The Cloning and Characterization of the Enolase2 Gene of Gekko japonicus and Its Polyclonal Antibody Preparation
Abstract
:1. Introduction
2. Results and Discussion
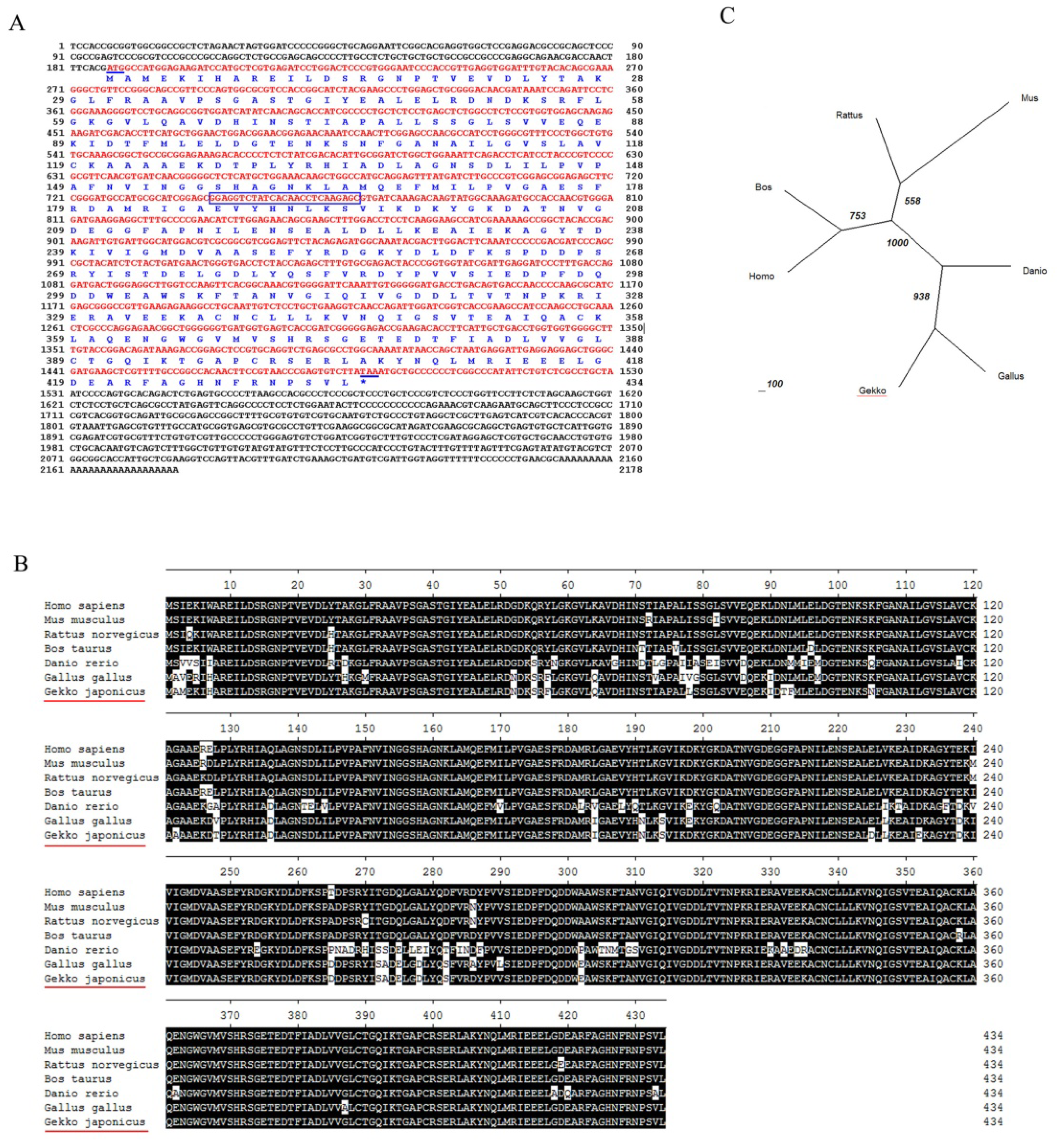
2.1. Molecular Cloning and Bioinformatic Analysis of NSE
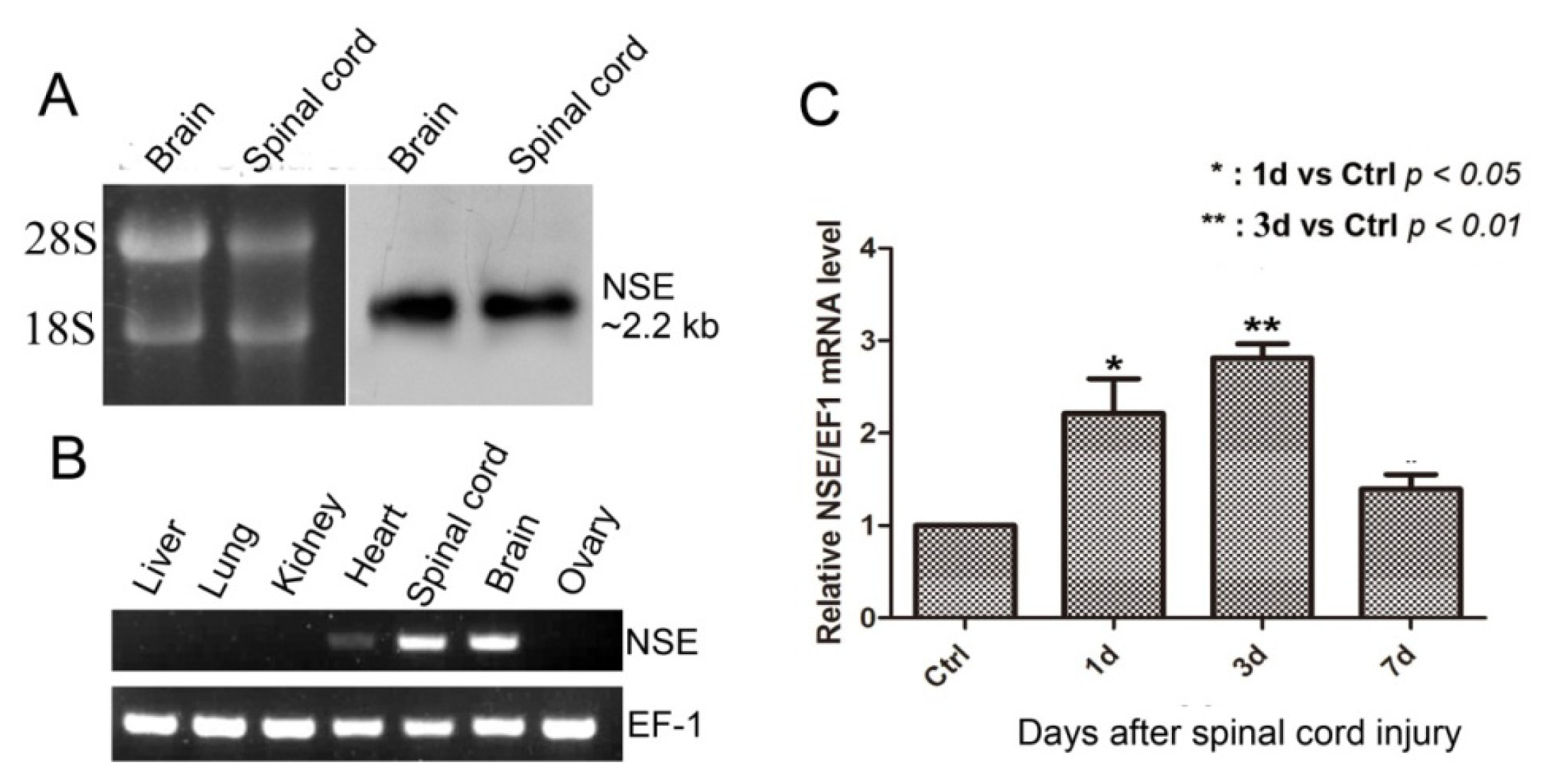
2.2. NSE mRNA Transcript in CNS, the Expression Pattern in Different Tissues and mRNA Expression after Spinal Cord Transection in Geckos
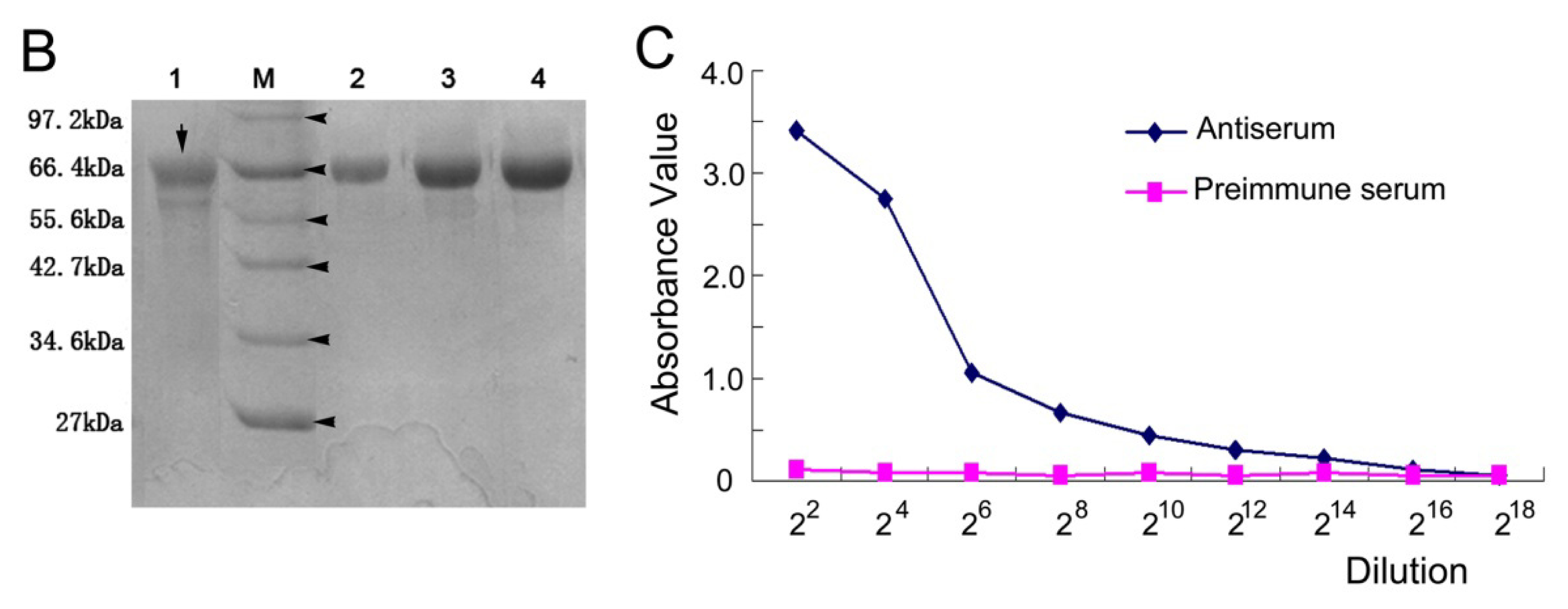
2.3. The Titer of the Prepared Antiserum
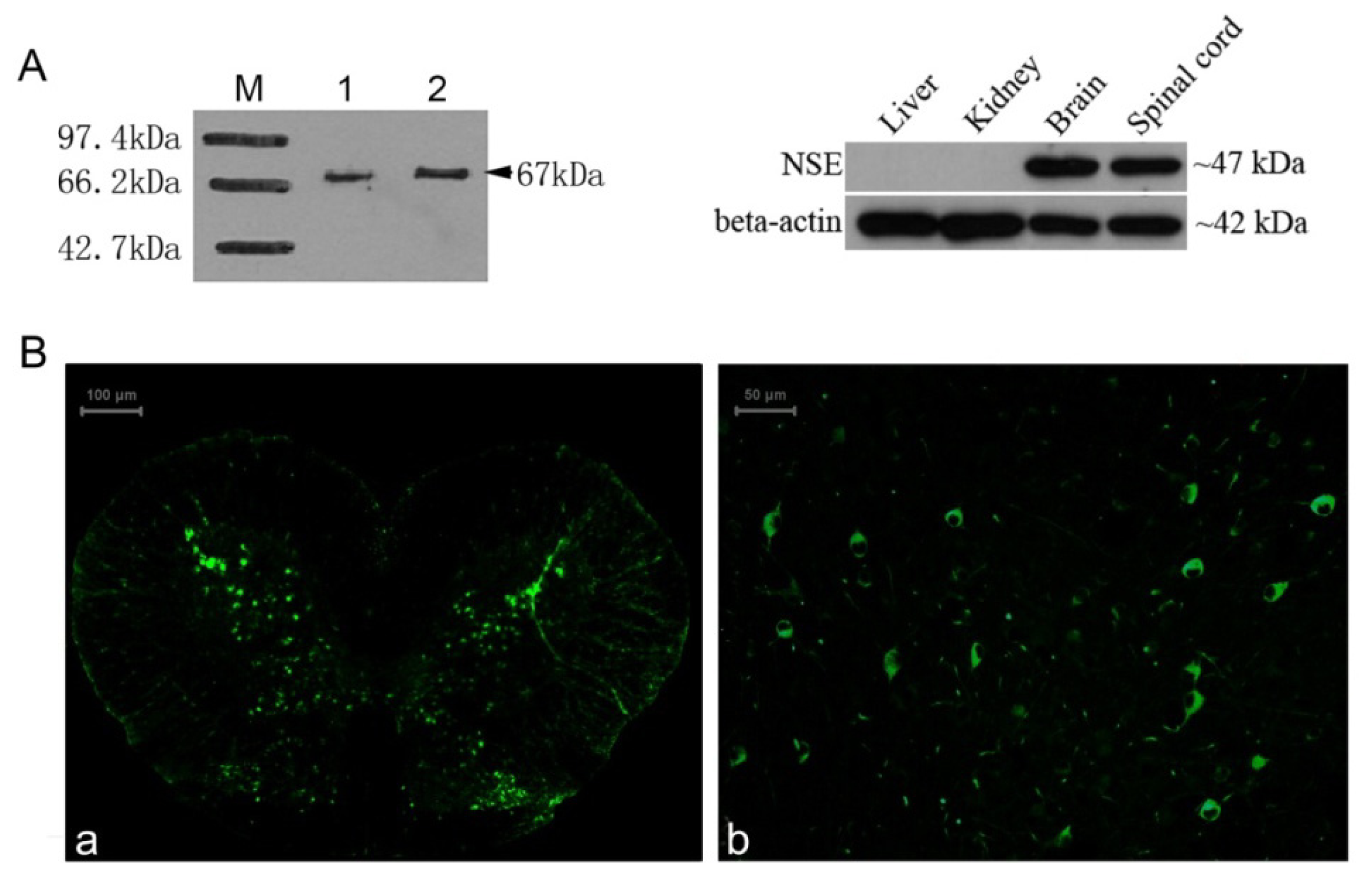
2.4. Identification of the NSE Polyclonal Antibody by Western Blot and Immunochemistry Analysis
3. Materials and Methods
3.1. Animals and Spinal Cord Injury
3.2. Molecular Cloning of Full-Length cDNA of NSE
3.3. Northern Blot Analysis
3.4. RT-PCR and Real-Time Quantitative PCR
3.5. Bioinformatic Analysis of NSE
3.6. Expression, Purification of His Fusion NSE Protein and Preparation of Anti NSE Polyclonal Antibody
3.7. Titer Determination of Anti-NSE Antiserum by ELISA
3.8. Western Blotting
3.9. Immunohistochemistry
4. Conclusions
Acknowledgments
Conflict of Interest
References
- Chernoff, E.A. Spinal cord regeneration: A phenomenon unique to urodeles? Int. J. Dev. Biol 1996, 40, 823–831. [Google Scholar]
- Goldshmit, Y.; Sztal, T.E.; Jusuf, P.R.; Hall, T.E.; Nguyen-Chi, M.; Currie, P.D. Fgf-dependent glial cell bridges facilitate spinal cord regeneration in zebrafish. J. Neurosci 2012, 32, 7477–7492. [Google Scholar]
- Romero-Aleman, M.M.; Monzon-Mayor, M.; Yanes, C.; Lang, D. Radial glial cells, proliferating periventricular cells, and microglia might contribute to successful structural repair in the cerebral cortex of the lizard Gallotia galloti. Exp. Neurol 2004, 188, 74–85. [Google Scholar]
- Dallimore, E.J.; Park, K.K.; Pollett, M.A.; Taylor, J.S.; Harvey, A.R. The life, death and regenerative ability of immature and mature rat retinal ganglion cells are influenced by their birthdate. Exp. Neurol 2010, 225, 353–365. [Google Scholar]
- Brockes, J.P. Amphibian limb regeneration: Rebuilding a complex structure. Science 1997, 276, 81–87. [Google Scholar]
- Gould, E. How widespread is adult neurogenesis in mammals? Nat. Rev. Neurosci 2007, 8, 481–488. [Google Scholar]
- Di Giovanni, S. Molecular targets for axon regeneration: Focus on the intrinsic pathways. Expert Opin. Ther. Targets 2009, 13, 1387–1398. [Google Scholar]
- Duffy, M.T.; Simpson, S.B., Jr; Liebich, D.R.; Davis, B.M. Origin of spinal cord axons in the lizard regenerated tail: Supernormal projections from local spinal neurons. J. Comp. Neurol 1990, 293, 208–222. [Google Scholar]
- Zhou, Y.; Xu, Q.; Li, D.; Zhao, L.; Wang, Y.; Liu, M.; Gu, X.; Liu, Y. Early neurogenesis during caudal spinal cord regeneration in adult Gekko japonicus. J. Mol. Histol. 2012. [Google Scholar] [CrossRef]
- McLean, K.E.; Vickaryous, M.K. A novel amniote model of epimorphic regeneration: The leopard gecko, Eublepharis macularius. BMC Dev. Biol 2011, 11, 50, :1–50:24.. [Google Scholar]
- Liu, Y.; Ding, F.; Liu, M.; Jiang, M.; Yang, H.; Feng, X.; Gu, X. EST-based identification of genes expressed in brain and spinal cord of Gekko japonicus, a species demonstrating intrinsic capacity of spinal cord regeneration. J. Mol. Neurosci 2006, 29, 21–28. [Google Scholar]
- Liu, Y.; Fan, Z.; Zhou, Y.; Liu, M.; Ding, F.; Gu, X. The molecular cloning of platelet-derived growth factor-C (PDGF-C) gene of Gekko japonicus and its expression change in the spinal cord after tail amputation. Cell Mol. Neurobiol 2009, 29, 263–271. [Google Scholar]
- Wang, Y.; Wang, R.; Jiang, S.; Zhou, W.; Liu, Y.; Gu, Q.; Gu, Y.; Dong, Y.; Liu, M.; Gu, X.; et al. Gecko CD59 is implicated in proximodistal identity during tail regeneration. PLoS One 2011, 6, e17878. [Google Scholar]
- Jiang, M.; Gu, X.; Feng, X.; Fan, Z.; Ding, F.; Liu, Y. The molecular characterization of the brain protein 44-like (Brp44l) gene of Gekko japonicus and its expression changes in spinal cord after tail amputation. Mol. Biol. Rep 2009, 36, 215–220. [Google Scholar]
- Liu, M.; Gu, Y.; Liu, Y.; Li, J.; He, J.; Lin, S.; Gu, X. Establishment and characterization of two cell lines derived from primary cultures of Gekko japonicus cerebral cortex. Cell Biol. Int 2010, 34, 153–161. [Google Scholar]
- Gao, D.; Wang, Y.; Liu, Y.; Ding, F.; Gu, X.; Li, Z. The molecular cloning of glial fibrillary acidic protein in Gekko japonicus and its expression changes after spinal cord transection. Cell Mol. Biol. Lett 2010, 15, 582–599. [Google Scholar]
- Li, J.; Qin, Y.; Gu, Y.; Liu, Y.; Liu, M. Molecular cloning of tubulin beta 3 (TUBB3) in Gekko japonicus and preparation of its polyclonal antibody. Dongwuxue Yanjiu 2012, 33, 395–401. [Google Scholar]
- Shimizu, A.; Suzuki, F.; Kato, K. Characterization of αα, ββ, γγ and αγ human enolase isozymes, and preparation of hybrid enolases (αγ, βγ and αβ) from homodimeric forms. Biochim. Biophys. Acta 1983, 748, 278–284. [Google Scholar]
- Marangos, P.J.; Zis, A.P.; Clark, R.L.; Goodwin, F.K. Neuronal, non-neuronal and hybrid forms of enolase in brain: Structural, immunological and functional comparisons. Brain Res 1978, 150, 117–133. [Google Scholar]
- Sakimura, K.; Kushiya, E.; Obinata, M.; Takahashi, Y. Molecular cloning and the nucleotide sequence of cDNA to mRNA for non-neuronal enolase (αα enolase) of rat brain and liver. Nucleic Acids Res 1985, 13, 4365–4378. [Google Scholar]
- Magro, G.; Grasso, S. Immunohistochemical identification and comparison of glial cell lineage in foetal, neonatal, adult and neoplastic human adrenal medulla. Histochem. J 1997, 29, 293–299. [Google Scholar]
- Schmechel, D.; Marangos, P.J.; Zis, A.P.; Brightman, M.; Goodwin, F.K. Brain endolases as specific markers of neuronal and glial cells. Science 1978, 199, 313–315. [Google Scholar]
- Schmechel, D.; Marangos, P.J.; Brightman, M. Neurone-specific enolase is a molecular marker for peripheral and central neuroendocrine cells. Nature 1978, 276, 834–836. [Google Scholar]
- Haimoto, H.; Takahashi, Y.; Koshikawa, T.; Nagura, H.; Kato, K. Immunohistochemical localization of gamma-enolase in normal human tissues other than nervous and neuroendocrine tissues. Lab. Invest 1985, 52, 257–263. [Google Scholar]
- Hafner, A.; Obermajer, N.; Kos, J. gamma-Enolase C-terminal peptide promotes cell survival and neurite outgrowth by activation of the PI3K/Akt and MAPK/ERK signalling pathways. Biochem. J 2012, 443, 439–450. [Google Scholar]
- Gross, J.; Zinsmeyer, J.; Lessing, A.; Wenzel, J.; Prenzlau, P.; Halle, H.; Grauel, E.L. Development of enolase isoenzymes in various regions of the human brain. Biomed. Biochim. Acta 1990, 49, 533–538. [Google Scholar]
- Cao, F.; Yang, X.F.; Liu, W.G.; Hu, W.W.; Li, G.; Zheng, X.J.; Shen, F.; Zhao, X.Q.; Lv, S.T. Elevation of neuron-specific enolase and S-100beta protein level in experimental acute spinal cord injury. J. Clin. Neurosci 2008, 15, 541–544. [Google Scholar]
- Dauberschmidt, R.; Marangos, P.J.; Zinsmeyer, J.; Bender, V.; Klages, G.; Gross, J. Severe head trauma and the changes of concentration of neuron-specific enolase in plasma and in cerebrospinal fluid. Clin. Chim. Acta 1983, 131, 165–170. [Google Scholar]
- Zhang, B.; Huang, Y.; Su, Z.; Wang, S.; Wang, J.; Wang, A.; Lai, X. Neurological, functional, and biomechanical characteristics after high-velocity behind armor blunt trauma of the spine. J. Trauma 2011, 71, 1680–1688. [Google Scholar]
- Ishiguro, Y.; Kato, K.; Ito, T.; Nagaya, M. Determination of three enolase isozymes and S-100 protein in various tumors in children. Cancer Res 1983, 43, 6080–6084. [Google Scholar]
- D’Alessandro, M.; Mariani, P.; Lomanto, D.; Carlei, F.; Lezoche, E.; Speranza, V. Serum neuron-specific enolase in diagnosis and follow-up of gastrointestinal neuroendocrine tumors. Tumour. Biol 1992, 13, 352–357. [Google Scholar]
- DeGiorgio, C.M.; Correale, J.D.; Gott, P.S.; Ginsburg, D.L.; Bracht, K.A.; Smith, T.; Boutros, R.; Loskota, W.J.; Rabinowicz, A.L. Serum neuron-specific enolase in human status epilepticus. Neurology 1995, 45, 1134–1137. [Google Scholar]
- Yamazaki, Y.; Yada, K.; Morii, S.; Kitahara, T.; Ohwada, T. Diagnostic significance of serum neuron-specific enolase and myelin basic protein assay in patients with acute head injury. Surg. Neurol 1995, 43, 267–271. [Google Scholar]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res 1997, 25, 3389–3402. [Google Scholar]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol 1999, 112, 531–552. [Google Scholar]
- Pearson, W.R.; Robins, G.; Zhang, T. Generalized neighbor-joining: More reliable phylogenetic tree reconstruction. Mol. Biol. Evol 1999, 16, 806–816. [Google Scholar]
- Haghi, F.; Peerayeh, S.N.; Siadat, S.D.; Montajabiniat, M. Cloning, expression and purification of outer membrane protein PorA of Neisseria meningitidis serogroup B. J. Infect. Dev. Ctries 2011, 5, 856–862. [Google Scholar]
- Engvall, E.; Perlmann, P. Enzyme-linked immunosorbent assay (ELISA). Quantitative assay of immunoglobulin G. Immunochemistry 1971, 8, 871–874. [Google Scholar]

© 2013 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Li, J.; Wu, R.; Chen, H.; Zhou, Y.; Li, Y.; Wang, Y.; Liu, Y.; Liu, M. The Cloning and Characterization of the Enolase2 Gene of Gekko japonicus and Its Polyclonal Antibody Preparation. Int. J. Mol. Sci. 2013, 14, 8787-8800. https://doi.org/10.3390/ijms14058787
Li J, Wu R, Chen H, Zhou Y, Li Y, Wang Y, Liu Y, Liu M. The Cloning and Characterization of the Enolase2 Gene of Gekko japonicus and Its Polyclonal Antibody Preparation. International Journal of Molecular Sciences. 2013; 14(5):8787-8800. https://doi.org/10.3390/ijms14058787
Chicago/Turabian StyleLi, Jing, Ronghua Wu, Haijiao Chen, Youlang Zhou, Yan Li, Yongjun Wang, Yan Liu, and Mei Liu. 2013. "The Cloning and Characterization of the Enolase2 Gene of Gekko japonicus and Its Polyclonal Antibody Preparation" International Journal of Molecular Sciences 14, no. 5: 8787-8800. https://doi.org/10.3390/ijms14058787



