Quantification of Maternal Serum Cell-Free Fetal DNA in Early-Onset Preeclampsia
Abstract
:1. Introduction
2. Results and Discussion
2.1. Results
2.1.1. Maternal Epidemiological Characteristics
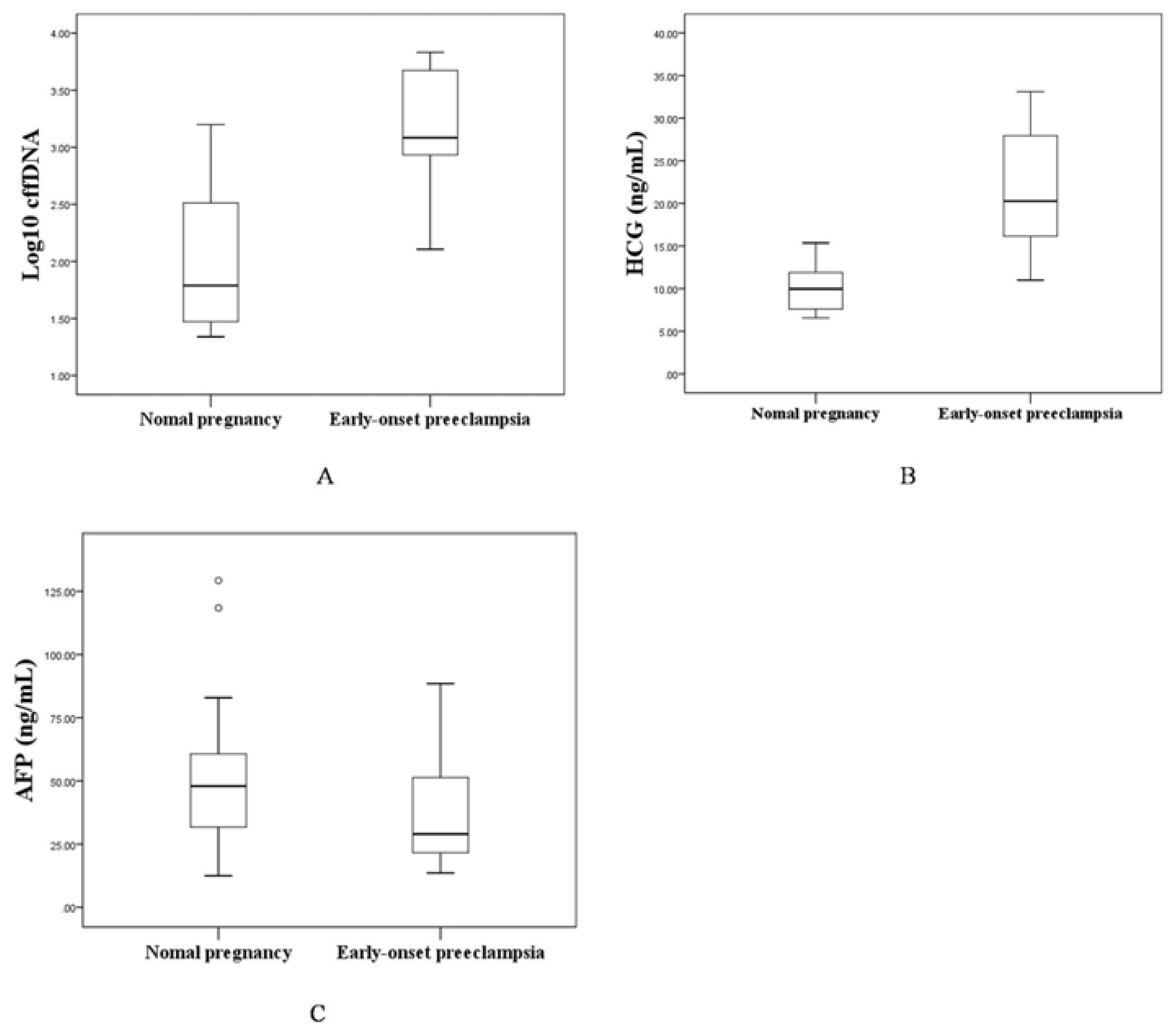
2.1.2. The Levels of Maternal Serum cffDNA, HCG and AFP in the EOPE and Control Groups
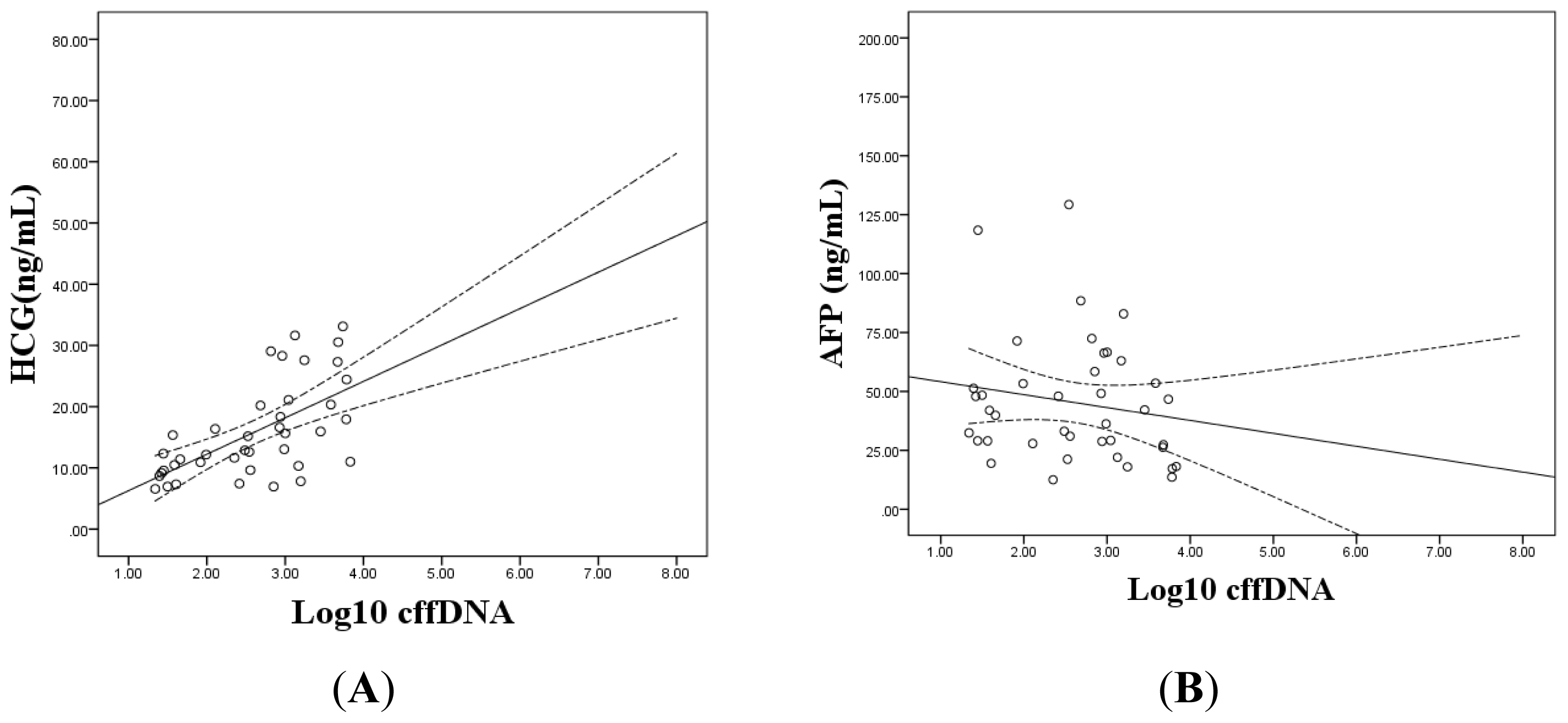
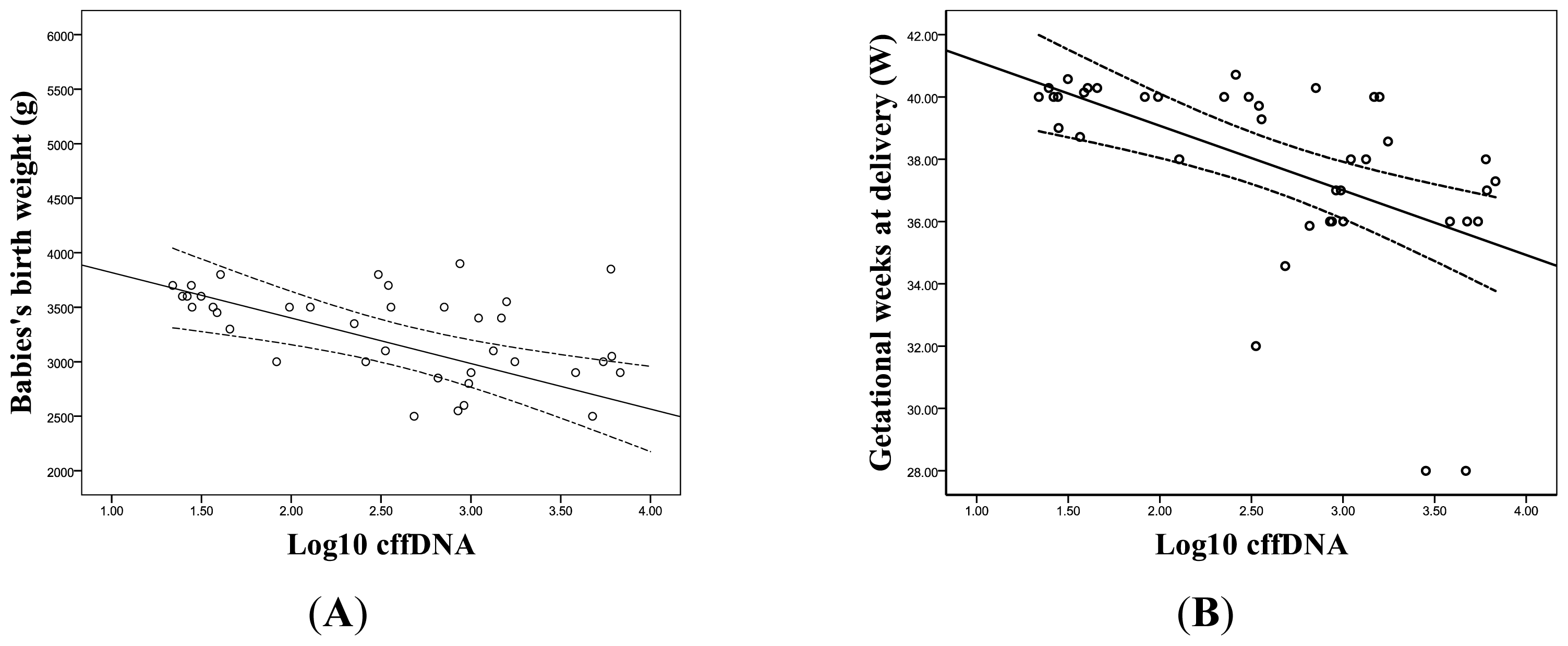
2.1.3. The Correlation between the Levels of cffDNA and Prenatal Screening Markers
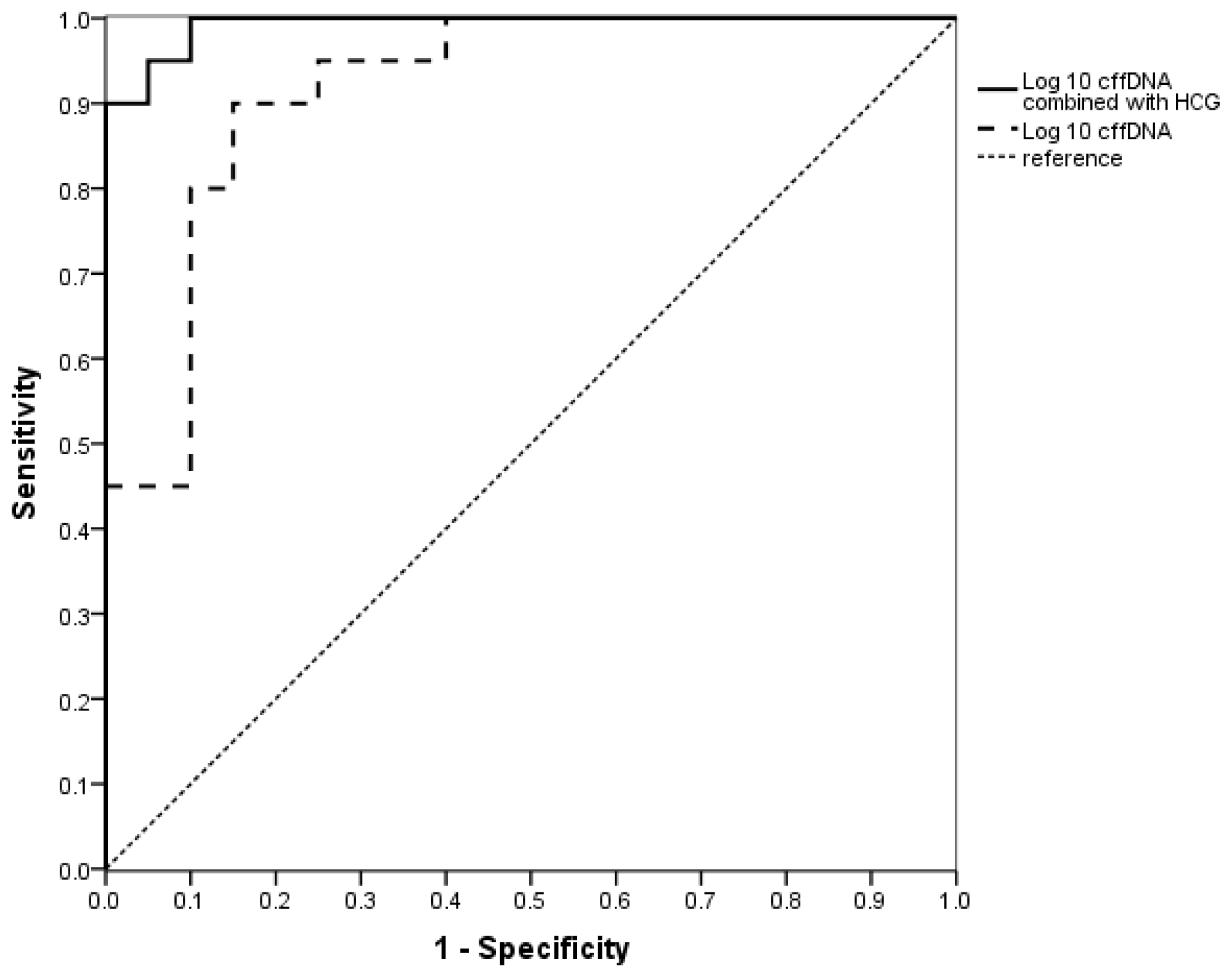
2.1.4. The Evaluation of the Value of cffDNA in Predicting EOPE
2.2. Discussion
3. Experimental Section
3.1. Study Population
3.2. Serum DNA Extraction
3.3. Real-Time Quantitative PCR Amplification
3.4. Statistical Analysis
4. Conclusions
Acknowledgments
Conflict of Interest
References
- Hahn, S.; Rusterholz, C.; Hosli, I.; Lapaire, O. Cell-free nucleic acids as potential markers for preeclampsia. Placenta 2011, 32, S17–S20. [Google Scholar]
- Redman, C.W.; Sargent, I.L. Placental stress and preeclampsia: A revised view. Placenta 2009, 30, S38–S42. [Google Scholar]
- Sibai, B.M.; Barton, J.R. Expectant management of severe preeclampsia remote from term: Patient selection, treatment, and delivery indications. Am. J. Obstet. Gynecol 2007, 196, 514, e1–9.. [Google Scholar]
- Lo, Y.M.; Corbetta, N.; Chamberlain, P.F.; Rai, V.; Sargent, I.L.; Redman, C.W.; Wainscoat, J.S. Presence of fetal DNA in maternal plasma and serum. Lancet 1997, 350, 485–487. [Google Scholar]
- Hyett, J.A.; Gardener, G.; Stojilkovic-Mikic, T.; Finning, K.M.; Martin, P.G.; Rodeck, C.H.; Chitty, L.S. Reduction in diagnostic and therapeutic interventions by non-invasive determination of fetal sex in early pregnancy. Prenat. Diagn 2005, 25, 1111–1116. [Google Scholar]
- Finning, K.; Martin, P.; Daniels, G. A clinical service in the UK to predict fetal Rh(Rhesus) D blood group using free fetal DNA in maternal plasma. Ann. N. Y. Acad. Sci 2004, 1022, 119–123. [Google Scholar]
- Daniels, G.; Finning, K.; Martin, P.; Massey, E. Noninvasive prenatal diagnosis of fetal blood group phenotypes: Current practice and future prospects. Prenat. Diagn 2009, 29, 101–107. [Google Scholar]
- Zhao, F.; Wang, J.; Liu, R.; Yang, J.; Cui, K.; Wu, Y.; Guo, J.; Mu, Y.; Wang, X. Quantification and application of the placental epigenetic signature of the RASSF1A gene in maternal plasma. Prenat. Diagn. 2010, 30, 778–782. [Google Scholar]
- Sifakis, S.; Zaravinos, A.; Maiz, N.; Spandidos, D.A.; Nicolaides, K.H. First-trimester maternal plasma cell-free fetal DNA and preeclampsia. Am. J. Obstet. Gynecol 2009, 201, 472, e1–7.. [Google Scholar]
- Lo, Y.M.; Leung, T.N.; Tein, M.S.; Sargent, I.L.; Zhang, J.; Lau, T.K.; Haines, C.J.; Redman, C.W. Quantitative abnormalities of fetal DNA in maternal serum in preeclampsia. Clin. Chem 1999, 45, 184–188. [Google Scholar]
- Zhong, X.Y.; Laivuori, H.; Livingston, J.C.; Ylikorkala, O.; Sibai, B.M.; Holzqreve, W.; Hahn, S. Elevation of both maternal and fetal extracellular circulating deoxyribonucleic acid concentrations in the plasma of pregnant women with preeclampsia. Am. J. Obstet. Gynecol 2001, 184, 414–419. [Google Scholar]
- Smid, M.; Vassallo, A.; Lagona, F.; Valsecchi, L.; Maniscalco, L.; Danti, L.; Lojacono, A.; Ferrar, A.; Ferrari, M.; Cremonesi, L. Quantitative analysis of fetal DNA in maternal plasma in pathological conditions associated with placental abnormalities. Ann. N. Y. Acad. Sci 2001, 945, 132–137. [Google Scholar]
- Swinkels, D.W.; de Kok, J.B.; Hendriks, J.C.; Wiegerinck, E.; Zusterzeel, P.L.; Steegers, E.A. Hemolysis, elevated liver enzymes, and low platelet count (HELLP) syndrome as a complication of preeclampsia in pregnant women increases the amount of cell-free fetal and maternal DNA inmaternal plasma and serum. Clin. Chem 2002, 48, 650–653. [Google Scholar]
- Lau, T.W.; Leung, T.N.; Chan, L.Y.; Lau, T.K.; Chan, K.C.; Tam, W.H.; Lo, YM. Fetal DNA clearance from maternal plasma is impaired in preeclampsia. Clin. Chem. 2002, 48, 2141–2146. [Google Scholar]
- Sekizawa, A.; Jimbo, M.; Saito, H.; Iwasaki, M.; Matsuoka, R.; Okai, T.; Farina, A. Cell-free fetal DNA in the plasma of pregnant women with severe fetal growth restriction. Am. J. Obstet. Gynecol 2003, 188, 480–484. [Google Scholar]
- Shimada, K.; Murakami, K.; Shozu, M.; Segawa, T.; Sumitani, H.; Inoue, M. Sex-determining region Y levels in maternal plasma: Evaluation in abnormal pregnancy. J. Obstet. Gynaecol. Res 2004, 30, 148–154. [Google Scholar]
- Farina, A.; Sekizawa, A.; Rizzo, N.; Concu, M.; Banzola, I.; Carinci, P.; Simonazzi, G.; Okai, T. Cell-free fetal DNA (SRY locus) concentration in maternal plasma is directly correlated to the time elapsed from the onset of preeclampsia to the collection of blood. Prenat. Diagn 2004, 24, 293–297. [Google Scholar]
- Zhong, X.Y.; Gebhardt, S.; Hillermann, R.; Tofa, K.C.; Holzgreve, W.; Hahn, S. Parallel assessment of circulatory fetal DNA and corticotropin-releasing hormone mRNA in early- and late-onset preeclampsia. Clin. Chem 2005, 51, 1730–1733. [Google Scholar]
- Smid, M.; Galbiati, S.; Lojacono, A.; Valsecchi, L.; Platto, C.; Cavoretto, P.; Calza, S.; Ferrari, A.; Ferrari, M.; Cremonesi, L. Correlation of fetal DNA levels in maternal plasma with Doppler status in pathological pregnancies. Prenat. Diagn. 2006, 26, 785–790. [Google Scholar]
- Leung, T.N.; Zhang, J.; Lau, T.K.; Lo, Y.M. Increased maternal plasma fetal DNA concentrations in women who eventually develop preeclampsia. Clin. Chem 2001, 47, 137–139. [Google Scholar]
- Zhong, X.Y.; Holzgreve, W.; Hahn, S. The levels of circulatory cell free fetal DNA in maternal plasma are elevated prior to the onset of preeclampsia. Hypertens. Pregnancy 2002, 21, 77–83. [Google Scholar]
- Levine, R.J.; Qian, C.; Leshane, E.S.; Yu, K.F.; Enqland, L.J.; Schisterman, E.F.; Wataqanara, T.; Romero, R.; Bianchi, D.W. Two-stage elevation of cell-free fetal DNA in maternal sera before onset of preeclampsia. Am. J. Obstet. Gynecol 2004, 190, 707–713. [Google Scholar]
- Farina, A.; Sekizawa, A.; Sugito, Y.; Iwasaki, M.; Jimbo, M.; Saito, H.; Okai, T. Fetal DNA in maternal plasma as a screening variable for preeclampsia: A preliminary nonparametric analysis of detection rate in low-risk nonsymptomatic patients. Prenat. Diagn 2004, 24, 83–86. [Google Scholar]
- Crowley, A.; Martin, C.; Fitzpatrick, P.; Sheils, O.; O’Herlihy, C.; O’Leary, J.J.; Byrne, B.M. Free fetal DNA is not increased before 22 weeks in intrauterine growth restriction or preeclampsia. Prenat. Diagn 2007, 27, 174–179. [Google Scholar]
- Lazar, L.; Rigó, J., Jr.; Nagy, B.; Balogh, K.; Makó, V.; Cervenak, L.; Mézes, M.; Prohászka, Z.; Molvarec, A. Relationship of circulating cell-free DNA levels to cell-free fetal DNA levels, clinical characteristics and laboratory parameters in preeclampsia. BMC Med. Genet. 2009, 10. [Google Scholar] [CrossRef]
- Morris, R.K.; Cnossen, J.S.; Langejans, M.; Robson, S.C.; Kleijnen, J.; TerRiet, G.; Mol, B.W.; van der Post, J.A.; Khan, K.S. Serum screening with Down’s syndrome markers to predict pre-eclampsia and small for gestational age: Systematic review and meta-analysis. BMC Pregnancy Childbirth 2008, 8. [Google Scholar] [CrossRef]
- Di Lorenzo, G.; Ceccarello, M.; Cecotti, V.; Ronfani, L.; Monasta, L.; Vecchi Brumatti, L.; Montico, M.; D’Ottavio, G. First trimester maternal serum PIGF, free b-hCG, PAPP-A, PP-13, uterine artery Doppler and maternal history for the prediction of preeclampsia. Placenta 2012, 33, 495–501. [Google Scholar]
- Audibert, F.; Boucoiran, I.; An, N.; Aleksandrov, N.; Delvin, E.; Bujold, E.; Rey, E. Screening for preeclampsia using first-trimester serummarkers and uterine artery Doppler in nulliparous women. Am. J. Obstet. Gynecol 2010, 203, 383, e1–8.. [Google Scholar]
- Tjoa, M.L.; Cindrova-Davies, T.; Spasic-Boskovic, O.; Bianchi, D.W.; Burton, G.J. Trophoblastic oxidative stress and the release of cell-free feto-placental DNA. Am. J. Pathol 2006, 169, 400–404. [Google Scholar]
- Ahn, H.; Park, J.; Gilman-Sachs, A.; Kwak-Kim, J. Immunologic characteristics of preeclampsia, a comprehensive review. Am. J. Reprod. Immunol 2011, 65, 377–394. [Google Scholar]
- Servitje, E.L.; Lopez, N.G.; Olson, D.M. An immunological insight into the origins of preeclampsia. Hum. Reprod. Update 2010, 16, 510–524. [Google Scholar]
- Uzan, J.; Carbonnel, M.; Piconne, O.; Asmar, R.; Ayoubi, J.M. Pre-eclampsia: Pathophysiology, diagnosis,and management. Vasc. Health Risk Manag 2011, 7, 467–474. [Google Scholar]
- Chen, J.Z.; Sheehan, P.M.; Brennecke, S.P.; Keogh, R.J. Vessel remodelling, pregnancy hormones and extravilloustrophoblast function. Mol. Cell. Endocrinol 2012, 349, 138–144. [Google Scholar]
- Jin, H.K.; Antonio, F.; Ji, H.P. Down syndrome biochemical markers and screening for preeclampsia at first and second trimester: Correlation with the week of onset and the severity. Prenat. Diagn 2008, 28, 704–709. [Google Scholar]
- Lambert-Messerlian, G.M.; Silver, H.M.; Petraglia, F.; Luisi, S.; Pezzani, I.; Maybruck, W.M.; Hogge, W.A.; Hanley-Yanez, K.; Roberts, J.M.; Neveux, L.M.; et al. Second-trimester levels of maternal serum human chorionic gonadotropin and inhibin a as predictiors of preeclampsia in the third trimester of pregnancy. J. Soc. Gynecol. Invest 2000, 7, 170–174. [Google Scholar]
- Davey, D.A.; MacGillivray, I. The classification and definition of the hypertensive disorders of pregnancy. Am. J. Obstet. Gynecol 1988, 158, 892–898. [Google Scholar]
| Case (n = 30) | Control (n = 867) | p-values | |
|---|---|---|---|
| Maternal age, y | 30.47 ± 5.31 | 27.48 ± 4.70 | 0.001 * |
| Gestational weeks at enrollment, w | 17.99 ± 1.17 | 17.87 ± 1.24 | 0.617 |
| Pregestational body mass index, kg/m2 | 23.49 ± 3.57 | 21.30 ± 2.76 | 0.002 * |
| Nulliparous | 80.0% | 88.5% | 0.158 |
| Previous abortion | 6.7% | 0.9% | 0.003 * |
| Family history of hypertension | 63.3% | 18.9% | <0.001 * |
| Medical history of hypertension, diabetes and nephritis | 10.0% | 0.3% | <0.001 * |
| Case (n = 20) | Control (n = 20) | p-values | |
|---|---|---|---|
| Maternal age, y | 29.54 ± 5.07 | 28.27 ± 4.78 | 0.421 |
| Gestational weeks at enrollment, w | 18.06 ± 1.37 | 17.80 ± 0.99 | 0.488 |
| Getational weeks at delivery, w | 35.66 ± 3.00 | 39.96 ± 0.48 | <0.001 * |
| Birth weight, kg | 2.79 ± 0.81 | 3.50 ± 0.22 | 0.001 * |
| Intrauterine growth restriction (IUGR) | 10% | 0% | 0.487 |
© 2013 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Yu, H.; Shen, Y.; Ge, Q.; He, Y.; Qiao, D.; Ren, M.; Zhang, J. Quantification of Maternal Serum Cell-Free Fetal DNA in Early-Onset Preeclampsia. Int. J. Mol. Sci. 2013, 14, 7571-7582. https://doi.org/10.3390/ijms14047571
Yu H, Shen Y, Ge Q, He Y, Qiao D, Ren M, Zhang J. Quantification of Maternal Serum Cell-Free Fetal DNA in Early-Onset Preeclampsia. International Journal of Molecular Sciences. 2013; 14(4):7571-7582. https://doi.org/10.3390/ijms14047571
Chicago/Turabian StyleYu, Hong, Yanting Shen, Qinyu Ge, Youji He, Dongyan Qiao, Mulan Ren, and Jianqiong Zhang. 2013. "Quantification of Maternal Serum Cell-Free Fetal DNA in Early-Onset Preeclampsia" International Journal of Molecular Sciences 14, no. 4: 7571-7582. https://doi.org/10.3390/ijms14047571




