Characterization of Rice NADPH Oxidase Genes and Their Expression under Various Environmental Conditions
Abstract
:1. Introduction
2. Results
2.1. Identification and Domain Composition of Nox Proteins in Rice
2.2. Evolution and Phylogenetic Distribution of Rice Nox Proteins
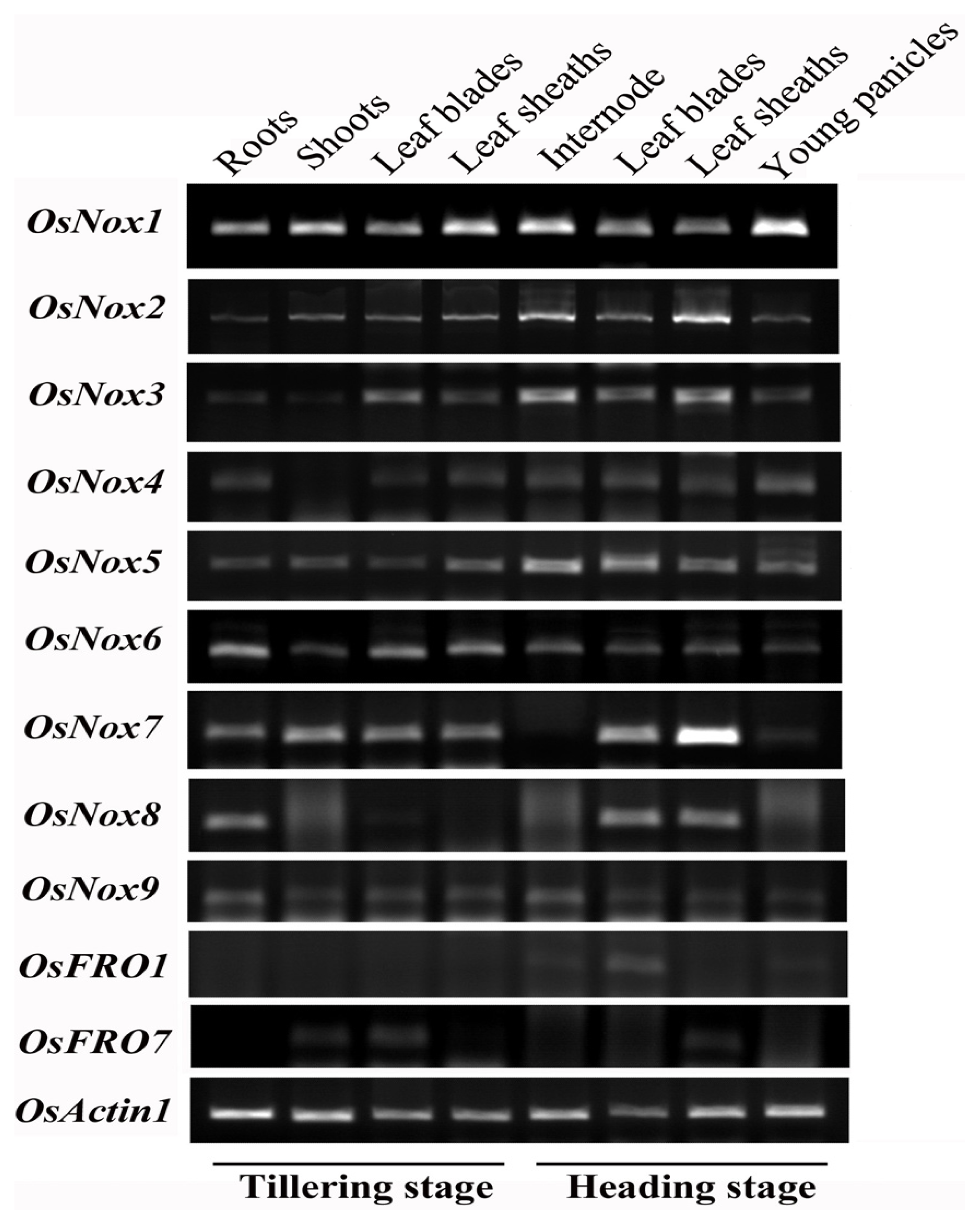
2.3. Expression Profiles of Rice Nox Genes in Different Tissues
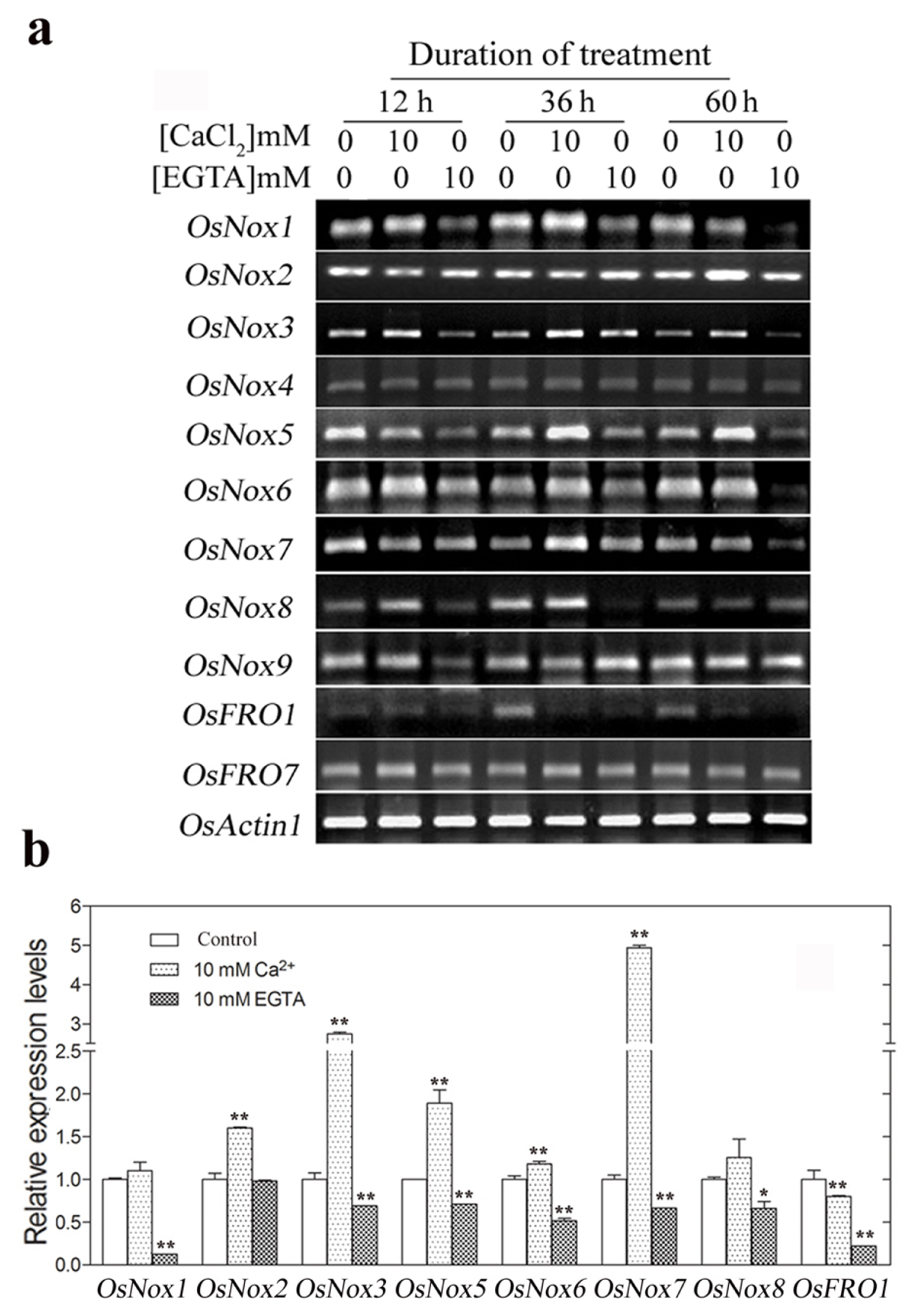
2.4. Expression of Rice Nox Genes under Reduced and Increased Calcium Conditions
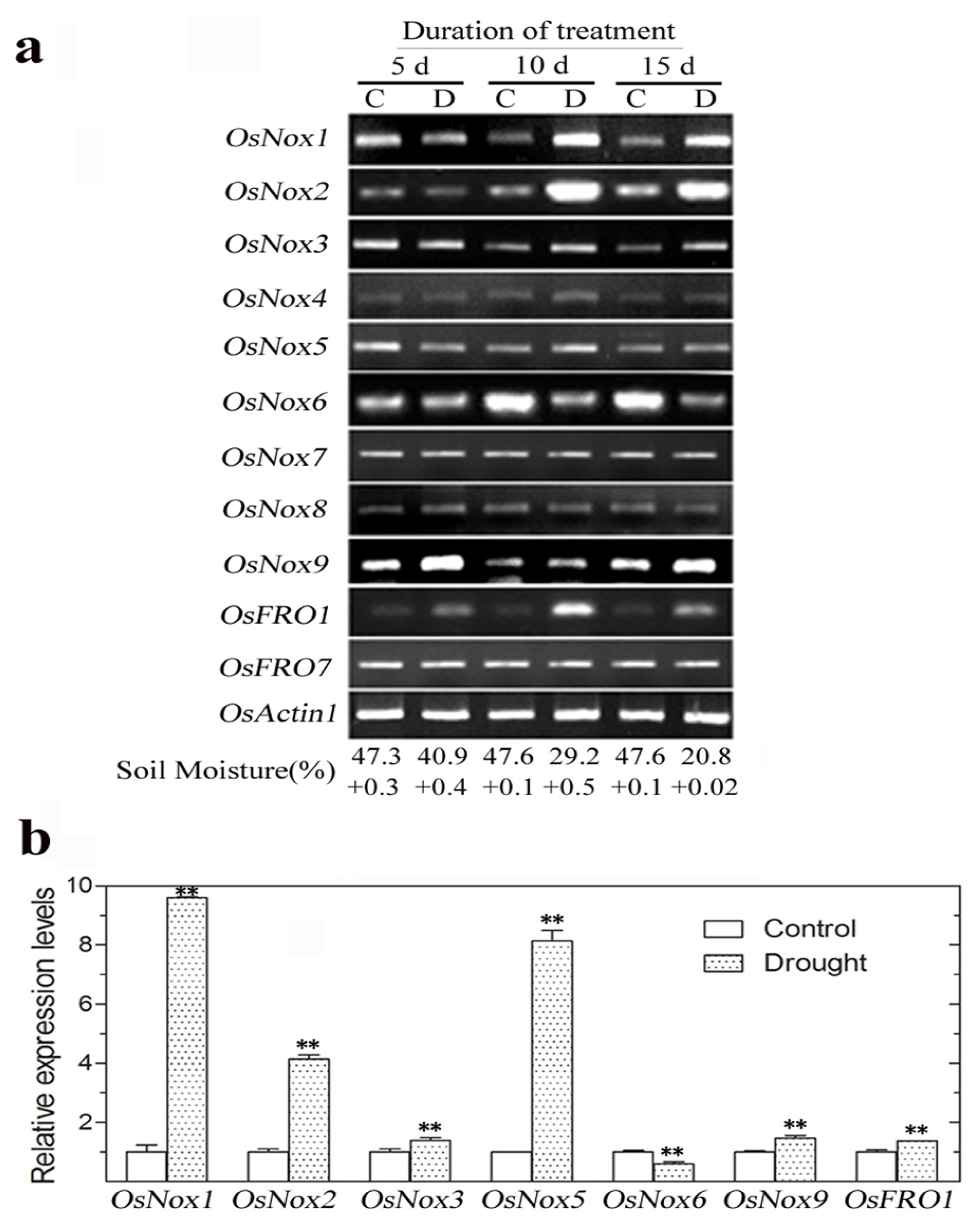
2.5. Expression of Rice Nox Genes under Drought Conditions
2.6. Expression of Rice Nox Genes at High Temperature
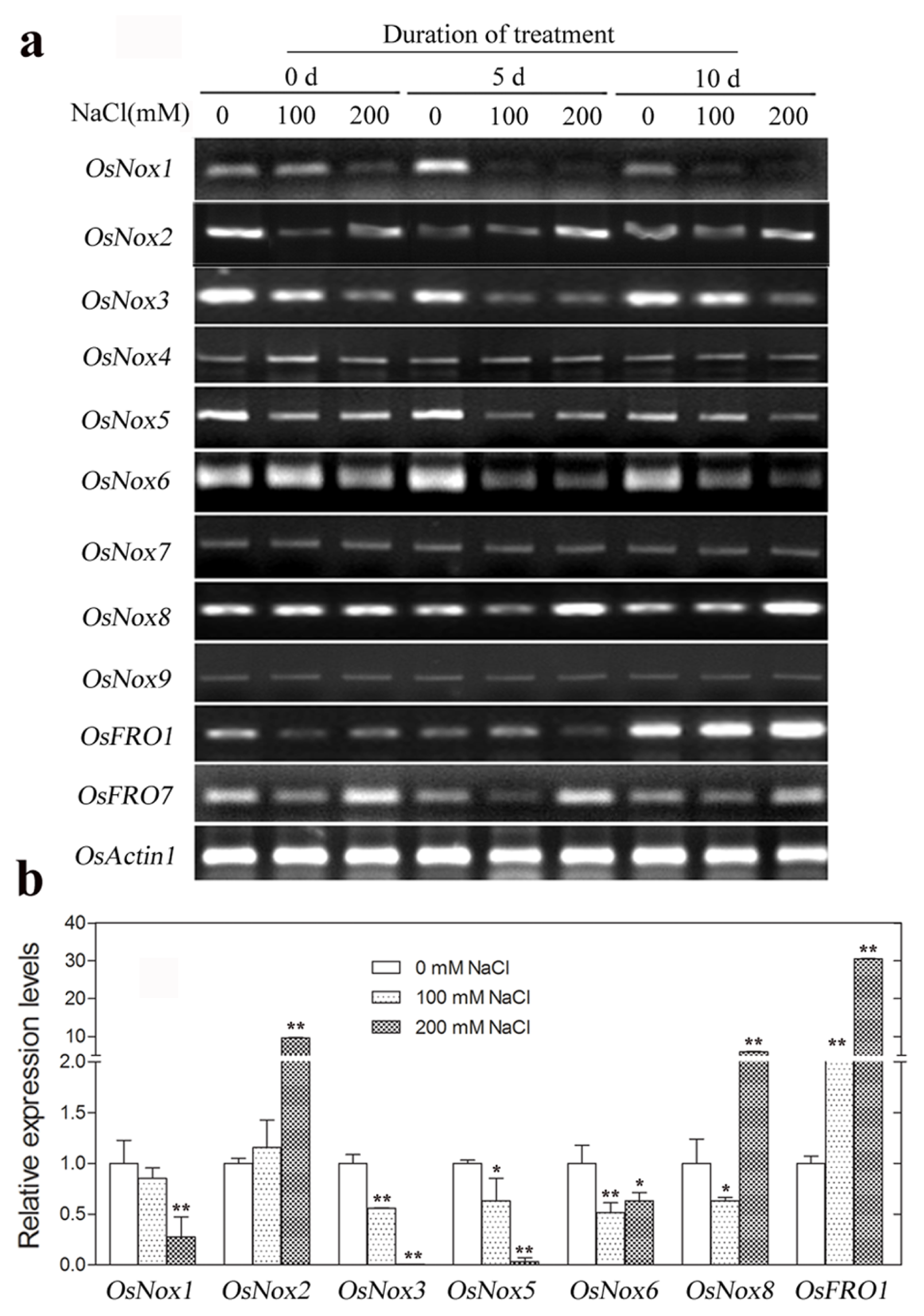
2.7. Expression of Rice Nox Genes under High NaCl Conditions
3. Discussion
4. Experimental Section
4.1. Plant Materials and Stress Treatments
4.2. Identification and Phylogenetic Analysis of Nox Family
4.3. Isolation of Total RNA and Semi-Quantitative RT-PCR Analysis
4.4. Real-Time qPCR Analysis
5. Conclusions
Supplementary Information
ijms-14-09440-s001.pdfAcknowledgments
Conflict of Interest
References
- Foreman, J.; Demidchik, V.; Bothwell, J.H.; Mylona, P.; Miedema, H.; Torres, M.A.; Linstead, P.; Costa, S.; Brownlee, C.; Jones, J.D.; et al. Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 2003, 422, 442–446. [Google Scholar]
- Sagi, M.; Fluhr, R. Production of reactive oxygen species by plant NADPH oxidases. Plant Physiol 2006, 141, 336–340. [Google Scholar]
- Bedard, K.; Lardy, B.; Krause, K.H. NOX family NADPH oxidases: Not just in mammals. Biochimie 2007, 89, 1107–1112. [Google Scholar]
- Geiszt, M. NADPH oxidases: New kids on the block. Cardiovasc. Res 2006, 71, 289–299. [Google Scholar]
- Del Rio, L.A.; Pastori, G.M.; Palma, J.M.; Sandalio, L.M.; Sevilla, F.; Corpas, F.J.; Jimenez, A.; Lopez-Huertas, E.; Hernandez, J.A. The activated oxygen role of peroxisomes in senescence. Plant Physiol 1998, 116, 1195–1200. [Google Scholar]
- Lamb, C.; Dixon, R.A. The oxidative burst in plant disease resistance. Annu. Rev. Plant Physiol. Plant Mol. Biol 1997, 48, 251–275. [Google Scholar]
- Pei, Z.M.; Murata, Y.; Benning, G.; Thomine, S.; Klusener, B.; Allen, G.J.; Grill, E.; Schroeder, J.I. Calcium channels activated by hydrogen peroxide mediate abscisic acid signalling in guard cells. Nature 2000, 406, 731–734. [Google Scholar]
- Potikha, T.S.; Collins, C.C.; Johnson, D.I.; Delmer, D.P.; Levine, A. The involvement of hydrogen peroxide in the differentiation of secondary walls in cotton fibers. Plant Physiol 1999, 119, 849–858. [Google Scholar]
- Jiang, M.; Zhang, J. Involvement of plasma-membrane NADPH oxidase in abscisic acid- and water stress-induced antioxidant defense in leaves of maize seedlings. Planta 2002, 215, 1022–1030. [Google Scholar]
- Laloi, C.; Apel, K.; Danon, A. Reactive oxygen signalling: The latest news. Curr. Opin. Plant Biol 2004, 7, 323–328. [Google Scholar]
- Overmyer, K.; Brosche, M.; Kangasjarvi, J. Reactive oxygen species and hormonal control of cell death. Trends Plant Sci 2003, 8, 335–342. [Google Scholar]
- Hao, F.; Wang, X.; Chen, J. Involvement of plasma-membrane NADPH oxidase in nickel-induced oxidative stress in roots of wheat seedlings. Plant Sci 2006, 170, 151–158. [Google Scholar]
- Torres, M.A.; Dangl, J.L.; Jones, J.D. Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc. Natl. Acad. Sci. USA 2002, 99, 517–522. [Google Scholar]
- Yoshioka, H.; Mase, K.; Yoshioka, M.; Kobayashi, M.; Asai, S. Regulatory mechanisms of nitric oxide and reactive oxygen species generation and their role in plant immunity. Nitric Oxide 2011, 25, 216–221. [Google Scholar]
- Liu, P.; Li, R.-L.; Zhang, L.; Wang, Q.-L.; Niehaus, K.; Baluška, F.; Šamaj, J.; Lin, J.-X. Lipid microdomain polarization is required for NADPH oxidase-dependent ROS signaling in Picea meyeri pollen tube tip growth. Plant J 2009, 60, 303–313. [Google Scholar]
- Monshausen, G.B.; Bibikova, T.N.; Weisenseel, M.H.; Gilroy, S. Ca2+ regulates reactive oxygen species production and pH during mechanosensing in Arabidopsis roots. Plant Cell 2009, 21, 2341–2356. [Google Scholar]
- Potocky, M.; Jones, M.A.; Bezvoda, R.; Smirnoff, N.; Zarsky, V. Reactive oxygen species produced by NADPH oxidase are involved in pollen tube growth. New Phytol 2007, 174, 742–751. [Google Scholar]
- Shin, L.-J.; Huang, H.-E.; Chang, H.; Lin, Y.-H.; Feng, T.-Y.; Ger, M.-J. Ectopic ferredoxin I protein promotes root hair growth through induction of reactive oxygen species in Arabidopsis thaliana. J. Plant Physiol 2011, 168, 434–440. [Google Scholar]
- Shi, Y.C.; Fu, Y.P.; Liu, W.Q. NADPH oxidase in plasma membrane is involved in stomatal closure induced by dehydroascorbate. Plant Physiol. Biochem 2012, 51, 26–30. [Google Scholar]
- Zhang, Y.; Zhu, H.; Zhang, Q.; Li, M.; Yan, M.; Wang, R.; Wang, L.; Welti, R.; Zhang, W.; Wang, X. Phospholipase dalpha1 and phosphatidic acid regulate NADPH oxidase activity and production of reactive oxygen species in ABA-mediated stomatal closure in Arabidopsis. Plant Cell 2009, 21, 2357–2377. [Google Scholar]
- Steffens, B.; Sauter, M. Epidermal cell death in rice is confined to cells with a distinct molecular identity and is mediated by ethylene and H2O2 through an autoamplified signal pathway. Plant Cell 2009, 21, 184–196. [Google Scholar]
- Tewari, R.K.; Watanabe, D.; Watanabe, M. Chloroplastic NADPH oxidase-like activity-mediated perpetual hydrogen peroxide generation in the chloroplast induces apoptotic-like death of Brassica napus leaf protoplasts. Planta 2012, 235, 99–110. [Google Scholar]
- Cano-Dominguez, N.; Alvarez-Delfin, K.; Hansberg, W.; Aguirre, J. NADPH oxidases NOX-1 and NOX-2 require the regulatory subunit NOR-1 to control cell differentiation and growth in Neurospora crassa. Eukaryot. Cell 2008, 7, 1352–1361. [Google Scholar]
- Asai, S.; Ohta, K.; Yoshioka, H. MAPK signaling regulates nitric oxide and NADPH oxidase-dependent oxidative bursts in Nicotiana benthamiana. Plant Cell 2008, 20, 1390–1406. [Google Scholar]
- Asano, T.; Hayashi, N.; Kobayashi, M.; Aoki, N.; Miyao, A.; Mitsuhara, I.; Ichikawa, H.; Komatsu, S.; Hirochika, H.; Kikuchi, S.; et al. A rice calcium-dependent protein kinase OsCPK12 oppositely modulates salt-stress tolerance and blast disease resistance. Plant J 2012, 69, 26–36. [Google Scholar]
- Kobayashi, M.; Ohura, I.; Kawakita, K.; Yokota, N.; Fujiwara, M.; Shimamoto, K.; Doke, N.; Yoshioka, H. Calcium-dependent protein kinases regulate the production of reactive oxygen species by potato NADPH oxidase. Plant Cell 2007, 19, 1065–1080. [Google Scholar]
- Nakashima, A.; Chen, L.; Thao, N.P.; Fujiwara, M.; Wong, H.L.; Kuwano, M.; Umemura, K.; Shirasu, K.; Kawasaki, T.; Shimamoto, K. RACK1 functions in rice innate immunity by interacting with the Rac1 immune complex. Plant Cell 2008, 20, 2265–2279. [Google Scholar]
- Kaye, Y.; Golani, Y.; Singer, Y.; Leshem, Y.; Cohen, G.; Ercetin, M.; Gillaspy, G.; Levine, A. Inositol polyphosphate 5-phosphatase7 regulates the production of reactive oxygen species and salt tolerance in Arabidopsis. Plant Physiol 2011, 157, 229–241. [Google Scholar]
- Evans, N.H.; McAinsh, M.R.; Hetherington, A.M.; Knight, M.R. ROS perception in Arabidopsis thaliana: The ozone-induced calcium response. Plant J 2005, 41, 615–626. [Google Scholar]
- Delledonne, M.; Murgia, I.; Ederle, D.; Sbicego, P.F.; Biondani, A.; Polverari, A.; Lamb, C. Reactive oxygen intermediates modulate nitric oxide signaling in the plant hypersensitive disease-resistance response. Plant Physiol. Biochem 2002, 40, 605–610. [Google Scholar]
- Li, J.; Wang, X.; Zhang, Y.; Jia, H.; Bi, Y. CGMP regulates hydrogen peroxide accumulation in calcium-dependent salt resistance pathway in Arabidopsis thaliana roots. Planta 2011, 234, 709–722. [Google Scholar]
- Song, C.J.; Steinebrunner, I.; Wang, X.; Stout, S.C.; Roux, S.J. Extracellular ATP induces the accumulation of superoxide via NADPH oxidases in Arabidopsis. Plant Physiol 2006, 140, 1222–1232. [Google Scholar]
- Marino, D.; Dunand, C.; Puppo, A.; Pauly, N. A burst of plant NADPH oxidases. Trends Plant Sci 2012, 17, 9–15. [Google Scholar]
- Groom, Q.J.; Torres, M.A.; Fordham-Skelton, A.P.; Hammond-Kosack, K.E.; Robinson, N.J.; Jones, J.D. RbohA, a rice homologue of the mammalian gp91phox respiratory burst oxidase gene. Plant J 1996, 10, 515–522. [Google Scholar]
- Wong, H.L.; Pinontoan, R.; Hayashi, K.; Tabata, R.; Yaeno, T.; Hasegawa, K.; Kojima, C.; Yoshioka, H.; Iba, K.; Kawasaki, T.; et al. Regulation of rice NADPH oxidase by binding of Rac GTPase to its N-terminal extension. Plant Cell 2007, 19, 4022–4034. [Google Scholar]
- Yoshie, Y.; Goto, K.; Takai, R.; Iwano, M.; Takayama, S.; Isogai, A.; Che, F.-S. Function of the rice gp91phox homologs OsrbohA and OsrbohE genes in ROS-dependent plant immune responses. Plant Biotechnol 2005, 22, 127–135. [Google Scholar]
- Marchler-Bauer, A.; Zheng, C.; Chitsaz, F.; Derbyshire, M.K.; Geer, L.Y.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Lanczycki, C.J.; et al. CDD: Conserved domains and protein three-dimensional structure. Nucleic Acids Res 2013, 41, D348–D352. [Google Scholar]
- Duan, Z.-Q.; Bai, L.; Zhao, Z.-G.; Zhang, G.-P.; Cheng, F.-M.; Jiang, L.-X.; Chen, K.-M. Drought-stimulated activity of plasma membrane nicotinamide adenine dinucleotide phosphate oxidase and its catalytic properties in rice. J. Integr. Plant Biol 2009, 51, 1104–1115. [Google Scholar]
- Jiang, M.Y.; Zhang, J.H. Cross-talk between calcium and reactive oxygen species originated from NADPH oxidase in abscisic acid-induced antioxidant defence in leaves of maize seedlings. Plant Cell Environ 2003, 26, 929–939. [Google Scholar]
- Takeda, S.; Gapper, C.; Kaya, H.; Bell, E.; Kuchitsu, K.; Dolan, L. Local positive feedback regulation determines cell shape in root hair cells. Science 2008, 319, 1241–1244. [Google Scholar]
- Muller, K.; Carstens, A.C.; Linkies, A.; Torres, M.A.; Leubner-Metzger, G. The NADPH-oxidase AtrbohB plays a role in Arabidopsis seed after-ripening. New Phytol 2009, 184, 885–897. [Google Scholar]
- Kimura, S.; Kaya, H.; Kawarazaki, T.; Hiraoka, G.; Senzaki, E.; Michikawa, M.; Kuchitsu, K. Protein phosphorylation is a prerequisite for the Ca2+-dependent activation of Arabidopsis NADPH oxidases and may function as a trigger for the positive feedback regulation of Ca2+ and reactive oxygen species. Biochim. Biophys. Acta 2012, 1823, 398–405. [Google Scholar]
- Lin, F.; Ding, H.; Wang, J.; Zhang, H.; Zhang, A.; Zhang, Y.; Tan, M.; Dong, W.; Jiang, M. Positive feedback regulation of maize NADPH oxidase by mitogen-activated protein kinase cascade in abscisic acid signalling. J. Exp. Bot 2009, 60, 3221–3238. [Google Scholar]
- Gross, J.S.; Stein, R.J.; Fett-Neto, A.G.; Fett, J.P. Iron homeostasis related genes in rice. Genet. Mol. Biol 2003, 4, 21. [Google Scholar]
- Ishimaru, Y.; Suzuki, M.; Tsukamoto, T.; Suzuki, K.; Nakazono, M.; Kobayashi, T.; Wada, Y.; Watanabe, S.; Matsuhashi, S.; Takahashi, M.; et al. Rice plants take up iron as an Fe3+-phytosiderophore and as Fe2+. Plant J 2006, 45, 335–346. [Google Scholar]
- Sperotto, R.A.; Boff, T.; Duarte, G.L.; Santos, L.S.; Grusak, M.A.; Fett, J.P. Identification of putative target genes to manipulate Fe and Zn concentrations in rice grains. J. Plant Physiol 2010, 167, 1500–1506. [Google Scholar]
- Schultz, J.; Milpetz, F.; Bork, P.; Ponting, C.P. SMART, a simple modular architecture research tool: Identification of signaling domains. Proc. Natl. Acad. Sci. USA 1998, 95, 5857–5864. [Google Scholar]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol 2010, 59, 307–321. [Google Scholar]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci 1992, 8, 275–282. [Google Scholar]
- Tamura, K.; Dudley, J.; Nei, M.; Kumar, S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol 2007, 24, 1596–1599. [Google Scholar]
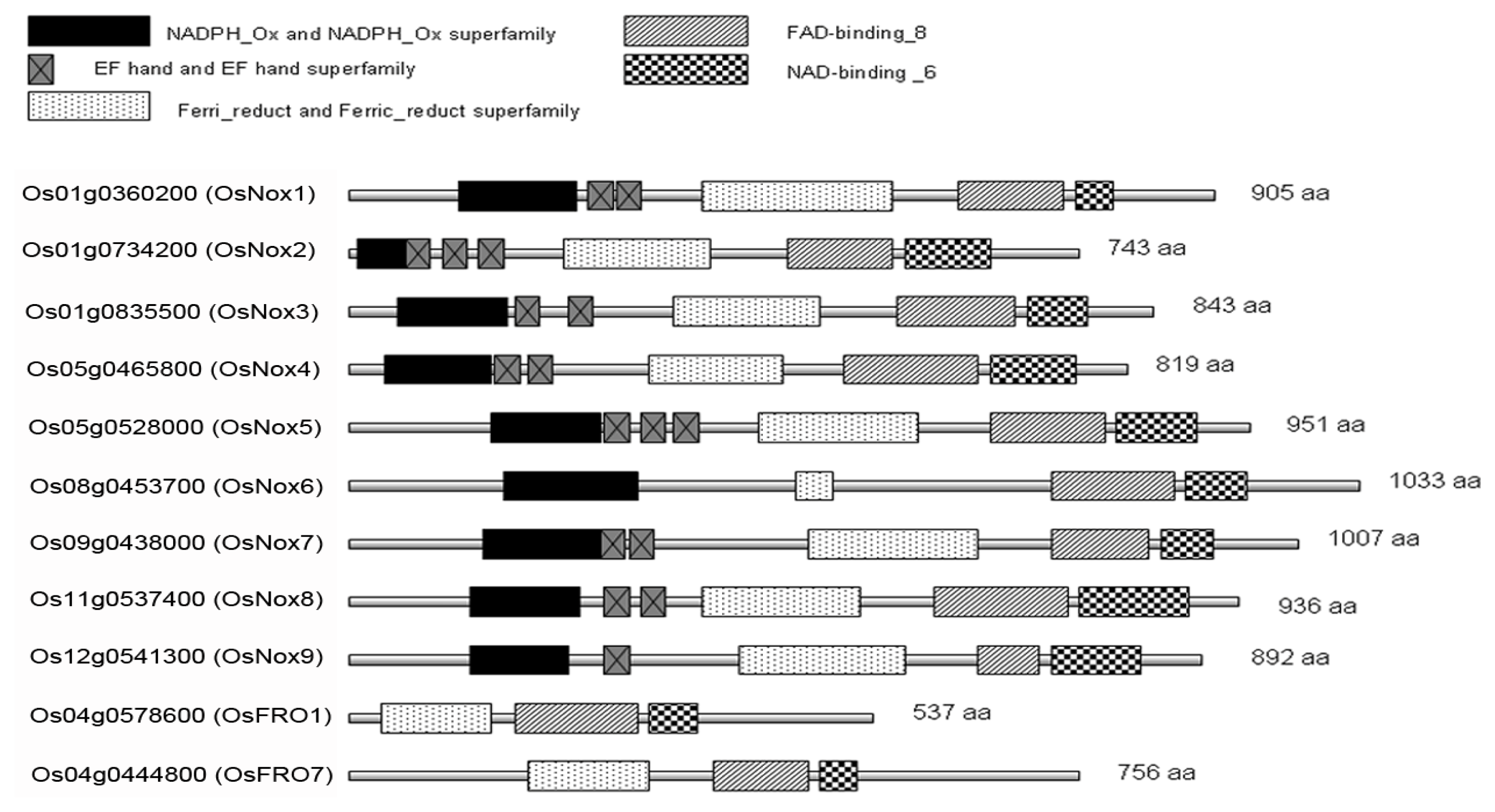


| Gene name | Other names | Accession numbers | Gene locus | Protein size (predicted, aa) | Molecular weight (predicted, kD) | Sources | |
|---|---|---|---|---|---|---|---|
| Os ID | MSU’s LOC_Os ID | ||||||
| OsNox1 | OsRbohB | AY603975 | Os01g0360200 | LOC_Os01g25820 | 905 | 101.759 | http://www.uniprot.org/uniprot/Q5ZAJ0 |
| OsNox2 | OsRbohA | NP_001044165.1 | Os01g0734200 | LOC_Os01g53294 | 745 | 85.336 | http://www.uniprot.org/uniprot/O48539 |
| OsNox3 | OsRbohE | AK100241 | Os01g0835500 | LOC_Os01g61880 | 843 | 94.79 | http://www.uniprot.org/uniprot/Q8S1T0 |
| OsNox4 | OsRbohD | AK072353 | Os05g0465800 | LOC_Os05g38980 | 819 | 92.35 | http://www.uniprot.org/uniprot/Q0DHH6 |
| OsNox5 | OsRbohC | AK120905 | Os05g0528000 | LOC_Os05g45210 | 951 | 107.171 | http://www.uniprot.org/uniprot/Q65XC8 |
| OsNox6 | RbohE | NP_001061956.1 | Os08g0453700 | LOC_Os08g35210 | 1033 | 115.014 | http://www.uniprot.org/uniprot/Q0J595 |
| OsNox7 | OsRbohG/OsRbohB | NP_001063267.1 | Os09g0438000 | LOC_Os09g26660 | 1007 | 112.134 | http://www.uniprot.org/uniprot/Q69LJ7 |
| OsNox8 | OsRbohI | AK063113 | Os11g0537400 | LOC_Os11g33120 | 936 | 72.025 | http://rapdblegacy.dna.affrc.go.jp/viewer/gbrowse_details/build5?name=Os11g0537400 |
| OsNox9 | OsRbohH | J075145A22 | Os12g0541300 | LOC_Os12g35610 | 892 | 99.893 | http://rice.plantbiology.msu.edu/cgi-bin/ORF_infopage.cgi?orf=LOC_Os12g35610.1 |
| OsFRO1 | AB126085 | Os04g0578600 | LOC_Os04g48930 | 537 | 58.095 | http://www.uniprot.org/uniprot/Q0JAT2 | |
| OsFRO7 | AK067009 | Os04g0444800 | LOC_Os04g36720 | 756 | 83.156 | http://www.uniprot.org/uniprot/Q0JCX7 | |
© 2013 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Wang, G.-F.; Li, W.-Q.; Li, W.-Y.; Wu, G.-L.; Zhou, C.-Y.; Chen, K.-M. Characterization of Rice NADPH Oxidase Genes and Their Expression under Various Environmental Conditions. Int. J. Mol. Sci. 2013, 14, 9440-9458. https://doi.org/10.3390/ijms14059440
Wang G-F, Li W-Q, Li W-Y, Wu G-L, Zhou C-Y, Chen K-M. Characterization of Rice NADPH Oxidase Genes and Their Expression under Various Environmental Conditions. International Journal of Molecular Sciences. 2013; 14(5):9440-9458. https://doi.org/10.3390/ijms14059440
Chicago/Turabian StyleWang, Gang-Feng, Wen-Qiang Li, Wen-Yan Li, Guo-Li Wu, Cong-Yi Zhou, and Kun-Ming Chen. 2013. "Characterization of Rice NADPH Oxidase Genes and Their Expression under Various Environmental Conditions" International Journal of Molecular Sciences 14, no. 5: 9440-9458. https://doi.org/10.3390/ijms14059440
APA StyleWang, G.-F., Li, W.-Q., Li, W.-Y., Wu, G.-L., Zhou, C.-Y., & Chen, K.-M. (2013). Characterization of Rice NADPH Oxidase Genes and Their Expression under Various Environmental Conditions. International Journal of Molecular Sciences, 14(5), 9440-9458. https://doi.org/10.3390/ijms14059440





