The Tumor Suppressor Roles of miR-433 and miR-127 in Gastric Cancer
Abstract
:1. Introduction
2. Results and Discussion
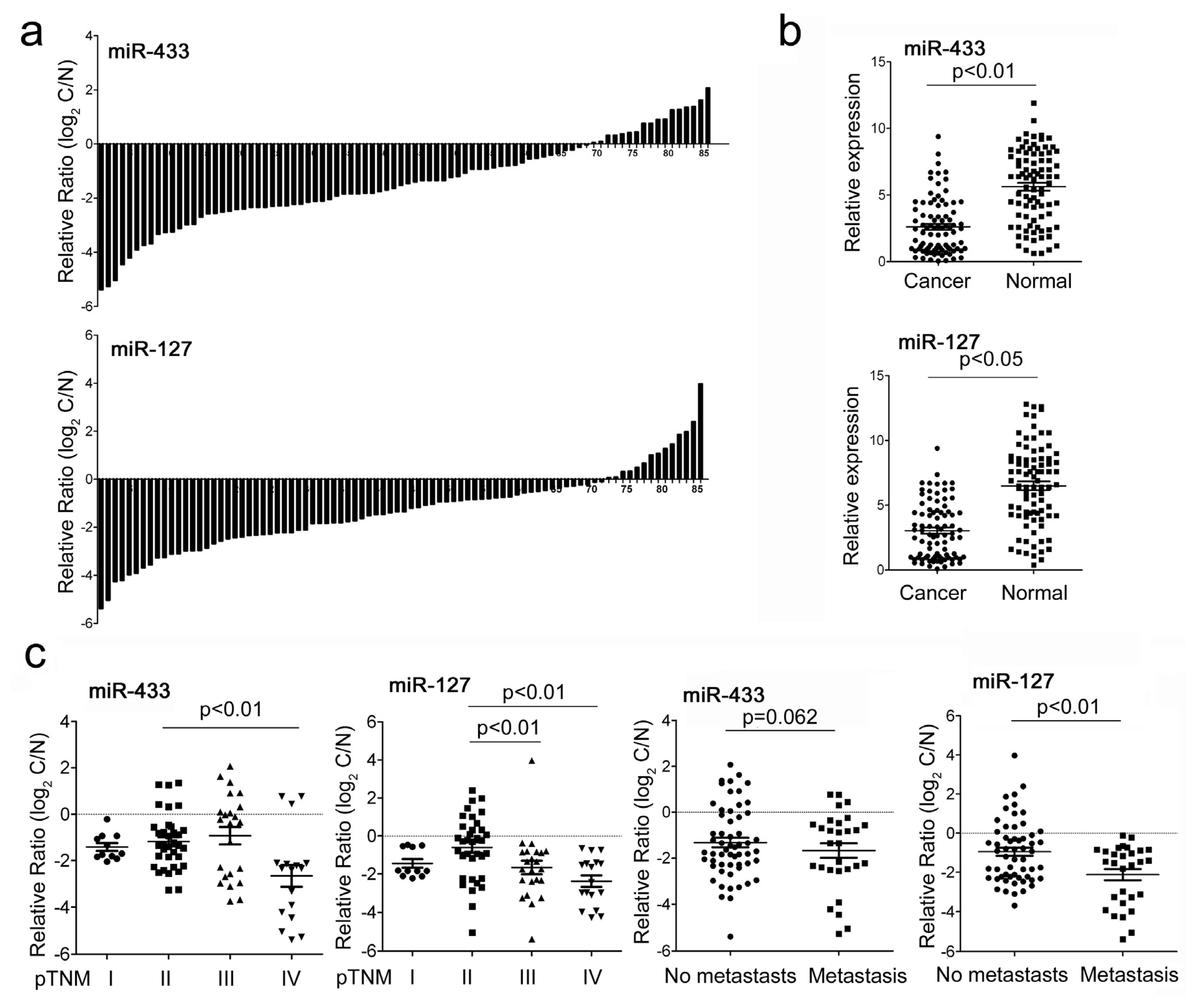
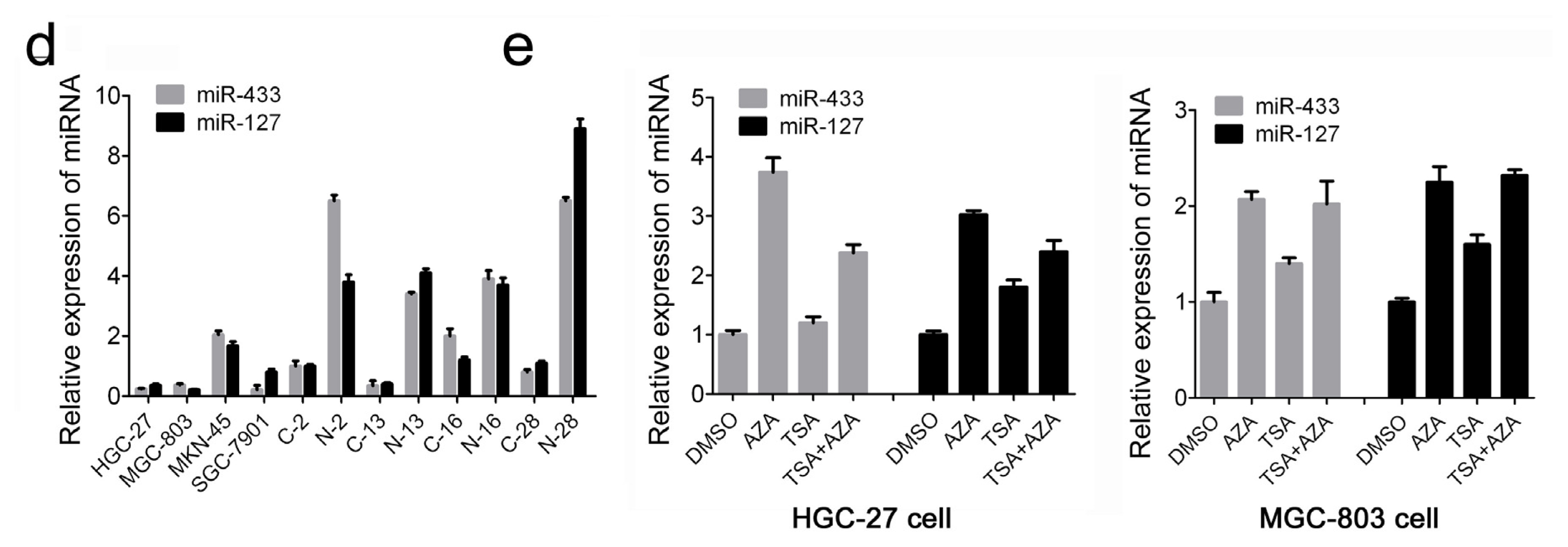
2.1. MiR-433 and miR-127 Were Down-Regulated in GC Patients and GC Cell Lines
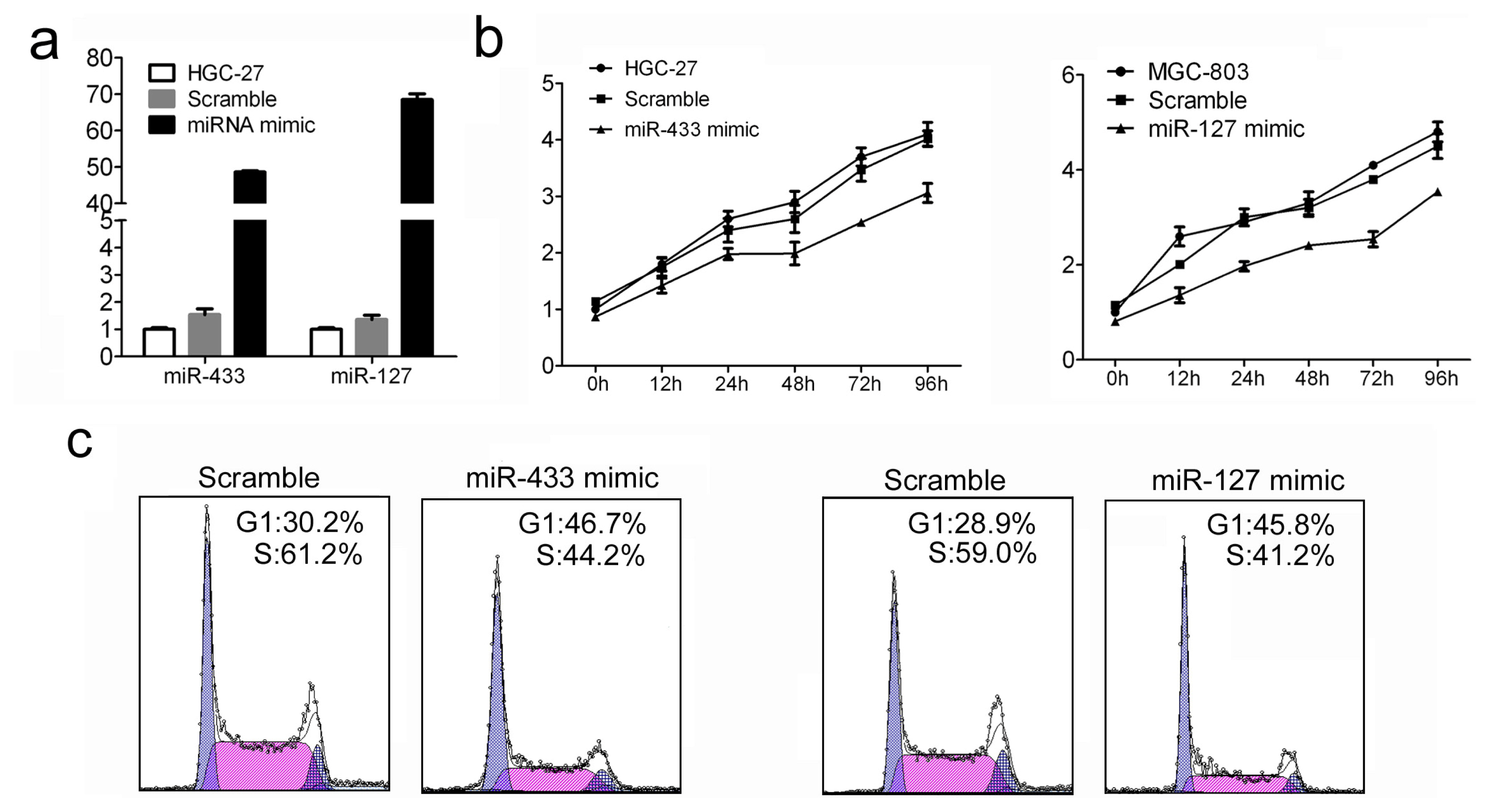
2.2. Increased miR-433 and miR-127 Inhibits Cell Proliferation and Cell Cycle Progression of GC Cells
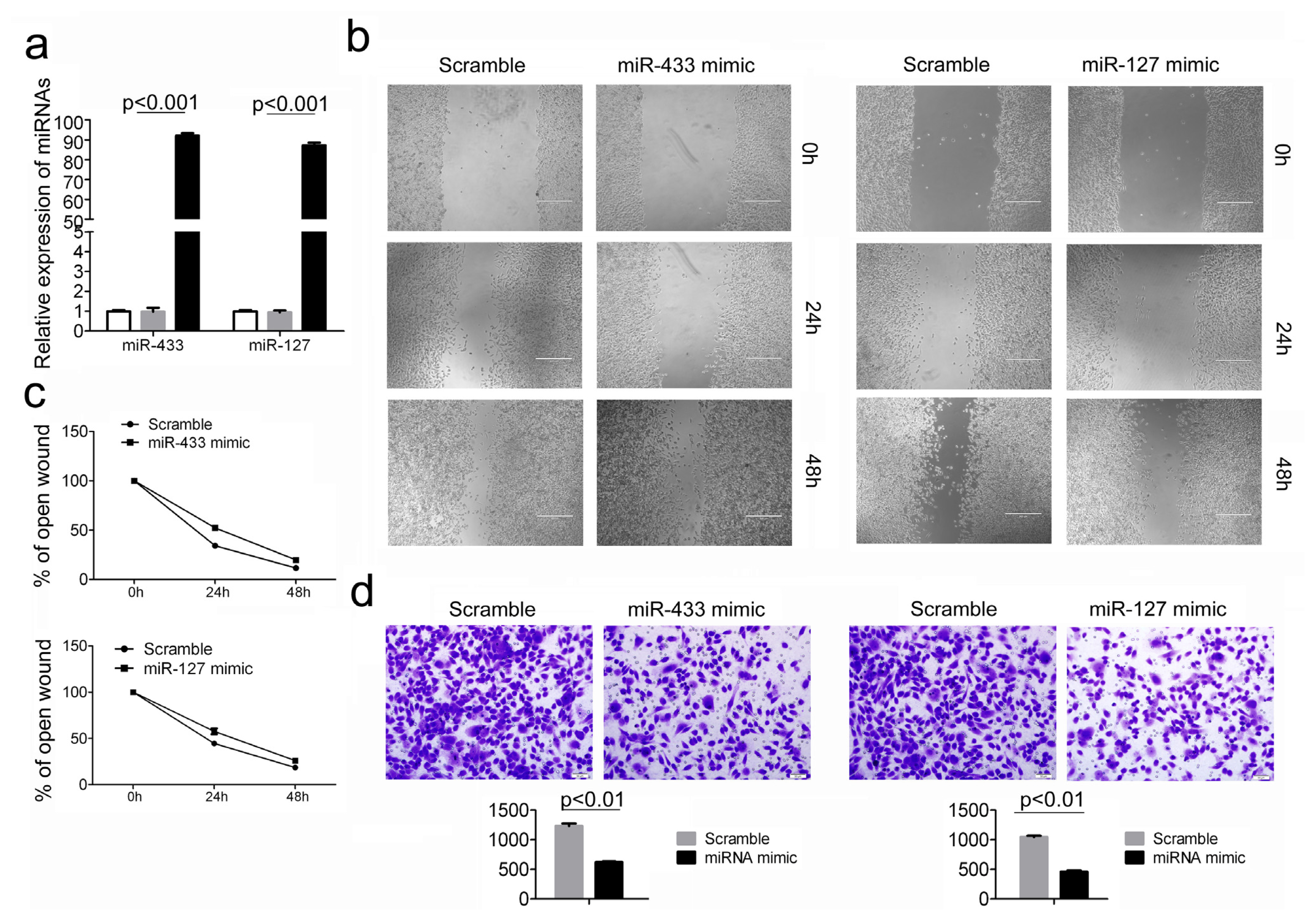
2.3. Over-Expression of miR-433 and miR-127 in GC Cells Inhibits Cell Migration and Invasion
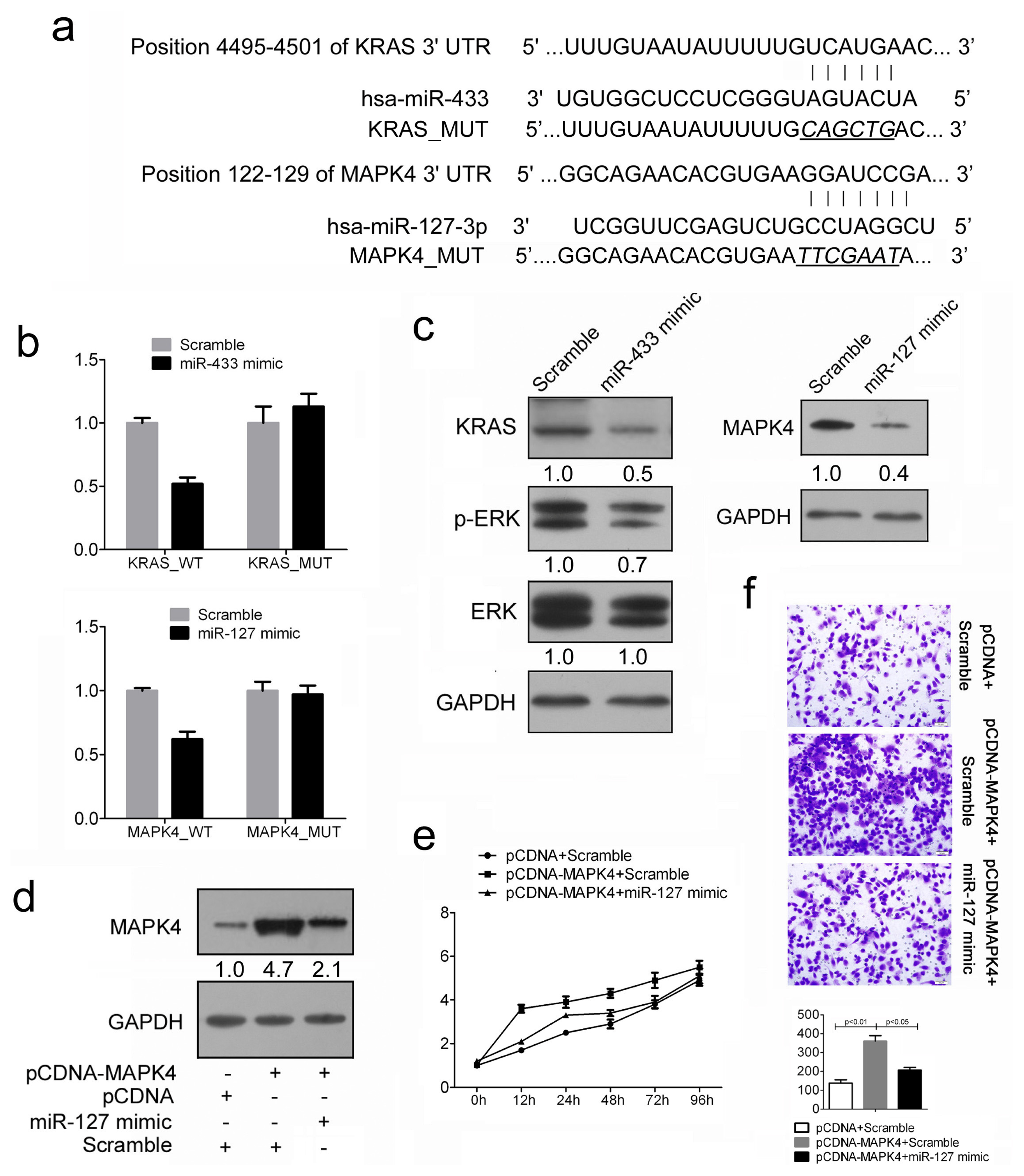
2.4. Restored miR-433 Expression Dampens KRAS Pathway in GC Cells
2.5. MiR-127 Suppresses the Expression of MAPK4 in GC Cells
2.6. Discussion
3. Experimental Section
3.1. Cell Cultures and Transfection
3.2. Tissue Specimens
3.3. TaqMan RT-PCR for miRNA Expression
3.4. 5-Aza-CdR and Trichostatin a Treatment of Cell Lines
3.5. Cell Proliferation and Cell Cycle Assay
3.6. Cell Migration and Invasion Assays
3.7. Protein Isolation and Western Blotting
3.8. Rescue Assays of MAPK4 Gene Expression
3.9. Statistics
4. Conclusions
Acknowledgments
Conflict of Interest
References
- Ezzati, M.; Henley, S.J.; Lopez, A.D.; Thun, M.J. Role of smoking in global and regional cancer epidemiology: Current patterns and data needs. Int. J. Cancer 2005, 116, 963–971. [Google Scholar]
- Calin, G.A.; Croce, C.M. MicroRNA signatures in human cancers. Nat. Rev. Cancer 2006, 6, 857–866. [Google Scholar]
- Kong, Y.W.; Ferland-McCollough, D.; Jackson, T.J.; Bushell, M. MicroRNAs in cancer management. Lancet Oncol 2012, 13, e249–e258. [Google Scholar]
- Gao, W.; He, H.W.; Wang, Z.M.; Zhao, H.; Lian, X.Q.; Wang, Y.S.; Zhu, J.; Yan, J.J.; Zhang, D.G.; Yang, Z.J.; et al. Plasma levels of lipometabolism-related miR-122 and miR-370 are increased in patients with hyperlipidemia and associated with coronary artery disease. Lipids Health Dis. 2012, 11. [Google Scholar] [CrossRef]
- Fernandez-Hernando, C.; Moore, K.J. MicroRNA modulation of cholesterol homeostasis. Arterioscler. Thromb. Vasc. Biol 2011, 31, 2378–2382. [Google Scholar]
- El Abiad, R.; Gerke, H. Gastric cancer: Endoscopic diagnosis and staging. Surg. Oncol. Clin. N. Am 2012, 21, 1–19. [Google Scholar]
- Rayner, K.J.; Esau, C.C.; Hussain, F.N.; McDaniel, A.L.; Marshall, S.M.; van Gils, J.M.; Ray, T.D.; Sheedy, F.J.; Goedeke, L.; Liu, X.; et al. Inhibition of miR-33a/b in non-human primates raises plasma HDL and lowers VLDL triglycerides. Nature 2011, 478, 404–407. [Google Scholar]
- Song, G.; Wang, L. A conserved gene structure and expression regulation of miR-433 and miR-127 in mammals. PLoS One 2009, 4, e7829. [Google Scholar]
- Yang, Z.; Zhang, Y.; Wang, L. A feedback inhibition between miRNA-127 and TGFβ/c-Jun cascade in HCC cell migration via MMP13. PLoS One 2013, 8, e65256. [Google Scholar]
- Luo, H.; Zhang, H.; Zhang, Z.; Zhang, X.; Ning, B.; Guo, J.; Nie, N.; Liu, B.; Wu, X. Down-regulated miR-9 and miR-433 in human gastric carcinoma. J. Exp. Clin. Cancer Res. 2009, 28. [Google Scholar] [CrossRef]
- Ueda, T.; Volinia, S.; Okumura, H.; Shimizu, M.; Taccioli, C.; Rossi, S.; Alder, H.; Liu, C.G.; Oue, N.; Yasui, W.; et al. Relation between microRNA expression and progression and prognosis of gastric cancer: A microRNA expression analysis. Lancet Oncol 2010, 11, 136–146. [Google Scholar]
- Lin, X.; Rice, K.L.; Buzzai, M.; Hexner, E.; Costa, F.F.; Kilpivaara, O.; Mullally, A.; Soares, M.B.; Ebert, B.L.; Levine, R.; et al. MiR-433 is aberrantly expressed in myeloproliferative neoplasms and suppresses hematopoietic cell growth and differentiation. Leukemia 2013, 27, 344–352. [Google Scholar]
- Tsai, K.W.; Wu, C.W.; Hu, L.Y.; Li, S.C.; Liao, Y.L.; Lai, C.H.; Kao, H.W.; Fang, W.L.; Huang, K.H.; Chan, W.C.; et al. Epigenetic regulation of miR-34b and miR-129 expression in gastric cancer. Int. J. Cancer 2011, 129, 2600–2610. [Google Scholar]
- Lukaszewicz-Zając, M.; Mroczko, B.; Szmitkowski, M. Gastric cancer—The role of matrix metalloproteinases in tumor progression. Clin. Chim. Acta 2011, 412, 1725–1730. [Google Scholar]
- Gurzu, S.; Szentirmay, Z.; Bara, T.; Turcu, M.; Toth, E.; Bara, T., Jr; Jung, I. Non-Epstein-Barr virus associated lymphoepithelioma-like carcinoma of the esophagogastric junction with microsatellite instability, K-ras wild type. Pathol. Res. Pract. 2013, 209, 128–131. [Google Scholar]
- Parikh, N.; Shuck, R.L.; Nguyen, T.A.; Herron, A.; Donehower, L.A. Mouse tissues that undergo neoplastic progression after K-Ras activation are distinguished by nuclear translocation of phospho-Erk1/2 and robust tumor suppressor responses. Mol. Cancer Res 2012, 10, 845–855. [Google Scholar]
- Betge, J.; Pollheimer, M.J.; Schlemmer, A.; Hoefler, G.; Langner, C. Gastric cancer and concomitant renal cancer: A systematic immunohistochemical and molecular analysis. Oncol. Rep 2011, 26, 567–575. [Google Scholar]
- Nana-Sinkam, S.P.; Croce, C.M. Clinical applications for microRNAs in cancer. Clin. Pharmacol. Ther 2013, 93, 98–104. [Google Scholar]
- Leonardo, T.R.; Schultheisz, H.L.; Loring, J.F.; Laurent, L.C. The functions of microRNAs in pluripotency and reprogramming. Nat. Cell Biol 2012, 14, 1114–1121. [Google Scholar]
- Van Rooij, E.; Olson, E.N. MicroRNA therapeutics for cardiovascular disease: Opportunities and obstacles. Nat. Rev. Drug Discov 2012, 11, 860–872. [Google Scholar]
- Cimmino, A.; Calin, G.A.; Fabbri, M.; Iorio, M.V.; Ferracin, M.; Shimizu, M.; Wojcik, S.E.; Aqeilan, R.I.; Zupo, S.; Dono, M.; et al. MiR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl. Acad. Sci. USA 2005, 102, 13944–13949. [Google Scholar]
- Kang, H.W.; Crawford, M.; Fabbri, M.; Nuovo, G.; Garofalo, M.; Nana-Sinkam, S.P.; Friedman, A. A mathematical model for microRNA in lung cancer. PLoS One 2013, 8, e53663. [Google Scholar]
- Fowler, A.; Thomson, D.; Giles, K.; Maleki, S.; Mreich, E.; Wheeler, H.; Leedman, P.; Biggs, M.; Cook, R.; Little, N.; et al. MiR-124a is frequently down-regulated in glioblastoma and is involved in migration and invasion. Eur. J. Cancer 2011, 47, 953–963. [Google Scholar]
- Matsubara, A.; Sekine, S.; Kushima, R.; Ogawa, R.; Taniguchi, H.; Tsuda, H.; Kanai, Y. Frequent GNAS and KRAS mutations in pyloric gland adenoma of the stomach and duodenum. J. Pathol 2013, 229, 579–587. [Google Scholar]
- Omar, M.F.; Ito, K.; Nga, M.E.; Soo, R.; Peh, B.K.; Ismail, T.M.; Thakkar, B.; Soong, R.; Ito, Y.; Salto-Tellez, M. RUNX3 downregulation in human lung adenocarcinoma is independent of p53, EGFR or KRASstatus. Pathol. Oncol. Res 2012, 18, 783–792. [Google Scholar]
- Fujimori, Y.; Inokuchi, M.; Takagi, Y.; Kato, K.; Kojima, K.; Sugihara, K. Prognostic value of RKIP and p-ERK in gastric cancer. J. Exp. Clin. Cancer Res 2012, 31, 30. [Google Scholar]
- Park, E.; Park, J.; Han, S.W.; Im, S.A.; Kim, T.Y.; Oh, D.Y.; Bang, Y.J. NVP-BKM120, a novel PI3K inhibitor, shows synergism with a STAT3 inhibitor in human gastric cancer cells harboring KRAS mutations. Int. J. Oncol 2012, 40, 1259–1266. [Google Scholar]
- Song, G.; Wang, L. Transcriptional mechanism for the paired miR-433 and miR-127 genes by nuclear receptors SHP and ERRgamma. Nucleic Acids Res 2008, 36, 5727–5735. [Google Scholar]
- Song, G.; Wang, L. MiR-433 and miR-127 arise from independent overlapping primary transcripts encoded by the miR-433-127 locus. PLoS One 2008, 3, e3574. [Google Scholar]
© 2013 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Guo, L.-H.; Li, H.; Wang, F.; Yu, J.; He, J.-S. The Tumor Suppressor Roles of miR-433 and miR-127 in Gastric Cancer. Int. J. Mol. Sci. 2013, 14, 14171-14184. https://doi.org/10.3390/ijms140714171
Guo L-H, Li H, Wang F, Yu J, He J-S. The Tumor Suppressor Roles of miR-433 and miR-127 in Gastric Cancer. International Journal of Molecular Sciences. 2013; 14(7):14171-14184. https://doi.org/10.3390/ijms140714171
Chicago/Turabian StyleGuo, Li-Hua, Hui Li, Fang Wang, Jia Yu, and Jin-Sheng He. 2013. "The Tumor Suppressor Roles of miR-433 and miR-127 in Gastric Cancer" International Journal of Molecular Sciences 14, no. 7: 14171-14184. https://doi.org/10.3390/ijms140714171
APA StyleGuo, L. -H., Li, H., Wang, F., Yu, J., & He, J. -S. (2013). The Tumor Suppressor Roles of miR-433 and miR-127 in Gastric Cancer. International Journal of Molecular Sciences, 14(7), 14171-14184. https://doi.org/10.3390/ijms140714171




