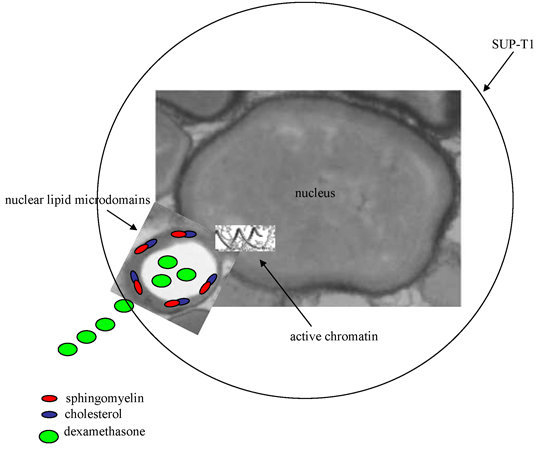
Nuclear Lipid Microdomain as Resting Place of Dexamethasone to Impair Cell Proliferation
Abstract
:1. Introduction
2. Results
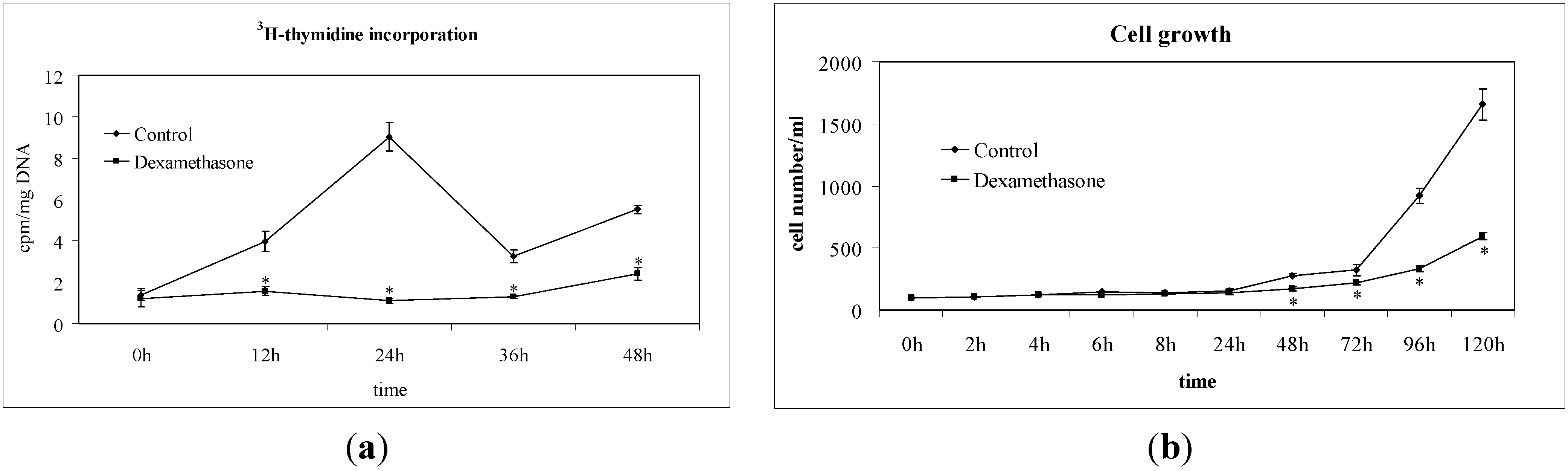
2.1. Non-Hodgkin’s T Cell Human Lymphoblastic Lymphoma Cell Growth Is Suppressed by Dexamethasone



2.2. Dexamethasone Localizes in Nuclear Lipid Microdomains


3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Cell Culture and Treatments
4.3. Preparation of Homogenate, Nuclei and Nuclear Lipid Microdomain Purification
4.4. 3H-Dexamethasone Incorporation
4.5. Sphingomyelinase and Sphingomyelin-Synthase Activity
4.6. 3H (Sphingomyelin and Phosphatidylcholine) Level
4.7. Electrophoresis and Western Blot Analysis
4.8. DNA Synthesis and Cell Growth
4.9. Morphological and Immunohistochemical Analysis
4.10. Reverse Transcription Quantitative PCR (RTqPCR)
4.11. Statistical Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Moalli, P.A.; Rosen, S.T. Glucocorticoid receptors and resistance to glucocorticoids in hematologic malignancies. Leuk. Lymphoma 1994, 15, 363–374. [Google Scholar]
- Schmidt, S.; Rainer, J.; Ploner, C.; Presul, E.; Riml, S.; Kofler, R. Glucocorticoid-induced apoptosis and glucocorticoid resistance: molecular mechanisms and clinical relevance. Cell Death Differ. 2004, 11, S45–S55. [Google Scholar]
- Laane, E.; Tamm, K.P.; Buentke, E.; Ito, K.; Kharaziha, P.; Oscarsson, J.; Corcoran, M.; Björklund, A.C.; Hultenby, K.; Lundin, J.; et al. Cell death induced by dexamethasone in lymphoid leukemia is mediated through initiation of autophagy. Cell Death Differ. 2009, 16, 1018–1029. [Google Scholar]
- Brunet, C.L.; Gunby, R.H.; Benson, R.S.P.; Hickman, J.A.; Watson, A.J.M.; Brady, G. Commitment to cell death measured by loss of clonogenicity is separable from the appearance of apoptotic markers. Cell Death Differ. 1998, 5, 107–115. [Google Scholar]
- Evans, R.M. The nuclear receptor superfamily: A Rosetta stone for physiology. Mol. Endocrinol. 2005, 19, 1429–1438. [Google Scholar]
- Kininis, M.; Kraus, W.L. A global view of transcriptional regulation by nuclear receptors: Gene expression, factor localization and DNA sequence analysis. Nucl. Recept. Signal. 2008, 6, e005. [Google Scholar]
- Banerjee, A.; Periyasamy, S.; Wolf, I.M.; Hinds, T.D.J.; Yong, W.; Shou, W.; Sanchez, E.R. Control of glucocorticoid and progesterone receptor subcellular localization by the ligand-binding domain is mediated by distinct interactions with tetratricopeptide repeat proteins. Biochemistry 2008, 47, 10471–10480. [Google Scholar]
- Burd, C.J.; Ward, J.M.; Crusselle-Davis, V.J.; Kissling, G.E.; Phadke, D.; Shah, R.R.; Archer, T.K. Analysis of chromatin dynamics during glucocorticoid receptor activation. Mol. Cell. Biol. 2012, 32, 1805–1817. [Google Scholar]
- Vandevyver, S.; Dejager, L.; Libert, C. On the trail of the glucocorticoid receptor: Into the nucleus and back. Traffic 2012, 13, 364–374. [Google Scholar]
- Newbold, A.; Salmon, J.M.; Martin, B.P.; Stanley, K.; Johnstone, R.W. The role of p21waf1/cip1 and p27Kip1 in HDACi-mediated tumor cell death and cell cycle arrest in the Eμ-myc model of B-cell lymphoma. Oncogene 2013. [Google Scholar] [CrossRef]
- Bustany, S.; Tchakarska, G.; Sola, B. Cyclin D1 regulates p27Kip1 stability in B cells. Cell Signal. 2011, 23, 171–179. [Google Scholar]
- Moskalev, A.A.; Smit-McBride, Z.; Shaposhnikov, M.V.; Plyusnina, E.N.; Zhavoronkov, A.; Budovsky, A.; Tacutu, R.; Fraifeld, V.E. Gadd45 proteins: Relevance to aging, longevity and age-related pathologies. Ageing Res. Rev. 2012, 11, 51–66. [Google Scholar]
- Xiong, H.; Zhang, Z.G.; Tian, X.Q.; Sun, D.F.; Liang, Q.C.; Zhang, Y.J.; Lu, R.; Chen, Y.X.; Fang, J.Y. Inhibition of JAK1, 2/STAT3 signaling induces apoptosis, cell cycle arrest and reduces tumor cell invasion in colorectal cancer cells. Neoplasia 2008, 10, 287–297. [Google Scholar]
- Reich, N.C. STATs get their move on. JAKSTAT 2013, 2, e27080. [Google Scholar]
- McDonnell, T.J.; Korsmeyer, S.J. Progression from lymphoid hyperplasia to high-grade malignant lymphoma in mice transgenic for the t(14; 18). Nature 1991, 349, 254–256. [Google Scholar]
- Albi, E.; Viola Magni, M. Sphingomyelin: A small-big molecule in the nucleus. In Sphingolipid and Cell Function; Albi, E., Ed.; Research Signpost: Kerale, India, 2006; Volume 37, pp. 211–227. [Google Scholar]
- Albi, E.; Viola Magni, M.P. Chromatin neutral sphingomyelinase and its role in hepatic regeneration. Biochim. Biophys. Res. Commun. 1997, 236, 29–33. [Google Scholar]
- Albi, E.; Lazzarini, R.; Viola Magni, M. Reverse sphingomyelin-synthase in rat liver chromatin. FEBS Lett. 2003, 549, 152–156. [Google Scholar]
- Albi, E.; Viola Magni, M.P. Sphingomyelin synthase in rat liver nuclear membrane and chromatin. FEBS Lett. 1999, 460, 369–372. [Google Scholar]
- Scassellati, C.; Albi, E.; Cmarko, D.; Tiberi, C.; Cmarkova, J.; Bouchet-Marquis, C.; Verschure, P.J.; Driel, R.; Magni, M.V.; Fakan, S. Intranuclear sphingomyelin is associated with transcriptionally active chromatin and plays a role in nuclear integrity. Biol. Cell 2010, 102, 361–735. [Google Scholar]
- Albi, E.; Lazzarini, A.; Lazzarini, R.; Floridi, A.; Damaskopoulou, E.; Curcio, F.; Cataldi, S. Nuclear lipid microdomain as place of interaction between sphingomyelin and DNA during liver regeneration. Int. J. Mol. Sci. 2013, 14, 6529–6541. [Google Scholar]
- Cascianelli, G.; Villani, M.; Tosti, M.; Marini, F.; Bartoccini, E.; Viola Magni, M.; Albi, E. Lipid microdomains in cell nucleus. Mol. Biol. Cell 2008, 19, 5289–5295. [Google Scholar]
- Albi, E.; Villani, M. Nuclear lipid microdomains regulate cell function. Commun. Integr. Biol. 2009, 2, 23–24. [Google Scholar]
- Bartoccini, E.; Marini, F.; Damaskopoulou, E.; Lazzarini, R.; Cataldi, S.; Cascianelli, G.; Gil Garcia, M.; Albi, E. Nuclear lipid microdomains regulate nuclear vitamin D3 uptake and influence embryonic hippocampal cell differentiation. Mol. Biol. Cell 2011, 22, 3022–3031. [Google Scholar]
- El Kadmiri, N.; Slassi, I.; el Moutawakil, B.; Nadifi, S.; Tadevosyan, A.; Hachem, A.; Soukri, A. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and Alzheimer’s disease. Pathol. Biol. 2014. [Google Scholar] [CrossRef]
- Cummings, M.; Sarveswaran, J.; Homer-Vanniasinkam, S.; Burke, D.; Orsi, N.M. Glyceraldehyde-3-phosphate dehydrogenase is an inappropriate housekeeping gene for normalising gene expression in sepsis. Inflammation 2014, 37, 1889–1894. [Google Scholar]
- Li, Q.Q.; Skinner, J.; Bennett, J.E. Evaluation of reference genes for real-time quantitative PCR studies in Candida glabrata following azole treatment. BMC Mol. Biol. 2012, 13, 22. [Google Scholar]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar]
- Gong, J.; Luo, Y.; Zhang, Z.; Wang, W.; Li, J. Effect of dexamethasone on expression of interleukin-21 and phospho-STAT3 in a murine model of chronic asthma. J. South Med. Univ. 2013, 3, 742–745. [Google Scholar]
- Clark, A.F.; Wilson, K.; McCartney, M.D.; Miggans, S.T.; Kunkle, M.; Howe, W. Glucocorticoid-induced formation of cross-linked actin networks in cultured human trabecular meshwork cells. Investig. Ophthalmol. Vis. Sci. 1994, 35, 281–294. [Google Scholar]
- Sanda, T.; Tyner, J.W.; Gutierrez, A.; Ngo, V.N.; Glover, J.; Chang, B.H.; Yost, A.; Ma, W.; Fleischman, A.G.; Zhou, W.; et al. TYK2-STAT1-BCL2 pathway dependence in T-cell acute lymphoblastic leukemia. Cancer Discov. 2013, 3, 564–577. [Google Scholar]
- Albi, E. The role of intranuclear lipids in health and disease. Clin. Lipidol. 2011, 6, 59–69. [Google Scholar]
- Takaoka, Y.; Goto, S.; Nakano, T.; Tseng, H.P.; Yang, S.M.; Kawamoto, S.; Ono, K.; Chen, C.L. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) prevents lipopolysaccharide (LPS)-induced, sepsis-related severe acute lung injury in mice. Sci. Rep. 2014, 4, 5204. [Google Scholar]
- Ren, R.; Oakley, R.H.; Cruz-Topete, D.; Cidlowski, J.A. Dual role for glucocorticoids in cardiomyocyte hypertrophy and apoptosis. Endocrinology 2012, 153, 5346–5360. [Google Scholar]
- Heessen, S.; Fornerod, M. The inner nuclear envelope as a transcription factor resting place. EMBO Rep. 2007, 8, 914–919. [Google Scholar]
- Pugliese, L.; Bernardini, I.; Pacifico, N.; Peverini, M.; Damaskopoulou, E.; Cataldi, S.; Albi, E. Severe hypocholesterolemia is often neglected in hematological malignancies. Eur. J. Cancer 2010, 46, 1735–1743. [Google Scholar]
- Marini, F.; Bartoccini, F.; Cascianelli, G.; Voccoli, V.; Caviglia, M.G.; Viola Magni, M.; Albi, E. Effect of 1α,25-Dihydroxyvitamin D3 in embryonic hippocampal cells. Hippocampus 2010, 20, 696–705. [Google Scholar]
- Albi, E.; la Porta, C.A.; Cataldi, S.; Magni, M.V. Nuclear sphingomyelin-synthase and protein kinase C delta in melanoma cells. Arch. Biochem. Biophys. 2005, 438, 156–161. [Google Scholar]
- Albi, E.; Cataldi, S.; Bartoccini, E.; Magni, M.V.; Marini, F.; Mazzoni, F.; Rainaldi, G.; Evangelista, M.; Garcia-Gil, M. Nuclear sphingomyelin pathway in serum deprivation-induced apoptosis of embryonic hippocampal cells. J. Cell Physiol. 2006, 206, 189–195. [Google Scholar]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cataldi, S.; Codini, M.; Cascianelli, G.; Tringali, S.; Tringali, A.R.; Lazzarini, A.; Floridi, A.; Bartoccini, E.; Garcia-Gil, M.; Lazzarini, R.; et al. Nuclear Lipid Microdomain as Resting Place of Dexamethasone to Impair Cell Proliferation. Int. J. Mol. Sci. 2014, 15, 19832-19846. https://doi.org/10.3390/ijms151119832
Cataldi S, Codini M, Cascianelli G, Tringali S, Tringali AR, Lazzarini A, Floridi A, Bartoccini E, Garcia-Gil M, Lazzarini R, et al. Nuclear Lipid Microdomain as Resting Place of Dexamethasone to Impair Cell Proliferation. International Journal of Molecular Sciences. 2014; 15(11):19832-19846. https://doi.org/10.3390/ijms151119832
Chicago/Turabian StyleCataldi, Samuela, Michela Codini, Giacomo Cascianelli, Sabina Tringali, Anna Rita Tringali, Andrea Lazzarini, Alessandro Floridi, Elisa Bartoccini, Mercedes Garcia-Gil, Remo Lazzarini, and et al. 2014. "Nuclear Lipid Microdomain as Resting Place of Dexamethasone to Impair Cell Proliferation" International Journal of Molecular Sciences 15, no. 11: 19832-19846. https://doi.org/10.3390/ijms151119832