Annealing Temperature (°C) | Product Length (bp) | Amplified Region |
|---|---|---|---|---|
| Primer pairs for measuring cloning and sequencing chicken CRBP IV gene | ||||
| CRBPIV-F | TGACTGTAACTCCTTCCCACAGTCT | 56 | 464 | 3–466 |
| CRBPIV-R | GAAGACAGCTTGCACCTTATGAC | |||
| Primer pairs for measuring chicken CRBP IV gene expression | ||||
| CRBPIV-F | CATACCACAAGCACATTCAGAGA | 58 | 125 | 1320–1444 |
| CRBPIV-R | AGTTTGTCATTGTCCCAGGTAAC | |||
| β-actin-F | GAGAAATTGTGCGTGACATCA | 60 | 152 | 685–836 |
| β-actin-R | CCTGAACCTCTCATTGCCA | |||
| Primer pairs for screening chicken CRBP IV gene polymorphisms | ||||
| P1-F | CTTCCCACAGTCTAGCAA | 56.6 | 236 | 3380706–3380919 |
| P1-R | ATCAGCTAACAAATATCCTTC | |||
| P2-F | TTCTTCTGTCAAAGGTATTG | 55.2 | 244 | 3381431–3381654 |
| P2-R | CTTCCTTCTGTAAGAACACAT | |||
| P3-F | CAGAGCCTGGTTACCTG | 56.6 | 180 | 3381893–3382052 |
| P3-R | AGCTTGCACCTTATGACTACA | |||
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Yin, H.-D.; Wang, Y.; Zhang, Z.-C.; Liu, Y.-P.; Chen, S.-Y.; Zhu, Q. Characterization of the Expression Profile and Genetic Polymorphism of the Cellular Retinol-Binding Protein (CRBP IV) Gene in Erlang Mountainous Chickens. Int. J. Mol. Sci. 2013, 14, 4432-4443. https://doi.org/10.3390/ijms14034432
Yin H-D, Wang Y, Zhang Z-C, Liu Y-P, Chen S-Y, Zhu Q. Characterization of the Expression Profile and Genetic Polymorphism of the Cellular Retinol-Binding Protein (CRBP IV) Gene in Erlang Mountainous Chickens. International Journal of Molecular Sciences. 2013; 14(3):4432-4443. https://doi.org/10.3390/ijms14034432
Chicago/Turabian StyleYin, Hua-Dong, Yan Wang, Zhi-Chao Zhang, Yi-Ping Liu, Shi-Yi Chen, and Qing Zhu. 2013. "Characterization of the Expression Profile and Genetic Polymorphism of the Cellular Retinol-Binding Protein (CRBP IV) Gene in Erlang Mountainous Chickens" International Journal of Molecular Sciences 14, no. 3: 4432-4443. https://doi.org/10.3390/ijms14034432




