Genome-Wide Analysis of the Cyclin Gene Family in Tomato
Abstract
:1. Introduction
2. Results
2.1. Identification of Tomato Cyclin Gene Family
2.2. Phylogenetic Analysis of Cyclin Family
2.3. Structure and Protein Sequence Analysis of Tomato Cyclins
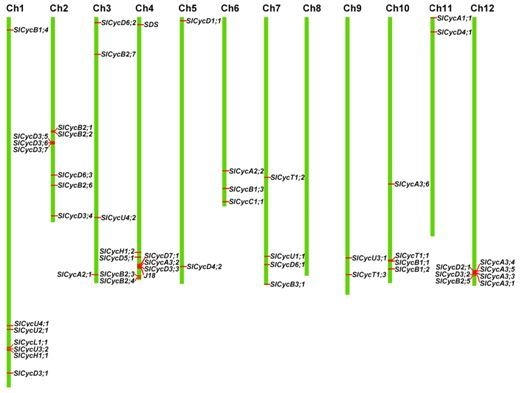
2.4. Chromosomal Localization and Gene Duplication
2.5. Organ-Specific Expressions of Tomato Cyclin Genes
2.6. Expression of Tomato Cyclin Genes in Response to Exogenous GA
3. Discussion
3.1. Cyclin Gene Family and Their Structures
3.2. Duplication of Tomato Cyclin Genes
3.3. Organ-Preferential and GA Responsive Expression Profiles of Tomato Cyclin Genes
4. Materials and Methods
4.1. Plant Materials
4.2. Identification of Putative Tomato Cyclin Genes
4.3. Phylogenetic Analysis of Tomato Cyclins
4.4. Structure Analysis of Cyclins in Tomato
4.5. Chromosomal Localization and Gene Duplication Analysis of Tomato Cyclins
4.6. RT-PCR and Real-Time qRT-PCR
5. Conclusions
Acknowledgments
Conflicts of Interest
Abbreviations
| RT-PCR | reverse transcription polymerase chain reaction |
| GA | gibberellic acid |
References
- Stals, H.; Inze, D. When plant cells decide to divide. Trends Plant Sci 2001, 6, 359–364. [Google Scholar]
- Rossi, V.; Varotto, S. Insights into the G1/S transition in plants. Planta 2002, 215, 345–356. [Google Scholar]
- Wang, G.; Kong, H.; Sun, Y.; Zhang, X.; Zhang, W.; Altman, N.; DePamphilis, C.W.; Ma, H. Genome-wide analysis of the cyclin family in Arabidopsis and comparative phylogenetic analysis of plant cyclin-like proteins. Plant Physiol 2004, 135, 1084–1099. [Google Scholar]
- La, H.; Li, J.; Ji, Z.; Cheng, Y.; Li, X.; Jiang, S.; Venkatesh, P.N.; Ramachandran, S. Genome-wide analysis of cyclin family in rice (Oryza Sativa L.). Mol. Genet. Genomics 2006, 275, 374–386. [Google Scholar]
- Booher, R.N.; Alfa, C.E.; Hyams, J.S.; Beach, D.H. The fission yeast cdc2/cdc13/suc1 protein kinase: regulation of catalytic activity and nuclear localization. Cell 1989, 58, 485–497. [Google Scholar]
- Hata, S.; Kouchi, H.; Suzuka, I.; Ishii, T. Isolation and characterization of cDNA clones for plant cyclins. EMBO J 1991, 10, 2681–2688. [Google Scholar]
- Dong, Q.; Zhao, Y.; Jiang, H.Y.; He, H.S.; Zhu, S.W.; Cheng, B.J.; Xiang, Y. Genome-wide identification and characterization of the cyclin gene family in Populus trichocarpa. Plant Cell Tiss. Org 2011, 107, 55–67. [Google Scholar]
- Jesty, J.H.F.; Francis, D. Cellular-responses of leaf explants of Cocos nucifera L. in vitro. Plant Cell Tiss. Org. Cult. 1992, 28, 235–244. [Google Scholar]
- Nugent, J.H.; Alfa, C.E.; Young, T.; Hyams, J.S. Conserved structural motifs in cyclins identified by sequence analysis. J. Cell Sci 1991, 99, 669–674. [Google Scholar]
- Horne, M.C.; Goolsby, G.L.; Donaldson, K.L.; Tran, D.; Neubauer, M.; Wahl, A.F. Cyclin G1 and cyclin G2 comprise a new family of cyclins with contrasting tissue-specific and cell cycle-regulated expression. J. Biol. Chem 1996, 271, 6050–6061. [Google Scholar]
- Lehner, C.F.; O’Farrell, P.H. The roles of Drosophila cyclins A and B in mitotic control. Cell 1990, 61, 535–547. [Google Scholar]
- Obaya, A.J.; Sedivy, J.M. Regulation of cyclin-Cdk activity in mammalian cells. Cell. Mol. Life Sci 2002, 59, 126–142. [Google Scholar]
- Pagano, M.; Pepperkok, R.; Verde, F.; Ansorge, W.; Draetta, G. Cyclin A is required at two points in the human cell cycle. EMBO J 1992, 11, 961–971. [Google Scholar]
- Yang, C.; Li, H.; Zhang, J.; Luo, Z.; Gong, P.; Zhang, C.; Li, J.; Wang, T.; Zhang, Y.; Lu, Y.; Ye, Z. A regulatory gene induces trichome formation and embryo lethality in tomato. Proc. Natl. Acad. Sci. USA 2011, 108, 11836–11841. [Google Scholar]
- Gutierrez, C.; Ramirez-Parra, E.; Castellano, M.M.; del Pozo, J.C. G(1) to S transition: more than a cell cycle engine switch. Curr. Opin. Plant Biol 2002, 5, 480–486. [Google Scholar]
- Shen, W.H. The plant E2F-Rb pathway and epigenetic control. Trends Plant Sci 2002, 7, 505–511. [Google Scholar]
- Trimarchi, J.M.; Lees, J.A. Sibling rivalry in the E2F family. Nat. Rev. Mol. Cell Biol 2002, 3, 11–20. [Google Scholar]
- Kvarnheden, A.; Yao, J.L.; Zhan, X.; O’Brien, I.; Morris, B.A. Isolation of three distinct CycD3 genes expressed during fruit development in tomato. J. Exp. Bot 2000, 51, 1789–1797. [Google Scholar]
- Huysman, M.J.; Fortunato, A.E.; Matthijs, M.; Costa, B.S.; Vanderhaeghen, R.; van den Daele, H.; Sachse, M.; Inze, D.; Bowler, C.; Kroth, P.G.; et al. AUREOCHROME1a-mediated induction of the diatom-specific cyclin dsCYC2 controls the onset of cell division in diatoms (Phaeodactylum tricornutum). Plant Cell 2013, 25, 215–228. [Google Scholar]
- Achard, P.; Gusti, A.; Cheminant, S.; Alioua, M.; Dhondt, S.; Coppens, F.; Beemster, G.T.; Genschik, P. Gibberellin signaling controls cell proliferation rate in Arabidopsis. Curr. Biol 2009, 19, 1188–1193. [Google Scholar]
- Sauter, M.; Mekhedov, S.L.; Kende, H. Gibberellin promotes histone H1 kinase activity and the expression of cdc2 and cyclin genes during the induction of rapid growth in deepwater rice internodes. Plant J 1995, 7, 623–632. [Google Scholar]
- Fabian, T.; Lorbiecke, R.; Umeda, M.; Sauter, M. The cell cycle genes cycA1;1 and cdc2Os-3 are coordinately regulated by gibberellin in planta. Planta 2000, 211, 376–383. [Google Scholar]
- Tomato Genome, C. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 2012, 485, 635–641. [Google Scholar] [Green Version]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant biol 2004, 4, 10. [Google Scholar]
- Joubes, J.; Walsh, D.; Raymond, P.; Chevalier, C. Molecular characterization of the expression of distinct classes of cyclins during the early development of tomato fruit. Planta 2000, 211, 430–439. [Google Scholar]
- Santner, A.; Estelle, M. Recent advances and emerging trends in plant hormone signalling. Nature 2009, 459, 1071–1078. [Google Scholar]
- Yamaguchi, S. Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol 2008, 59, 225–251. [Google Scholar]
- Nakamura, T.; Sanokawa, R.; Sasaki, Y.F.; Ayusawa, D.; Oishi, M.; Mori, N. Cyclin-I—A new cyclin encoded by a gene isolated from human brain. Exp. Cell Res 1995, 221, 534–542. [Google Scholar]
- Pines, J. Cyclins and Cyclin-dependent kinases—Theme and variations. Adv. Cancer Res 1995, 66, 181–212. [Google Scholar]
- Yamaguchi, S.; Okayama, H.; Nurse, P. Fission yeast Fizzy-related protein srw1p is a G(1)-specific promoter of mitotic cyclin B degradation. EMBO J 2000, 19, 3968–3977. [Google Scholar]
- Barroco, R.M.; de Veylder, L.; Magyar, Z.; Engler, G.; Inze, D.; Mironov, V. Novel complexes of cyclin-dependent kinases and a cyclin-like protein from Arabidopsis thaliana with a function unrelated to cell division. Cell. Mol. Life Sci 2003, 60, 401–412. [Google Scholar]
- Renaudin, J.P.; Doonan, J.H.; Freeman, D.; Hashimoto, J.; Hirt, H.; Inze, D.; Jacobs, T.; Kouchi, H.; Rouze, P.; Sauter, M.; et al. Plant cyclins: A unified nomenclature for plant A-, B- and D-type cyclins based on sequence organization. Plant Mol. Biol 1996, 32, 1003–1018. [Google Scholar]
- Hu, X.; Cheng, X.; Jiang, H.; Zhu, S.; Cheng, B.; Xiang, Y. Genome-wide analysis of cyclins in maize (Zea mays). Genet. Mol. Res 2010, 9, 1490–1503. [Google Scholar]
- Moore, R.C.; Purugganan, M.D. The early stages of duplicate gene evolution. Proc. Natl. Acad. Sci. USA 2003, 100, 15682–15687. [Google Scholar]
- Vision, T.J.; Brown, D.G.; Tanksley, S.D. The origins of genomic duplications in Arabidopsis. Science 2000, 290, 2114–2117. [Google Scholar]
- Blanc, G.; Barakat, A.; Guyot, R.; Cooke, R.; Delseny, M. Extensive duplication and reshuffling in the Arabidopsis genome. Plant Cell 2000, 12, 1093–1101. [Google Scholar]
- Ku, H.M.; Vision, T.; Liu, J.; Tanksley, S.D. Comparing sequenced segments of the tomato and Arabidopsis genomes: Large-scale duplication followed by selective gene loss creates a network of synteny. Proc. Natl. Acad. Sci. USA 2000, 97, 9121–9126. [Google Scholar]
- Grant, D.; Cregan, P.; Shoemaker, R.C. Genome organization in dicots: genome duplication in Arabidopsis and synteny between soybean and Arabidopsis. Proc. Natl. Acad. Sci. USA 2000, 97, 4168–4173. [Google Scholar]
- Achard, P.; Genschik, P. Releasing the brakes of plant growth: How GAs shutdown DELLA proteins. J. Exp. Bot 2009, 60, 1085–1092. [Google Scholar]
- Gocal, G.F.W.; King, R.W. Early increased expression of a cyclin-dependant protein kinase (LtCDKA1;1) during inflorescence initiation of the long day grass Lolium temulentum. Funct. Plant Biol 2013, 40, 986–995. [Google Scholar]
- SOL Genomics Network. Available online: http://solgenomics.net (accessed on 9 March 2012).
- National Center for Biotechnology Information (NCBI). Available online: http://www.ncbi.nlm.nih.gov/ (accessed on 8 March 2012).
- DFCI. Available online: http://compbio.dfci.harvard.edu/tgi/plant.html (accessed on 8 March 2012).
- Punta, M.; Coggill, P.C.; Eberhardt, R.Y.; Mistry, J.; Tate, J.; Boursnell, C.; Pang, N.; Forslund, K.; Ceric, G.; Clements, J.; et al. The Pfam protein families database. Nucleic Acids Res 2012, 40, D290–D301. [Google Scholar]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol 2011, 28, 2731–2739. [Google Scholar]
- Guo, A.Y.; Zhu, Q.H.; Chen, X.; Luo, J.C. GSDS: A gene structure display server. Yi Chuan 2007, 29, 1023–1026. [Google Scholar]
- MEME (Multiple Em for Motif Elicitation). Available online: http://meme.nbcr.net/meme/cgi-bin/meme.cgi (accessed on 6 August 2013).
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res 2012, 40, e49. [Google Scholar]








| Gene name | Annotated CDS | Chr. No. | Protein length (aa) |
|---|---|---|---|
| SlCycA1;1 | Solyc11g005090.1.1 | 11 | 490 |
| SlCycA2;1 | Solyc03g120440.1.1 | 3 | 458 |
| SlCycA2;2 | Solyc06g065680.2.1 | 6 | 475 |
| SlCycA3;1 | Solyc12g088530.1.1 | 12 | 354 |
| SlCycA3;2 | Solyc04g078310.2.1 | 4 | 371 |
| SlCycA3;3 | Solyc12g088520.1.1 | 12 | 355 |
| SlCycA3;4 | Solyc12g088470.1.1 | 12 | 360 |
| SlCycA3;5 | Solyc12g088500.1.1 | 12 | 354 |
| SlCycA3;6 | Solyc10g049360.1.1 | 10 | 378 |
| SlCycB1;1 | Solyc10g078330.1.1 | 10 | 423 |
| SlCycB1;2 | Solyc10g080950.1.1 | 10 | 462 |
| SlCycB1;3 | Solyc06g073610.2.1 | 6 | 440 |
| SlCycB1;4 | Solyc01g009040.2.1 | 1 | 435 |
| SlCycB2;1 | Solyc02g061650.1.1 | 2 | 351 |
| SlCycB2;2 | Solyc02g061690.1.1 | 2 | 162 |
| SlCycB2;3 | Solyc04g081660.1.1 | 4 | 142 |
| SlCycB2;4 | Solyc04g082430.2.1 | 4 | 393 |
| SlCycB2;5 | Solyc12g094600.1.1 | 12 | 425 |
| SlCycB2;6 | Solyc02g082820.2.1 | 2 | 434 |
| SlCycB2;7 | Solyc03g032190.2.1 | 3 | 431 |
| SlCycB3;1 | Solyc07g066660.2.1 | 7 | 739 |
| SlCycC1;1 | Solyc06g083260.2.1 | 6 | 250 |
| SlCycD1;1 | Solyc05g006050.2.1 | 5 | 337 |
| SlCycD2;1 | Solyc12g087900.1.1 | 12 | 316 |
| SlCycD3;1 | Solyc01g107730.2.1 | 1 | 364 |
| SlCycD3;2 | Solyc12g088650.1.1 | 12 | 325 |
| SlCycD3;3 | Solyc04g078470.2.1 | 4 | 336 |
| SlCycD3;4 | Solyc02g092980.2.1 | 2 | 359 |
| SlCycD3;5 | Solyc02g064860.1.1 | 2 | 235 |
| SlCycD3;6 | Solyc02g064900.1.1 | 2 | 346 |
| SlCycD3;7 | Solyc02g064910.1.1 | 2 | 235 |
| SlCycD4;1 | Solyc11g010460.1.1 | 11 | 312 |
| SlCycD4;2 | Solyc05g051410.2.1 | 5 | 344 |
| SlCycD5;1 | Solyc04g076040.2.1 | 4 | 355 |
| SlCycD6;1 | Solyc07g054950.1.1 | 7 | 310 |
| SlCycD6;2 | Solyc03g006670.2.1 | 3 | 311 |
| SlCycD6;3 | Solyc02g079370.2.1 | 2 | 297 |
| SlCycD7;1 | Solyc04g077880.1.1 | 4 | 298 |
| SlCycH1;1 | Solyc01g099310.2.1 | 1 | 328 |
| SlCycH1;2 | Solyc04g072880.2.1 | 4 | 327 |
| SlCycL1;1 | Solyc01g098350.2.1 | 1 | 426 |
| SlCycU1;1 | Solyc07g052610.2.1 | 7 | 241 |
| SlCycU2;1 | Solyc01g090800.2.1 | 1 | 220 |
| SlCycU3;1 | Solyc09g065200.2.1 | 9 | 237 |
| SlCycU3;2 | Solyc01g098930.2.1 | 1 | 226 |
| SlCycU4;1 | Solyc01g089850.2.1 | 1 | 206 |
| SlCycU4;2 | Solyc03g093790.2.1 | 3 | 196 |
| SlCycT1;1 | Solyc10g078180.1.1 | 10 | 621 |
| SlCycT1;2 | Solyc07g032480.2.1 | 7 | 368 |
| SlCycT1;3 | Solyc09g075760.2.1 | 9 | 413 |
| SDS | Solyc04g008070.1.1 | 4 | 641 |
| J18 | Solyc04g078500.1.1 | 4 | 231 |
| Motif | Motif length (AA) | Best possible match |
|---|---|---|
| 1 | 21 | LQLLGVTCLLLAAKYEEIxVP |
| 2 | 41 | QKDVNESMRGILVDWLVEVHDKYKLxPETLYLAVNYIDRFL |
| 3 | 33 | VLRMEKLVLNTLKWRMTVPTPYTFLRRFLKAAQ |
| 4 | 15 | PSxIAAAAIYLARFT |
| 5 | 29 | HPWSLTLEHHTGYSESQLKECVLLIVDLH |
| 6 | 29 | DDAKNPLACVEYVEDIYAYYKKMEIEKRR |
| 7 | 21 | LEFLSFYLAELCLLEYECLKF |
| 8 | 23 | KLTAVRRKYSSHKFKCVALLGPP |
| 9 | 15 | VEDFCYITDNAYTKK |
| 10 | 29 | PPVRVTRPATRKFAAQMASQLQQPNKKRV |
| 11 | 47 | AYYAKVGGITTREMNKLEVDFLFGLGFQLHVNVTTFESYCSYLEKEM |
| 12 | 50 | CKEDPLDEGDLGGGYHSDERNWNVKKISPLLECDMFWEDGEVETLLSKEK |
| 13 | 50 | NSPSRKDGIDVEEEQHLRKFYCFFLQDLGIRLKFPQKTIATALILCHRFY |
| 14 | 29 | GVIDAYFSSESSNDSWVVASSVSSLPEPQ |
| 15 | 15 | DLQVEDAKFVFEAKT |
| 16 | 20 | GGERGRNRRALGDINQNLVG |
| 17 | 21 | FHGLRAPNISIQSYLERIFKY |
| 18 | 49 | MADFVTSTHRTKWIFTPQDIKHKYKVANHRAKQALEKYGTTRMEVDIDG |
| 19 | 21 | LLRRAEQLILSTITDIRFLEY |
| 20 | 29 | VLSEIQDLCNVGINQIEDKVFVSEPLRPK |
| Gene | Forward primer sequence | Reverse primer sequence |
|---|---|---|
| SlCycA1;1 | ATGACAACCCAGAATAAGCG | ACCGAGTTCCGAGCAGAG |
| SlCycA2;4 | TTGGGTTCATCAGGAGGACT | CTTCGCTTAGGCTGTTGAGA |
| SlCycA3;1 | CTAAGAAAAGAGCAGCAGAAGCA | GATTCCTTATCTTTTTCAGCAACAG |
| SlCycA3;2 | GCTCCACAGGAACAACGAA | CCACAACCCTTTTCACCTTC |
| SlCycA3;4 | AACCCACGAATCCAAACG | CCAAGGAGTGAAGATGCTGA |
| SlCycB1;1 | GTATCTCGCCCCGTAACAAG | TCTCCTCAGGTTTTGGCTTT |
| SlCycC1;1 | AAGTTCTGGAAGCCCTAAACTATTA | GAATTAGGTCCATCTTATAGGTGTCA |
| SlCycD1;1 | CTGCTACTTCTTCTTCAAATCCA | CGGAGAAACAGTCGGAGTAA |
| SlCycD2;1 | GTGATGGCTCAGGGGTTG | GTTATGTGTTTGTGGGGTGC |
| SlCycD3;1 | CTGCCAAAGCCTCAAGCG | CAGTGGAGCTAGTGTCATTCGC |
| SlCycD4;1 | GCAGTGGCAATGTCTGTTTC | CCATTGGGAGTTTGAGGC |
| SlCycD6;1 | GATGGAGACGACGATGACGA | TGATTGCGAATGGAAGAGTGA |
| SlCycD7;1 | AGATGGAGAGTTTGCTTTGTGAT | AAAATCTTAAAAGCCTCTTCACAAT |
| SlCycH1;1 | ACCTGGGCAGTTGGCATT | CGTCTTTGGAAGTCGGAGTC |
| SlCycL1;1 | CGTAGATTTCAAGTGCCCCTC | TATTTTTGCTTTTGGAAGACTGTAA |
| SlCycU1;1 | CGGACGATGTAGCCACCC | CCTGCCCCGTTGCTCAT |
| SlCycU3;1 | ATTGTCTTCTTGCCCCTTTT | TCAGGTTCAGGTGCCAAAG |
| SlCycU4;1 | TCAAAATCTGATGCCGAAAC | CGAAACAACAAGGGCTACAA |
| J18 | GATCGAGAATGAAGAAGGAGGT | GATAGAATCGGTTGGCAAAAGT |
| Actin | GTCCTCTTCCAGCCATCCAT | ACCACTGAGCACAATGTTACCG |
© 2014 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Zhang, T.; Wang, X.; Lu, Y.; Cai, X.; Ye, Z.; Zhang, J. Genome-Wide Analysis of the Cyclin Gene Family in Tomato. Int. J. Mol. Sci. 2014, 15, 120-140. https://doi.org/10.3390/ijms15010120
Zhang T, Wang X, Lu Y, Cai X, Ye Z, Zhang J. Genome-Wide Analysis of the Cyclin Gene Family in Tomato. International Journal of Molecular Sciences. 2014; 15(1):120-140. https://doi.org/10.3390/ijms15010120
Chicago/Turabian StyleZhang, Tingyan, Xin Wang, Yongen Lu, Xiaofeng Cai, Zhibiao Ye, and Junhong Zhang. 2014. "Genome-Wide Analysis of the Cyclin Gene Family in Tomato" International Journal of Molecular Sciences 15, no. 1: 120-140. https://doi.org/10.3390/ijms15010120
APA StyleZhang, T., Wang, X., Lu, Y., Cai, X., Ye, Z., & Zhang, J. (2014). Genome-Wide Analysis of the Cyclin Gene Family in Tomato. International Journal of Molecular Sciences, 15(1), 120-140. https://doi.org/10.3390/ijms15010120