Principles of miRNA-Target Regulation in Metazoan Models
Abstract
:1. Introduction
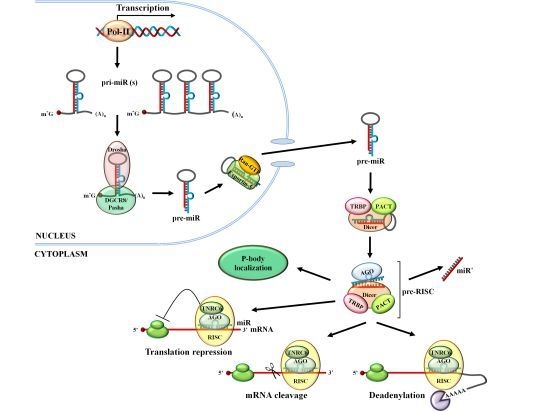
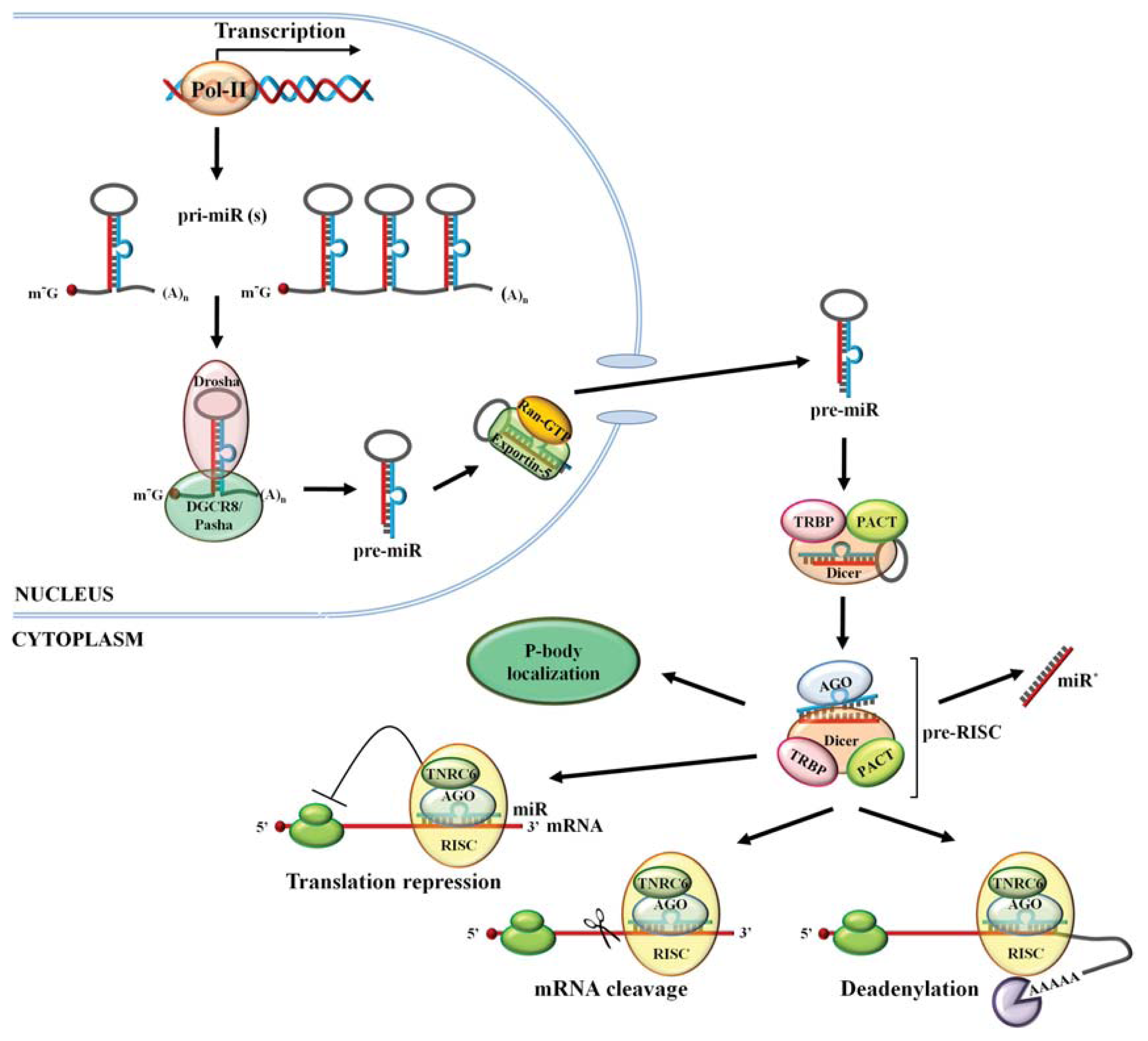
2. miR Biogenesis
3. Efficacy of miR Repression
3.1. The miR Nucleotide Sequence
3.2. Target Site Features
3.3. Target Accessibility and Polyadenylation
3.4. miR and Target RNA Levels
3.5. RNA Binding Proteins
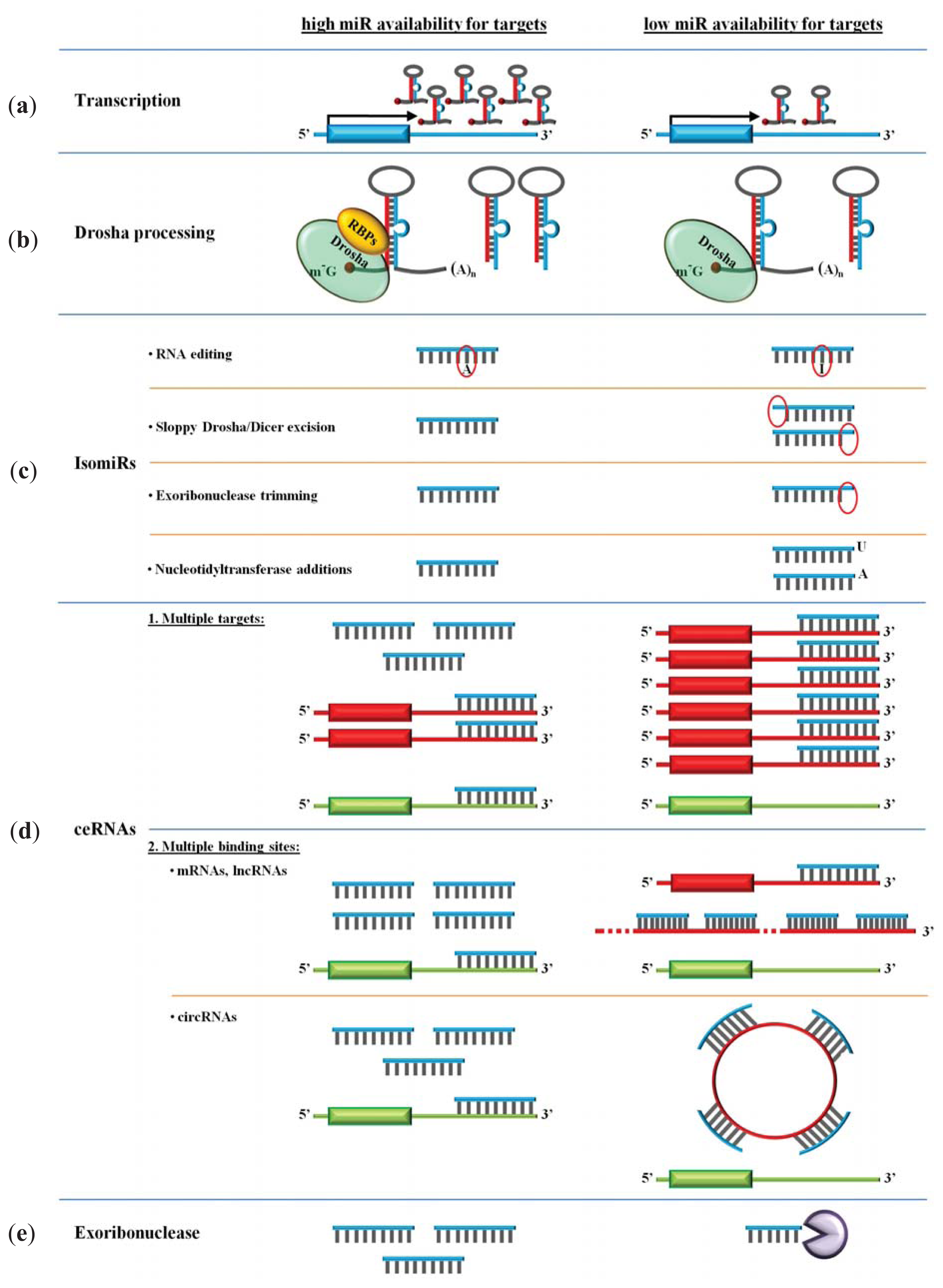
4. Availability of miRs
4.1. Transcription
4.2. Drosha Processing
4.3. miR Polymorphism and isomiRs
4.4. ceRNAs and miR Degrading Enzymes
5. Non-Canonical miR Activities
5.1. Promoter Activation
5.2. Target Activation
5.3. Receptors’ Ligands
6. Conclusions
Acknowledgments
Conflict of Interest
References
- Kozomara, A.; Griffiths-Jones, S. miRBase: Integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res 2011, 39, D152–D157. [Google Scholar]
- Chiang, H.R.; Schoenfeld, L.W.; Ruby, J.G.; Auyeung, V.C.; Spies, N.; Baek, D.; Johnston, W.K.; Russ, C.; Luo, S.; Babiarz, J.E.; et al. Mammalian microRNAs: Experimental evaluation of novel and previously annotated genes. Genes Dev 2010, 24, 992–1009. [Google Scholar]
- Meunier, J.; Lemoine, F.; Soumillon, M.; Liechti, A.; Weier, M.; Guschanski, K.; Hu, H.; Khaitovich, P.; Kaessmann, H. Birth and expression evolution of mammalian microRNA genes. Genome Res 2013, 23, 34–45. [Google Scholar]
- Friedman, R.C.; Farh, K.K.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 2009, 19, 92–105. [Google Scholar]
- Beuvink, I.; Kolb, F.A.; Budach, W.; Garnier, A.; Lange, J.; Natt, F.; Dengler, U.; Hall, J.; Filipowicz, W.; Weiler, J. A novel microarray approach reveals new tissue-specific signatures of known and predicted mammalian microRNAs. Nucleic Acids Res 2007, 35, e52. [Google Scholar]
- Landgraf, P.; Rusu, M.; Sheridan, R.; Sewer, A.; Iovino, N.; Aravin, A.; Pfeffer, S.; Rice, A.; Kamphorst, A.O.; Landthaler, M.; et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 2007, 129, 1401–1414. [Google Scholar]
- Liang, Y.; Ridzon, D.; Wong, L.; Chen, C. Characterization of microRNA expression profiles in normal human tissues. BMC Genomics 2007, 8, 166. [Google Scholar]
- Moreau, M.P.; Bruse, S.E.; Jornsten, R.; Liu, Y.; Brzustowicz, L.M. Chronological changes in microRNA expression in the developing human brain. PLoS One 2013, 8, e60480. [Google Scholar]
- Lu, J.; Getz, G.; Miska, E.A.; Alvarez-Saavedra, E.; Lamb, J.; Peck, D.; Sweet-Cordero, A.; Ebert, B.L.; Mak, R.H.; Ferrando, A.A.; et al. microRNA expression profiles classify human cancers. Nature 2005, 435, 834–838. [Google Scholar]
- Aldrich, B.T.; Frakes, E.P.; Kasuya, J.; Hammond, D.L.; Kitamoto, T. Changes in expression of sensory organ-specific microRNAs in rat dorsal root ganglia in association with mechanical hypersensitivity induced by spinal nerve ligation. Neuroscience 2009, 164, 711–723. [Google Scholar]
- Elmen, J.; Lindow, M.; Schutz, S.; Lawrence, M.; Petri, A.; Obad, S.; Lindholm, M.; Hedtjarn, M.; Hansen, H.F.; Berger, U.; et al. LNA-mediated microRNA silencing in non-human primates. Nature 2008, 452, 896–899. [Google Scholar]
- Elmen, J.; Lindow, M.; Silahtaroglu, A.; Bak, M.; Christensen, M.; Lind-Thomsen, A.; Hedtjarn, M.; Hansen, J.B.; Hansen, H.F.; Straarup, E.M.; et al. Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Res 2008, 36, 1153–1162. [Google Scholar]
- Krutzfeldt, J.; Rajewsky, N.; Braich, R.; Rajeev, K.G.; Tuschl, T.; Manoharan, M.; Stoffel, M. Silencing of microRNAs in vivo with “antagomirs”. Nature 2005, 438, 685–689. [Google Scholar]
- Corcoran, D.L.; Pandit, K.V.; Gordon, B.; Bhattacharjee, A.; Kaminski, N.; Benos, P.V. Features of mammalian microRNA promoters emerge from polymerase II chromatin immunoprecipitation data. PLoS One 2009, 4, e5279. [Google Scholar]
- Marson, A.; Levine, S.S.; Cole, M.F.; Frampton, G.M.; Brambrink, T.; Johnstone, S.; Guenther, M.G.; Johnston, W.K.; Wernig, M.; Newman, J.; et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell 2008, 134, 521–533. [Google Scholar]
- Ozsolak, F.; Poling, L.L.; Wang, Z.; Liu, H.; Liu, X.S.; Roeder, R.G.; Zhang, X.; Song, J.S.; Fisher, D.E. Chromatin structure analyses identify miRNA promoters. Genes Dev 2008, 22, 3172–3183. [Google Scholar]
- Saini, H.K.; Griffiths-Jones, S.; Enright, A.J. Genomic analysis of human microRNA transcripts. Proc. Natl. Acad. Sci. USA 2007, 104, 17719–17724. [Google Scholar]
- Wang, X.; Xuan, Z.; Zhao, X.; Li, Y.; Zhang, M.Q. High-resolution human core-promoter prediction with CoreBoost_HM. Genome Res 2009, 19, 266–275. [Google Scholar]
- Zhou, X.; Ruan, J.; Wang, G.; Zhang, W. Characterization and identification of microRNA core promoters in four model species. PLoS Comput. Biol 2007, 3, e37. [Google Scholar]
- Rodriguez, A.; Griffiths-Jones, S.; Ashurst, J.L.; Bradley, A. Identification of mammalian microRNA host genes and transcription units. Genome Res 2004, 14, 1902–1910. [Google Scholar]
- Han, J.; Lee, Y.; Yeom, K.H.; Nam, J.W.; Heo, I.; Rhee, J.K.; Sohn, S.Y.; Cho, Y.; Zhang, B.T.; Kim, V.N. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell 2006, 125, 887–901. [Google Scholar]
- Zeng, Y.; Cullen, B.R. Efficient processing of primary microRNA hairpins by Drosha requires flanking nonstructured RNA sequences. J. Biol. Chem 2005, 280, 27595–27603. [Google Scholar]
- Basyuk, E.; Suavet, F.; Doglio, A.; Bordonne, R.; Bertrand, E. Human let-7 stem-loop precursors harbor features of RNase III cleavage products. Nucleic Acids Res 2003, 31, 6593–6597. [Google Scholar]
- Denli, A.M.; Tops, B.B.; Plasterk, R.H.; Ketting, R.F.; Hannon, G.J. Processing of primary microRNAs by the Microprocessor complex. Nature 2004, 432, 231–235. [Google Scholar]
- Gregory, R.I.; Yan, K.P.; Amuthan, G.; Chendrimada, T.; Doratotaj, B.; Cooch, N.; Shiekhattar, R. The Microprocessor complex mediates the genesis of microRNAs. Nature 2004, 432, 235–240. [Google Scholar]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Radmark, O.; Kim, S.; et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar]
- Bernstein, E.; Caudy, A.A.; Hammond, S.M.; Hannon, G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 2001, 409, 363–366. [Google Scholar]
- Chakravarthy, S.; Sternberg, S.H.; Kellenberger, C.A.; Doudna, J.A. Substrate-specific kinetics of Dicer-catalyzed RNA processing. J. Mol. Biol 2010, 404, 392–402. [Google Scholar]
- Haase, A.D.; Jaskiewicz, L.; Zhang, H.; Laine, S.; Sack, R.; Gatignol, A.; Filipowicz, W. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep 2005, 6, 961–967. [Google Scholar]
- Hutvagner, G.; McLachlan, J.; Pasquinelli, A.E.; Balint, E.; Tuschl, T.; Zamore, P.D. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 2001, 293, 834–838. [Google Scholar]
- Lee, H.Y.; Zhou, K.; Smith, A.M.; Noland, C.L.; Doudna, J.A. Differential roles of human Dicer-binding proteins TRBP and PACT in small RNA processing. Nucleic Acids Res. 2013. [Google Scholar] [CrossRef]
- Lee, Y.; Hur, I.; Park, S.Y.; Kim, Y.K.; Suh, M.R.; Kim, V.N. The role of PACT in the RNA silencing pathway. EMBO J 2006, 25, 522–532. [Google Scholar]
- MacRae, I.J.; Zhou, K.; Doudna, J.A. Structural determinants of RNA recognition and cleavage by Dicer. Nat. Struct. Mol. Biol 2007, 14, 934–940. [Google Scholar]
- Chendrimada, T.P.; Gregory, R.I.; Kumaraswamy, E.; Norman, J.; Cooch, N.; Nishikura, K.; Shiekhattar, R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 2005, 436, 740–744. [Google Scholar]
- Khvorova, A.; Reynolds, A.; Jayasena, S.D. Functional siRNAs and miRNAs exhibit strand bias. Cell 2003, 115, 209–216. [Google Scholar]
- Krol, J.; Sobczak, K.; Wilczynska, U.; Drath, M.; Jasinska, A.; Kaczynska, D.; Krzyzosiak, W.J. Structural features of microRNA (miRNA) precursors and their relevance to miRNA biogenesis and small interfering RNA/short hairpin RNA design. J. Biol. Chem 2004, 279, 42230–42239. [Google Scholar]
- Schwarz, D.S.; Hutvagner, G.; Du, T.; Xu, Z.; Aronin, N.; Zamore, P.D. Asymmetry in the assembly of the RNAi enzyme complex. Cell 2003, 115, 199–208. [Google Scholar]
- Czech, B.; Zhou, R.; Erlich, Y.; Brennecke, J.; Binari, R.; Villalta, C.; Gordon, A.; Perrimon, N.; Hannon, G.J. Hierarchical rules for Argonaute loading in Drosophila. Mol. Cell 2009, 36, 445–456. [Google Scholar]
- Hu, H.Y.; Yan, Z.; Xu, Y.; Hu, H.; Menzel, C.; Zhou, Y.H.; Chen, W.; Khaitovich, P. Sequence features associated with microRNA strand selection in humans and flies. BMC Genomics 2009, 10, 413. [Google Scholar]
- Noland, C.L.; Doudna, J.A. Multiple sensors ensure guide strand selection in human RNAi pathways. RNA 2013, 19, 639–648. [Google Scholar]
- Okamura, K.; Liu, N.; Lai, E.C. Distinct mechanisms for microRNA strand selection by Drosophila Argonautes. Mol. Cell 2009, 36, 431–444. [Google Scholar]
- Ameres, S.L.; Martinez, J.; Schroeder, R. Molecular basis for target RNA recognition and cleavage by human RISC. Cell 2007, 130, 101–112. [Google Scholar]
- Fabian, M.R.; Sonenberg, N. The mechanics of miRNA-mediated gene silencing: A look under the hood of miRISC. Nat. Struct. Mol. Biol 2012, 19, 586–593. [Google Scholar]
- Huntzinger, E.; Kuzuoglu-Ozturk, D.; Braun, J.E.; Eulalio, A.; Wohlbold, L.; Izaurralde, E. The interactions of GW182 proteins with PABP and deadenylases are required for both translational repression and degradation of miRNA targets. Nucleic Acids Res 2013, 41, 978–994. [Google Scholar]
- Zekri, L.; Kuzuoglu-Ozturk, D.; Izaurralde, E. GW182 proteins cause PABP dissociation from silenced miRNA targets in the absence of deadenylation. EMBO J 2013, 32, 1052–1065. [Google Scholar]
- Brennecke, J.; Stark, A.; Russell, R.B.; Cohen, S.M. Principles of microRNA-target recognition. PLoS Biol 2005, 3, e85. [Google Scholar]
- Krek, A.; Grun, D.; Poy, M.N.; Wolf, R.; Rosenberg, L.; Epstein, E.J.; MacMenamin, P.; da Piedade, I.; Gunsalus, K.C.; Stoffel, M.; et al. Combinatorial microRNA target predictions. Nat. Genet 2005, 37, 495–500. [Google Scholar]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005, 120, 15–20. [Google Scholar]
- Lewis, B.P.; Shih, I.H.; Jones-Rhoades, M.W.; Bartel, D.P.; Burge, C.B. Prediction of mammalian microRNA targets. Cell 2003, 115, 787–798. [Google Scholar]
- Elkayam, E.; Kuhn, C.D.; Tocilj, A.; Haase, A.D.; Greene, E.M.; Hannon, G.J.; Joshua-Tor, L. The structure of human argonaute-2 in complex with miR-20a. Cell 2012, 150, 100–110. [Google Scholar]
- Schirle, N.T.; MacRae, I.J. The crystal structure of human Argonaute 2. Science 2012, 336, 1037–1040. [Google Scholar]
- Wang, Y.; Sheng, G.; Juranek, S.; Tuschl, T.; Patel, D.J. Structure of the guide-strand-containing argonaute silencing complex. Nature 2008, 456, 209–213. [Google Scholar]
- Chi, S.W.; Hannon, G.J.; Darnell, R.B. An alternative mode of microRNA target recognition. Nat. Struct. Mol. Biol 2012, 19, 321–327. [Google Scholar]
- Miranda, K.C.; Huynh, T.; Tay, Y.; Ang, Y.S.; Tam, W.L.; Thomson, A.M.; Lim, B.; Rigoutsos, I. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell 2006, 126, 1203–1217. [Google Scholar]
- Vella, M.C.; Choi, E.Y.; Lin, S.Y.; Reinert, K.; Slack, F.J. The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3′ UTR. Genes Dev 2004, 18, 132–137. [Google Scholar]
- Reinhart, B.J.; Slack, F.J.; Basson, M.; Pasquinelli, A.E.; Bettinger, J.C.; Rougvie, A.E.; Horvitz, H.R.; Ruvkun, G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 2000, 403, 901–906. [Google Scholar]
- Shin, C.; Nam, J.W.; Farh, K.K.; Chiang, H.R.; Shkumatava, A.; Bartel, D.P. Expanding the microRNA targeting code: functional sites with centered pairing. Mol. Cell 2010, 38, 789–802. [Google Scholar]
- Lal, A.; Navarro, F.; Maher, C.A.; Maliszewski, L.E.; Yan, N.; O’Day, E.; Chowdhury, D.; Dykxhoorn, D.M.; Tsai, P.; Hofmann, O.; et al. miR-24 inhibits cell proliferation by targeting E2F2, MYC, and other cell-cycle genes via binding to “seedless” 3′ UTR microRNA recognition elements. Mol. Cell 2009, 35, 610–625. [Google Scholar]
- Tay, Y.; Zhang, J.; Thomson, A.M.; Lim, B.; Rigoutsos, I. microRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature 2008, 455, 1124–1128. [Google Scholar]
- Didiano, D.; Hobert, O. Perfect seed pairing is not a generally reliable predictor for miRNA-target interactions. Nat. Struct. Mol. Biol 2006, 13, 849–851. [Google Scholar]
- Grimson, A.; Farh, K.K.; Johnston, W.K.; Garrett-Engele, P.; Lim, L.P.; Bartel, D.P. microRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol. Cell 2007, 27, 91–105. [Google Scholar]
- Hon, L.S.; Zhang, Z. The roles of binding site arrangement and combinatorial targeting in microRNA repression of gene expression. Genome Biol 2007, 8, R166. [Google Scholar]
- Saetrom, P.; Heale, B.S.; Snove, O., Jr; Aagaard, L.; Alluin, J.; Rossi, J.J. Distance constraints between microRNA target sites dictate efficacy and cooperativity. Nucleic Acids Res. 2007, 35, 2333–2342. [Google Scholar]
- Doxakis, E. Post-transcriptional regulation of alpha-synuclein expression by mir-7 and mir-153. J. Biol. Chem 2010, 285, 12726–12734. [Google Scholar]
- Paschou, M.; Doxakis, E. Neurofibromin 1 is a miRNA target in neurons. PLoS One 2012, 7, e46773. [Google Scholar]
- Chi, S.W.; Zang, J.B.; Mele, A.; Darnell, R.B. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature 2009, 460, 479–486. [Google Scholar]
- Hafner, M.; Landthaler, M.; Burger, L.; Khorshid, M.; Hausser, J.; Berninger, P.; Rothballer, A.; Ascano, M., Jr; Jungkamp, A.C.; Munschauer, M.; et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 2010, 141, 129–141. [Google Scholar]
- Leung, A.K.; Vyas, S.; Rood, J.E.; Bhutkar, A.; Sharp, P.A.; Chang, P. Poly(ADP-ribose) regulates stress responses and microRNA activity in the cytoplasm. Mol. Cell 2011, 42, 489–499. [Google Scholar]
- Rigoutsos, I. New tricks for animal microRNAS: Targeting of amino acid coding regions at conserved and nonconserved sites. Cancer Res 2009, 69, 3245–3248. [Google Scholar]
- Zisoulis, D.G.; Lovci, M.T.; Wilbert, M.L.; Hutt, K.R.; Liang, T.Y.; Pasquinelli, A.E.; Yeo, G.W. Comprehensive discovery of endogenous Argonaute binding sites in Caenorhabditis elegans. Nat. Struct. Mol. Biol 2010, 17, 173–179. [Google Scholar]
- Baek, D.; Villen, J.; Shin, C.; Camargo, F.D.; Gygi, S.P.; Bartel, D.P. The impact of microRNAs on protein output. Nature 2008, 455, 64–71. [Google Scholar]
- Selbach, M.; Schwanhausser, B.; Thierfelder, N.; Fang, Z.; Khanin, R.; Rajewsky, N. Widespread changes in protein synthesis induced by microRNAs. Nature 2008, 455, 58–63. [Google Scholar]
- Hausser, J.; Syed, A.P.; Bilen, B.; Zavolan, M. Analysis of CDS-located miRNA target sites suggests that they can effectively inhibit translation. Genome Res 2013, 23, 604–615. [Google Scholar]
- Schnall-Levin, M.; Rissland, O.S.; Johnston, W.K.; Perrimon, N.; Bartel, D.P.; Berger, B. Unusually effective microRNA targeting within repeat-rich coding regions of mammalian mRNAs. Genome Res 2011, 21, 1395–1403. [Google Scholar] [Green Version]
- Fang, Z.; Rajewsky, N. The impact of miRNA target sites in coding sequences and in 3′ UTRs. PLoS One 2011, 6, e18067. [Google Scholar]
- Wang, W.X.; Wilfred, B.R.; Xie, K.; Jennings, M.H.; Hu, Y.H.; Stromberg, A.J.; Nelson, P.T. Individual microRNAs (miRNAs) display distinct mRNA targeting “rules”. RNA Biol 2010, 7, 373–380. [Google Scholar]
- Kertesz, M.; Iovino, N.; Unnerstall, U.; Gaul, U.; Segal, E. The role of site accessibility in microRNA target recognition. Nat. Genet 2007, 39, 1278–1284. [Google Scholar]
- Long, D.; Lee, R.; Williams, P.; Chan, C.Y.; Ambros, V.; Ding, Y. Potent effect of target structure on microRNA function. Nat. Struct. Mol. Biol 2007, 14, 287–294. [Google Scholar]
- Paschou, M.; Paraskevopoulou, M.D.; Vlachos, I.S.; Koukouraki, P.; Hatzigeorgiou, A.G.; Doxakis, E. miRNA regulons associated with synaptic function. PLoS One 2012, 7, e46189. [Google Scholar]
- Bergauer, T.; Krueger, U.; Lader, E.; Pilk, S.; Wolter, I.; Bielke, W. Analysis of putative miRNA binding sites and mRNA 3′ ends as targets for siRNA-mediated gene knockdown. Oligonucleotides 2009, 19, 41–52. [Google Scholar]
- Ji, Z.; Lee, J.Y.; Pan, Z.; Jiang, B.; Tian, B. Progressive lengthening of 3′ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc. Natl. Acad. Sci. USA 2009, 106, 7028–7033. [Google Scholar]
- Sandberg, R.; Neilson, J.R.; Sarma, A.; Sharp, P.A.; Burge, C.B. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science 2008, 320, 1643–1647. [Google Scholar]
- Arvey, A.; Larsson, E.; Sander, C.; Leslie, C.S.; Marks, D.S. Target mRNA abundance dilutes microRNA and siRNA activity. Mol. Syst. Biol 2010, 6, 363. [Google Scholar]
- Karreth, F.A.; Tay, Y.; Perna, D.; Ala, U.; Tan, S.M.; Rust, A.G.; de Nicola, G.; Webster, K.A.; Weiss, D.; Perez-Mancera, P.A.; et al. In vivo identification of tumor-suppressive PTEN ceRNAs in an oncogenic BRAF-induced mouse model of melanoma. Cell 2011, 147, 382–395. [Google Scholar]
- Sumazin, P.; Yang, X.; Chiu, H.S.; Chung, W.J.; Iyer, A.; Llobet-Navas, D.; Rajbhandari, P.; Bansal, M.; Guarnieri, P.; Silva, J.; et al. An extensive microRNA-mediated network of RNA-RNA interactions regulates established oncogenic pathways in glioblastoma. Cell 2011, 147, 370–381. [Google Scholar]
- Ameres, S.L.; Horwich, M.D.; Hung, J.H.; Xu, J.; Ghildiyal, M.; Weng, Z.; Zamore, P.D. Target RNA-directed trimming and tailing of small silencing RNAs. Science 2010, 328, 1534–1539. [Google Scholar]
- Rybak, A.; Fuchs, H.; Hadian, K.; Smirnova, L.; Wulczyn, E.A.; Michel, G.; Nitsch, R.; Krappmann, D.; Wulczyn, F.G. The let-7 target gene mouse lin-41 is a stem cell specific E3 ubiquitin ligase for the miRNA pathway protein Ago2. Nat. Cell Biol 2009, 11, 1411–1420. [Google Scholar]
- Pullmann, R., Jr; Kim, H.H.; Abdelmohsen, K.; Lal, A.; Martindale, J.L.; Yang, X.; Gorospe, M. Analysis of turnover and translation regulatory RNA-binding protein expression through binding to cognate mRNAs. Mol. Cell. Biol. 2007, 27, 6265–6278. [Google Scholar]
- Jacobsen, A.; Wen, J.; Marks, D.S.; Krogh, A. Signatures of RNA binding proteins globally coupled to effective microRNA target sites. Genome Res 2010, 20, 1010–1019. [Google Scholar]
- Larsson, E.; Sander, C.; Marks, D. mRNA turnover rate limits siRNA and microRNA efficacy. Mol. Syst. Biol 2010, 6, 433. [Google Scholar]
- Lebedeva, S.; Jens, M.; Theil, K.; Schwanhausser, B.; Selbach, M.; Landthaler, M.; Rajewsky, N. Transcriptome-wide analysis of regulatory interactions of the RNA-binding protein HuR. Mol. Cell 2011, 43, 340–352. [Google Scholar]
- Mukherjee, N.; Corcoran, D.L.; Nusbaum, J.D.; Reid, D.W.; Georgiev, S.; Hafner, M.; Ascano, M., Jr; Tuschl, T.; Ohler, U.; Keene, J.D. Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Mol. Cell 2011, 43, 327–339. [Google Scholar]
- Srikantan, S.; Tominaga, K.; Gorospe, M. Functional interplay between RNA-binding protein HuR and microRNAs. Curr. Protein Pept. Sci 2012, 13, 372–379. [Google Scholar]
- Abdelmohsen, K.; Srikantan, S.; Kuwano, Y.; Gorospe, M. miR-519 reduces cell proliferation by lowering RNA-binding protein HuR levels. Proc. Natl. Acad. Sci. USA 2008, 105, 20297–20302. [Google Scholar]
- Guo, X.; Wu, Y.; Hartley, R.S. microRNA-125a represses cell growth by targeting HuR in breast cancer. RNA Biol 2009, 6, 575–583. [Google Scholar]
- Young, L.E.; Moore, A.E.; Sokol, L.; Meisner-Kober, N.; Dixon, D.A. The mRNA stability factor HuR inhibits microRNA-16 targeting of COX-2. Mol. Cancer Res 2012, 10, 167–180. [Google Scholar]
- Borchert, G.M.; Lanier, W.; Davidson, B.L. RNA polymerase III transcribes human microRNAs. Nat. Struct. Mol. Biol 2006, 13, 1097–1101. [Google Scholar]
- Dieci, G.; Fiorino, G.; Castelnuovo, M.; Teichmann, M.; Pagano, A. The expanding RNA polymerase III transcriptome. Trends Genet 2007, 23, 614–622. [Google Scholar]
- Lee, Y.; Kim, M.; Han, J.; Yeom, K.H.; Lee, S.; Baek, S.H.; Kim, V.N. microRNA genes are transcribed by RNA polymerase II. EMBO J 2004, 23, 4051–4060. [Google Scholar]
- Carlson, D.P.; Ross, J. Human beta-globin promoter and coding sequences transcribed by RNA polymerase III. Cell 1983, 34, 857–864. [Google Scholar]
- Chung, J.; Sussman, D.J.; Zeller, R.; Leder, P. The c-myc gene encodes superimposed RNA polymerase II and III promoters. Cell 1987, 51, 1001–1008. [Google Scholar]
- Listerman, I.; Bledau, A.S.; Grishina, I.; Neugebauer, K.M. Extragenic accumulation of RNA polymerase II enhances transcription by RNA polymerase III. PLoS Genet 2007, 3, e212. [Google Scholar]
- Felton-Edkins, Z.A.; Kenneth, N.S.; Brown, T.R.; Daly, N.L.; Gomez-Roman, N.; Grandori, C.; Eisenman, R.N.; White, R.J. Direct regulation of RNA polymerase III transcription by RB, p53 and c-Myc. Cell Cycle 2003, 2, 181–184. [Google Scholar]
- Baskerville, S.; Bartel, D.P. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA 2005, 11, 241–247. [Google Scholar]
- Fujita, S.; Ito, T.; Mizutani, T.; Minoguchi, S.; Yamamichi, N.; Sakurai, K.; Iba, H. miR-21 gene expression triggered by AP-1 is sustained through a double-negative feedback mechanism. J. Mol. Biol 2008, 378, 492–504. [Google Scholar]
- He, L.; Thomson, J.M.; Hemann, M.T.; Hernando-Monge, E.; Mu, D.; Goodson, S.; Powers, S.; Cordon-Cardo, C.; Lowe, S.W.; Hannon, G.J.; et al. A microRNA polycistron as a potential human oncogene. Nature 2005, 435, 828–833. [Google Scholar]
- O’Donnell, K.A.; Wentzel, E.A.; Zeller, K.I.; Dang, C.V.; Mendell, J.T. c-Myc-regulated microRNAs modulate E2F1 expression. Nature 2005, 435, 839–843. [Google Scholar]
- Monteys, A.M.; Spengler, R.M.; Wan, J.; Tecedor, L.; Lennox, K.A.; Xing, Y.; Davidson, B.L. Structure and activity of putative intronic miRNA promoters. RNA 2010, 16, 495–505. [Google Scholar]
- Yuan, X.; Liu, C.; Yang, P.; He, S.; Liao, Q.; Kang, S.; Zhao, Y. Clustered microRNAs’ coordination in regulating protein-protein interaction network. BMC Syst. Biol 2009, 3, 65. [Google Scholar]
- Gusev, Y. Computational methods for analysis of cellular functions and pathways collectively targeted by differentially expressed microRNA. Methods 2008, 44, 61–72. [Google Scholar]
- Davis, B.N.; Hilyard, A.C.; Lagna, G.; Hata, A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature 2008, 454, 56–61. [Google Scholar]
- Davis, B.N.; Hilyard, A.C.; Nguyen, P.H.; Lagna, G.; Hata, A. Smad proteins bind a conserved RNA sequence to promote microRNA maturation by Drosha. Mol. Cell 2010, 39, 373–384. [Google Scholar]
- Suzuki, H.I.; Yamagata, K.; Sugimoto, K.; Iwamoto, T.; Kato, S.; Miyazono, K. Modulation of microRNA processing by p53. Nature 2009, 460, 529–533. [Google Scholar]
- Guil, S.; Caceres, J.F. The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nat. Struct. Mol. Biol 2007, 14, 591–596. [Google Scholar]
- Michlewski, G.; Caceres, J.F. Antagonistic role of hnRNP A1 and KSRP in the regulation of let-7a biogenesis. Nat. Struct. Mol. Biol 2010, 17, 1011–1018. [Google Scholar]
- Trabucchi, M.; Briata, P.; Garcia-Mayoral, M.; Haase, A.D.; Filipowicz, W.; Ramos, A.; Gherzi, R.; Rosenfeld, M.G. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature 2009, 459, 1010–1014. [Google Scholar]
- Chen, K.; Rajewsky, N. Natural selection on human microRNA binding sites inferred from SNP data. Nat. Genet 2006, 38, 1452–1456. [Google Scholar]
- Muinos-Gimeno, M.; Montfort, M.; Bayes, M.; Estivill, X.; Espinosa-Parrilla, Y. Design and evaluation of a panel of single-nucleotide polymorphisms in microRNA genomic regions for association studies in human disease. Eur. J. Hum. Genet 2010, 18, 218–226. [Google Scholar]
- Saunders, M.A.; Liang, H.; Li, W.H. Human polymorphism at microRNAs and microRNA target sites. Proc. Natl. Acad. Sci. USA 2007, 104, 3300–3305. [Google Scholar]
- Zorc, M.; Skok, D.J.; Godnic, I.; Calin, G.A.; Horvat, S.; Jiang, Z.; Dovc, P.; Kunej, T. Catalog of microRNA seed polymorphisms in vertebrates. PLoS One 2012, 7, e30737. [Google Scholar]
- Ryan, B.M.; Robles, A.I.; Harris, C.C. Genetic variation in microRNA networks: The implications for cancer research. Nat. Rev. Cancer 2010, 10, 389–402. [Google Scholar]
- Marti, E.; Pantano, L.; Banez-Coronel, M.; Llorens, F.; Minones-Moyano, E.; Porta, S.; Sumoy, L.; Ferrer, I.; Estivill, X. A myriad of miRNA variants in control and Huntington’s disease brain regions detected by massively parallel sequencing. Nucleic Acids Res 2010, 38, 7219–7235. [Google Scholar]
- Morin, R.D.; O’Connor, M.D.; Griffith, M.; Kuchenbauer, F.; Delaney, A.; Prabhu, A.L.; Zhao, Y.; McDonald, H.; Zeng, T.; Hirst, M.; et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res 2008, 18, 610–621. [Google Scholar]
- Nishikura, K. Functions and regulation of RNA editing by ADAR deaminases. Annu. Rev. Biochem 2010, 79, 321–349. [Google Scholar]
- De Hoon, M.J.; Taft, R.J.; Hashimoto, T.; Kanamori-Katayama, M.; Kawaji, H.; Kawano, M.; Kishima, M.; Lassmann, T.; Faulkner, G.J.; Mattick, J.S.; et al. Cross-mapping and the identification of editing sites in mature microRNAs in high-throughput sequencing libraries. Genome Res 2010, 20, 257–264. [Google Scholar]
- Kawahara, Y.; Megraw, M.; Kreider, E.; Iizasa, H.; Valente, L.; Hatzigeorgiou, A.G.; Nishikura, K. Frequency and fate of microRNA editing in human brain. Nucleic Acids Res 2008, 36, 5270–5280. [Google Scholar]
- Peng, Z.; Cheng, Y.; Tan, B.C.; Kang, L.; Tian, Z.; Zhu, Y.; Zhang, W.; Liang, Y.; Hu, X.; Tan, X.; et al. Comprehensive analysis of RNA-Seq data reveals extensive RNA editing in a human transcriptome. Nat. Biotechnol 2012, 30, 253–260. [Google Scholar]
- Choudhury, Y.; Tay, F.C.; Lam, D.H.; Sandanaraj, E.; Tang, C.; Ang, B.T.; Wang, S. Attenuated adenosine-to-inosine editing of microRNA-376a* promotes invasiveness of glioblastoma cells. J. Clin. Invest 2012, 122, 4059–4076. [Google Scholar]
- Liu, N.; Abe, M.; Sabin, L.R.; Hendriks, G.J.; Naqvi, A.S.; Yu, Z.; Cherry, S.; Bonini, N.M. The exoribonuclease Nibbler controls 3′ end processing of microRNAs in Drosophila. Curr. Biol 2011, 21, 1888–1893. [Google Scholar]
- Ota, H.; Sakurai, M.; Gupta, R.; Valente, L.; Wulff, B.E.; Ariyoshi, K.; Iizasa, H.; Davuluri, R.V.; Nishikura, K. ADAR1 Forms a complex with Dicer to spromote microRNA processing and RNA-induced gene silencing. Cell 2013, 153, 575–589. [Google Scholar]
- Ruby, J.G.; Jan, C.; Player, C.; Axtell, M.J.; Lee, W.; Nusbaum, C.; Ge, H.; Bartel, D.P. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell 2006, 127, 1193–1207. [Google Scholar]
- Ruby, J.G.; Stark, A.; Johnston, W.K.; Kellis, M.; Bartel, D.P.; Lai, E.C. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res 2007, 17, 1850–1864. [Google Scholar]
- Fukunaga, R.; Han, B.W.; Hung, J.H.; Xu, J.; Weng, Z.; Zamore, P.D. Dicer partner proteins tune the length of mature miRNAs in flies and mammals. Cell 2012, 151, 533–546. [Google Scholar]
- Lee, H.Y.; Doudna, J.A. TRBP alters human precursor microRNA processing in vitro. RNA 2012, 18, 2012–2019. [Google Scholar]
- Han, B.W.; Hung, J.H.; Weng, Z.; Zamore, P.D.; Ameres, S.L. The 3′-to-5′ exoribonuclease Nibbler shapes the 3′ ends of microRNAs bound to Drosophila Argonaute1. Curr. Biol 2011, 21, 1878–1887. [Google Scholar]
- Martin, G.; Keller, W. RNA-specific ribonucleotidyl transferases. RNA 2007, 13, 1834–1849. [Google Scholar]
- Wyman, S.K.; Knouf, E.C.; Parkin, R.K.; Fritz, B.R.; Lin, D.W.; Dennis, L.M.; Krouse, M.A.; Webster, P.J.; Tewari, M. Post-transcriptional generation of miRNA variants by multiple nucleotidyl transferases contributes to miRNA transcriptome complexity. Genome Res 2011, 21, 1450–1461. [Google Scholar]
- Fernandez-Valverde, S.L.; Taft, R.J.; Mattick, J.S. Dynamic isomiR regulation in Drosophila development. RNA 2010, 16, 1881–1888. [Google Scholar]
- Katoh, T.; Sakaguchi, Y.; Miyauchi, K.; Suzuki, T.; Kashiwabara, S.; Baba, T. Selective stabilization of mammalian microRNAs by 3′ adenylation mediated by the cytoplasmic poly(A) polymerase GLD-2. Genes Dev 2009, 23, 433–438. [Google Scholar]
- Burroughs, A.M.; Ando, Y.; de Hoon, M.J.; Tomaru, Y.; Nishibu, T.; Ukekawa, R.; Funakoshi, T.; Kurokawa, T.; Suzuki, H.; Hayashizaki, Y.; et al. A comprehensive survey of 3′ animal miRNA modification events and a possible role for 3′ adenylation in modulating miRNA targeting effectiveness. Genome Res 2010, 20, 1398–1410. [Google Scholar]
- Jones, M.R.; Quinton, L.J.; Blahna, M.T.; Neilson, J.R.; Fu, S.; Ivanov, A.R.; Wolf, D.A.; Mizgerd, J.P. Zcchc11-dependent uridylation of microRNA directs cytokine expression. Nat. Cell Biol 2009, 11, 1157–1163. [Google Scholar]
- Guo, L.; Yang, Q.; Lu, J.; Li, H.; Ge, Q.; Gu, W.; Bai, Y.; Lu, Z. A comprehensive survey of miRNA repertoire and 3′ addition events in the placentas of patients with pre-eclampsia from high-throughput sequencing. PLoS One 2011, 6, e21072. [Google Scholar]
- Kuchenbauer, F.; Morin, R.D.; Argiropoulos, B.; Petriv, O.I.; Griffith, M.; Heuser, M.; Yung, E.; Piper, J.; Delaney, A.; Prabhu, A.L.; et al. In-depth characterization of the microRNA transcriptome in a leukemia progression model. Genome Res 2008, 18, 1787–1797. [Google Scholar]
- Cesana, M.; Daley, G.Q. Deciphering the rules of ceRNA networks. Proc. Natl. Acad. Sci. USA 2013, 110, 7112–7113. [Google Scholar]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar]
- Jeyapalan, Z.; Deng, Z.; Shatseva, T.; Fang, L.; He, C.; Yang, B.B. Expression of CD44 3′-untranslated region regulates endogenous microRNA functions in tumorigenesis and angiogenesis. Nucleic Acids Res 2011, 39, 3026–3041. [Google Scholar]
- Lee, D.Y.; Shatseva, T.; Jeyapalan, Z.; Du, W.W.; Deng, Z.; Yang, B.B. A 3′-untranslated region (3′ UTR) induces organ adhesion by regulating miR-199a* functions. PLoS One 2009, 4, e4527. [Google Scholar]
- Tay, Y.; Kats, L.; Salmena, L.; Weiss, D.; Tan, S.M.; Ala, U.; Karreth, F.; Poliseno, L.; Provero, P.; di Cunto, F.; et al. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell 2011, 147, 344–357. [Google Scholar]
- Cesana, M.; Cacchiarelli, D.; Legnini, I.; Santini, T.; Sthandier, O.; Chinappi, M.; Tramontano, A.; Bozzoni, I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 2011, 147, 358–369. [Google Scholar]
- Poliseno, L.; Salmena, L.; Zhang, J.; Carver, B.; Haveman, W.J.; Pandolfi, P.P. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 2010, 465, 1033–1038. [Google Scholar]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar]
- Bail, S.; Swerdel, M.; Liu, H.; Jiao, X.; Goff, L.A.; Hart, R.P.; Kiledjian, M. Differential regulation of microRNA stability. RNA 2010, 16, 1032–1039. [Google Scholar]
- Das, S.K.; Sokhi, U.K.; Bhutia, S.K.; Azab, B.; Su, Z.Z.; Sarkar, D.; Fisher, P.B. Human polynucleotide phosphorylase selectively and preferentially degrades microRNA-221 in human melanoma cells. Proc. Natl. Acad. Sci. USA 2010, 107, 11948–11953. [Google Scholar]
- Janowski, B.A.; Younger, S.T.; Hardy, D.B.; Ram, R.; Huffman, K.E.; Corey, D.R. Activating gene expression in mammalian cells with promoter-targeted duplex RNAs. Nat. Chem. Biol 2007, 3, 166–173. [Google Scholar]
- Li, L.C.; Okino, S.T.; Zhao, H.; Pookot, D.; Place, R.F.; Urakami, S.; Enokida, H.; Dahiya, R. Small dsRNAs induce transcriptional activation in human cells. Proc. Natl. Acad. Sci. USA 2006, 103, 17337–17342. [Google Scholar]
- Place, R.F.; Li, L.C.; Pookot, D.; Noonan, E.J.; Dahiya, R. microRNA-373 induces expression of genes with complementary promoter sequences. Proc. Natl. Acad. Sci. USA 2008, 105, 1608–1613. [Google Scholar]
- Majid, S.; Dar, A.A.; Saini, S.; Yamamura, S.; Hirata, H.; Tanaka, Y.; Deng, G.; Dahiya, R. microRNA-205-directed transcriptional activation of tumor suppressor genes in prostate cancer. Cancer 2010, 116, 5637–5649. [Google Scholar]
- Tsai, N.P.; Lin, Y.L.; Wei, L.N. microRNA mir-346 targets the 5′-untranslated region of receptor-interacting protein 140 (RIP140) mRNA and up-regulates its protein expression. Biochem. J 2009, 424, 411–418. [Google Scholar]
- Cordes, K.R.; Sheehy, N.T.; White, M.P.; Berry, E.C.; Morton, S.U.; Muth, A.N.; Lee, T.H.; Miano, J.M.; Ivey, K.N.; Srivastava, D. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature 2009, 460, 705–710. [Google Scholar]
- Ma, F.; Liu, X.; Li, D.; Wang, P.; Li, N.; Lu, L.; Cao, X. microRNA-466l upregulates IL-10 expression in TLR-triggered macrophages by antagonizing RNA-binding protein tristetraprolin-mediated IL-10 mRNA degradation. J. Immunol 2010, 184, 6053–6059. [Google Scholar]
- Heil, F.; Hemmi, H.; Hochrein, H.; Ampenberger, F.; Kirschning, C.; Akira, S.; Lipford, G.; Wagner, H.; Bauer, S. Species-specific recognition of single-stranded RNA via Toll-like receptor 7 and 8. Science 2004, 303, 1526–1529. [Google Scholar]
- Lund, J.M.; Alexopoulou, L.; Sato, A.; Karow, M.; Adams, N.C.; Gale, N.W.; Iwasaki, A.; Flavell, R.A. Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proc. Natl. Acad. Sci. USA 2004, 101, 5598–5603. [Google Scholar]
- Fabbri, M.; Paone, A.; Calore, F.; Galli, R.; Gaudio, E.; Santhanam, R.; Lovat, F.; Fadda, P.; Mao, C.; Nuovo, G.J.; et al. microRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc. Natl. Acad. Sci. USA 2012, 109, E2110–E2116. [Google Scholar]

© 2013 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Doxakis, E. Principles of miRNA-Target Regulation in Metazoan Models. Int. J. Mol. Sci. 2013, 14, 16280-16302. https://doi.org/10.3390/ijms140816280
Doxakis E. Principles of miRNA-Target Regulation in Metazoan Models. International Journal of Molecular Sciences. 2013; 14(8):16280-16302. https://doi.org/10.3390/ijms140816280
Chicago/Turabian StyleDoxakis, Epaminondas. 2013. "Principles of miRNA-Target Regulation in Metazoan Models" International Journal of Molecular Sciences 14, no. 8: 16280-16302. https://doi.org/10.3390/ijms140816280
APA StyleDoxakis, E. (2013). Principles of miRNA-Target Regulation in Metazoan Models. International Journal of Molecular Sciences, 14(8), 16280-16302. https://doi.org/10.3390/ijms140816280