Characterisation of Indica Special Protein (ISP), a Marker Protein for the Differentiation of Oryza sativa Subspecies indica and japonica
Abstract
:1. Introduction
2. Results
2.1. Comparative Analysis by Two-Dimensional Gel Electrophoresis (2-DE)
2.2. Mass Spectrometry Data Analysis
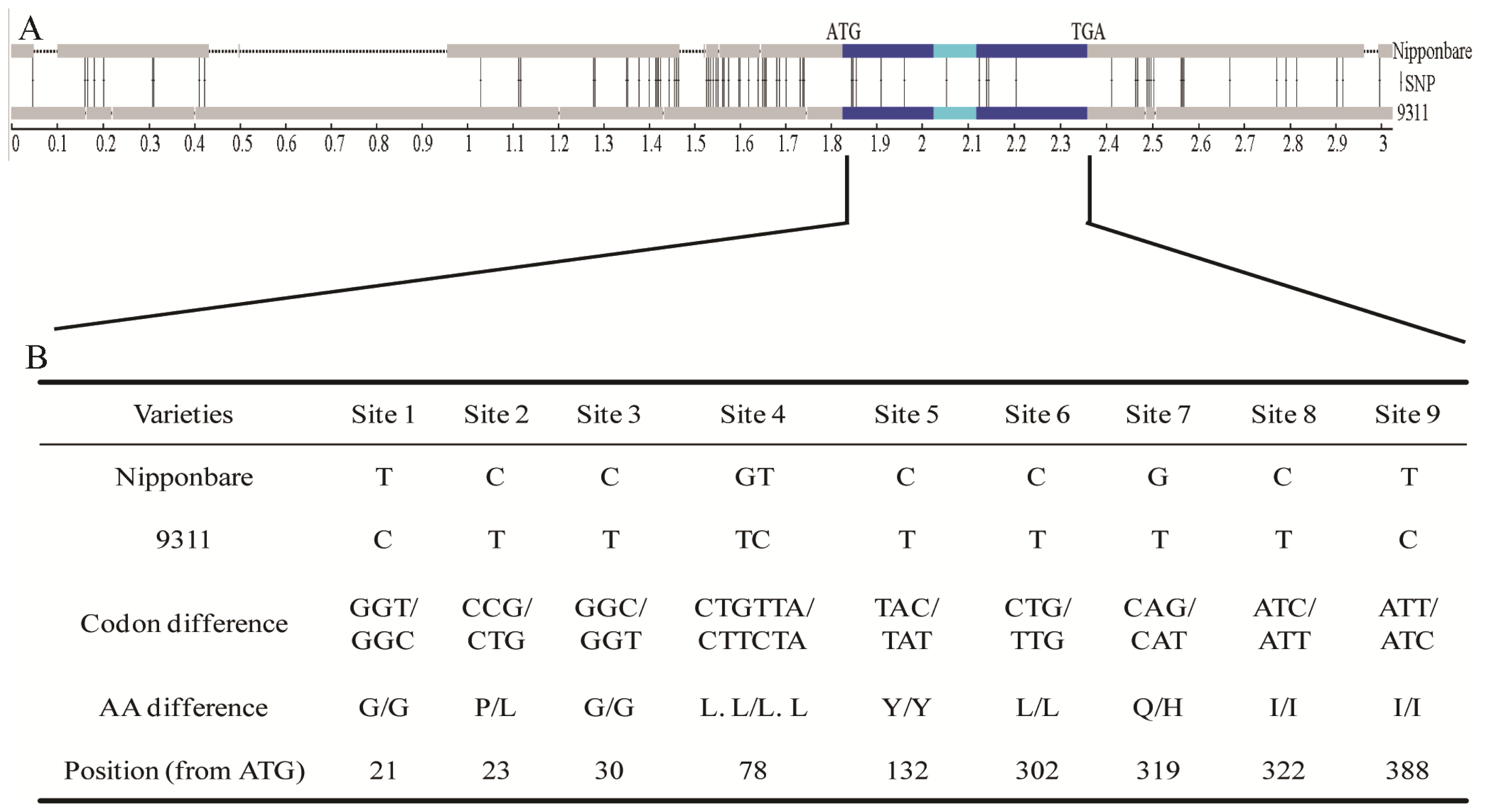
2.3. Nucleotide Variations and Protein Divergences in Indica Special Protein (ISP) Region
2.4. Expression of the ISP Gene
3. Discussion
4. Experimental Section
4.1. Plant Material
4.2. Protein Sample Preparation
4.3. Two-Dimensional Electrophoresis (2-DE)
4.4. In-Gel Digestion and Mass Spectrometry Analysis
4.5. Protein Identification
4.6. Gene Expression Analysis
5. Conclusions
Supplementary Information
ijms-15-07332-s001.pdfAcknowledgments
Conflicts of Interest
- Author ContributionsK.Z. performed most of the experiments and wrote the manuscript; C.M. helped K.Z. with analysis of 2-DE; H.X. and Y.Y. helped K.Z. with analysis of MS; B.W. collected the rice seedlings; K.C. supervised the study and designed the experiments. All the authors discussed the results and contributed to the manuscript.
References
- Goff, S.A.; Ricke, D.; Lan, T.-H.; Presting, G.; Wang, R.; Dunn, M.; Glazebrook, J.; Sessions, A.; Oeller, P.; Varma, H. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 2002, 296, 92–100. [Google Scholar]
- Sasaki, T.; Burr, B. International rice genome sequencing project: The effort to completely sequence the rice genome. Curr. Opin. Plant Biol 2000, 3, 138–142. [Google Scholar]
- Yu, J.; Hu, S.; Wang, J.; Wong, G.K.-S.; Li, S.; Liu, B.; Deng, Y.; Dai, L.; Zhou, Y.; Zhang, X.; et al. A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 2002, 296, 79–92. [Google Scholar]
- Shimamoto, K.; Kyozuka, J. Rice as a model for comparative genomics of plants. Annu. Rev. Plant Biol 2002, 53, 399–419. [Google Scholar]
- Kato, S.; Kosaka, H.; Hara, S. On the affinity of rice varieties as shown by fertility of hybrid plants. Bull. Sci. Fac. Agric. Kyushu Univ 1928, 3, 132–147. [Google Scholar]
- Oka, H.I. Origin of Cultivated Rice; Japan Scientific Societies Press: Tokyo, Japan, 1988. [Google Scholar]
- Zhang, Q.; Maroof, M.S.; Lu, T.; Shen, B. Genetic diversity and differentiation of indica and japonica rice detected by RFLP analysis. Theor. Appl. Genet 1992, 83, 495–499. [Google Scholar]
- Zhang, Q.; Liu, K.; Yang, G.; Maroof, M.S.; Xu, C.; Zhou, Z. Molecularmarker diversity and hybrid sterility in indica–japonica rice crosses. Theor. Appl. Genet 1997, 95, 112–118. [Google Scholar]
- Ting, Y. The origin and evolution of cultivated rice in China. Acta Agron. Sin 1957, 8, 243–260. [Google Scholar]
- Chang, T.-T. The origin, evolution, cultivation, dissemination, and diversification of Asian and African rices. Euphytica 1976, 25, 425–441. [Google Scholar]
- Second, G. Origin of the genic diversity of cultivated rice (Oryza spp.): Study of the polymorphism scored at 40 isozyme loci. Jpn. J. Genet 1982, 57, 25–57. [Google Scholar]
- Bautista, N.S.; Solis, R.; Kamijima, O.; Ishii, T. RAPD, RFLP and SSLP analyses of phylogenetic relationships between cultivated and wild species of rice. Genes Genet. Syst 2001, 76, 71–79. [Google Scholar]
- Cheng, C.; Motohashi, R.; Tsuchimoto, S.; Fukuta, Y.; Ohtsubo, H.; Ohtsubo, E. Polyphyletic origin of cultivated rice: Based on the interspersion pattern of SINEs. Mol. Biol. Evol 2003, 20, 67–75. [Google Scholar]
- Negrao, S.; Oliveira, M.; Jena, K.; Mackill, D. Integration of genomic tools to assist breeding in the japonica subspecies of rice. Mol. Breed 2008, 22, 159–168. [Google Scholar]
- Chang, T.; Oka, H. Genetic variousness in the climatic adaptation of rice cultivars. Proceedings of the International Symposium on Climate and Rice, Los Banos, Philippines, 1976; pp. 87–111.
- Cheng, K.; Wang, X.; Lu, Y. The synthetical research and utilization of Yunnan rice resource: The reorganization of the Asian cultivated rice classification. Acta Agron. Sin 1984, 10, 271–279. [Google Scholar]
- Harushima, Y.; Nakagahra, M.; Yano, M.; Sasaki, T.; Kurata, N. Diverse variation of reproductive barriers in three intraspecific rice crosses. Genetics 2002, 160, 313–322. [Google Scholar]
- McCouch, S.; Kochert, G.; Yu, Z.; Wang, Z.; Khush, G.; Coffman, W.; Tanksley, S. Molecular mapping of rice chromosomes. Theor. Appl. Genet 1988, 76, 815–829. [Google Scholar]
- Wang, Z.; Tanksley, S. Restriction fragment length polymorphism in Oryza sativa L. Genome 1989, 32, 1113–1118. [Google Scholar]
- Matsumoto, T.; Wu, J.; Kanamori, H.; Katayose, Y.; Fujisawa, M.; Namiki, N.; Mizuno, H.; Yamamoto, K.; Antonio, B.A.; Baba, T. The map-based sequence of the rice genome. Nature 2005, 436, 793–800. [Google Scholar]
- Kovach, M.J.; Sweeney, M.T.; McCouch, S.R. New insights into the history of rice domestication. Trends Genet 2007, 23, 578–587. [Google Scholar]
- Du, H.; Ouyang, Y.; Zhang, C.; Zhang, Q. Complex evolution of S5, a major reproductive barrier regulator, in the cultivated rice Oryza sativa and its wild relatives. New Phytol 2011, 191, 275–287. [Google Scholar]
- Huffaker, R.C. Biochemistry and physiology of leaf proteins. Nucleic Acids Proteins Plants I 1982, 14, 370–400. [Google Scholar]
- Sharma, S.; Tripathy, S.; Biswal, J. Origin of O. sativa and its ecotypes. In Rice Breeding and Genetics: Research Priorities and Challenges; Science Publishers, Enfield/Oxford and IBH: New Delhi, India, 2000; pp. 349–369. [Google Scholar]
- Chang, T.-T. Origin, domestication, and diversification. In Rice Origin, History, Technology, and Production; J. Wiley: Hoboken, NJ, USA, 2002; pp. 3–25. [Google Scholar]
- Sweeney, M.; McCouch, S. The complex history of the domestication of rice. Ann. Bot 2007, 100, 951–957. [Google Scholar]
- Claes, B.; Dekeyser, R.; Villarroel, R.; van den Bulcke, M.; Bauw, G.; van Montagu, M.; Caplan, A. Characterization of a rice gene showing organ-specific expression in response to salt stress and drought. Plant Cell 1990, 2, 19–27. [Google Scholar]
- Chen, Z.; Zhong, W.; Yang, J.; Huang, Z. Evaluation of salt tolerance of rice (Oryza sativa L.) germplasm. J. Plant Genet. Res 2004, 5, 351–355. (In Chinese) [Google Scholar]
- Zhang, Y.; IH, S. Salt-tolerant variation in Asian cultivated rice analyzed from classificatory and geographical differences. Xinan Nongye Xuebao 2000, 13, 6–11. (In Chinese) [Google Scholar]
- Londo, J.P.; Chiang, Y.-C.; Hung, K.-H.; Chiang, T.-Y.; Schaal, B.A. Phylogeography of Asian wild rice, Oryza rufipogon, reveals multiple independent domestications of cultivated rice, Oryza sativa. Proc. Natl. Acad. Sci. USA 2006, 103, 9578–9583. [Google Scholar]
- Kawasaki, S.; Borchert, C.; Deyholos, M.; Wang, H.; Brazille, S.; Kawai, K.; Galbraith, D.; Bohnert, H.J. Gene expression profiles during the initial phase of salt stress in rice. Plant Cell 2001, 13, 889–906. [Google Scholar]
- Garcia, A.B.; Engler, J.A.; Claes, B.; Villarroel, R.; van Montagu, M.; Gerats, T.; Caplan, A. The expression of the salt-responsive gene sal T from rice is regulated by hormonal and developmental cues. Planta 1998, 207, 172–180. [Google Scholar]
- De Souza Filho, G.A.; Ferreira, B.S.; Dias, J.M.; Queiroz, K.S.; Branco, A.T.; Bressan-Smith, R.E.; Oliveira, J.G.; Garcia, A.B. Accumulation of SALT protein in rice plants as a response to environmental stresses. Plant Sci 2003, 164, 623–628. [Google Scholar]
- Kim, S.T.; Kim, S.G.; Hwang, D.H.; Kang, S.Y.; Koo, S.C.; Cho, M.J.; Kang, K.Y. Expression of a salt-induced protein (SALT) in suspension-cultured cells and leaves of rice following exposure to fungal elicitor and phytohormones. Plant Cell Rep 2004, 23, 256–262. [Google Scholar]
- Ting, C.-T.; Tsaur, S.-C.; Wu, M.-L.; Wu, C.-I. A rapidly evolving homeobox at the site of a hybrid sterility gene. Science 1998, 282, 1501–1504. [Google Scholar]
- Presgraves, D.C.; Balagopalan, L.; Abmayr, S.M.; Orr, H.A. Adaptive evolution drives divergence of a hybrid inviability gene between two species of Drosophila. Nature 2003, 423, 715–719. [Google Scholar]
- Barbash, D.A.; Awadalla, P.; Tarone, A.M. Functional divergence caused by ancient positive selection of a Drosophila hybrid incompatibility locus. PLoS Biol 2004, 2, e142. [Google Scholar]
- Brideau, N.J.; Flores, H.A.; Wang, J.; Maheshwari, S.; Wang, X.; Barbash, D.A. Two Dobzhansky-Muller genes interact to cause hybrid lethality in Drosophila. Science 2006, 314, 1292–1295. [Google Scholar]
- Oliver, P.L.; Goodstadt, L.; Bayes, J.J.; Birtle, Z.; Roach, K.C.; Phadnis, N.; Beatson, S.A.; Lunter, G.; Malik, H.S.; Ponting, C.P. Accelerated evolution of the Prdm9 speciation gene across diverse metazoan taxa. PLoS Genet 2009, 5, e1000753. [Google Scholar]
- Cilia, M.; Fish, T.; Yang, X.; McLaughlin, M.; Thannhauser, T.W.; Gray, S. A comparison of protein extraction methods suitable for gel-based proteomic studies of aphid proteins. JBT 2009, 20, 201–215. [Google Scholar]
- Kim, Y.; Nandakumar, M.P.; Marten, M.R. Proteome map of Aspergillus nidulans during osmoadaptation. Fungal Genet. Biol 2007, 44, 886–895. [Google Scholar]
- Liang, Y.U.; Chen, H.U.I.; Tang, M.; Shen, S. Proteome analysis of an ectomycorrhizal fungus Boletus edulis under salt shock. Mycol. Res 2007, 111, 939–946. [Google Scholar]
- Zhou, Z.; Yang, H.; Chen, M.; Lou, C.; Zhang, Y.; Chen, K.; Wang, Y.; Yu, M.; Yu, F.; Li, J. Comparative proteomic analysis between the domesticated silkworm (Bombyx mori) reared on fresh mulberry leaves and on artificial diet. J. Proteome Res 2008, 7, 5103–5111. [Google Scholar]
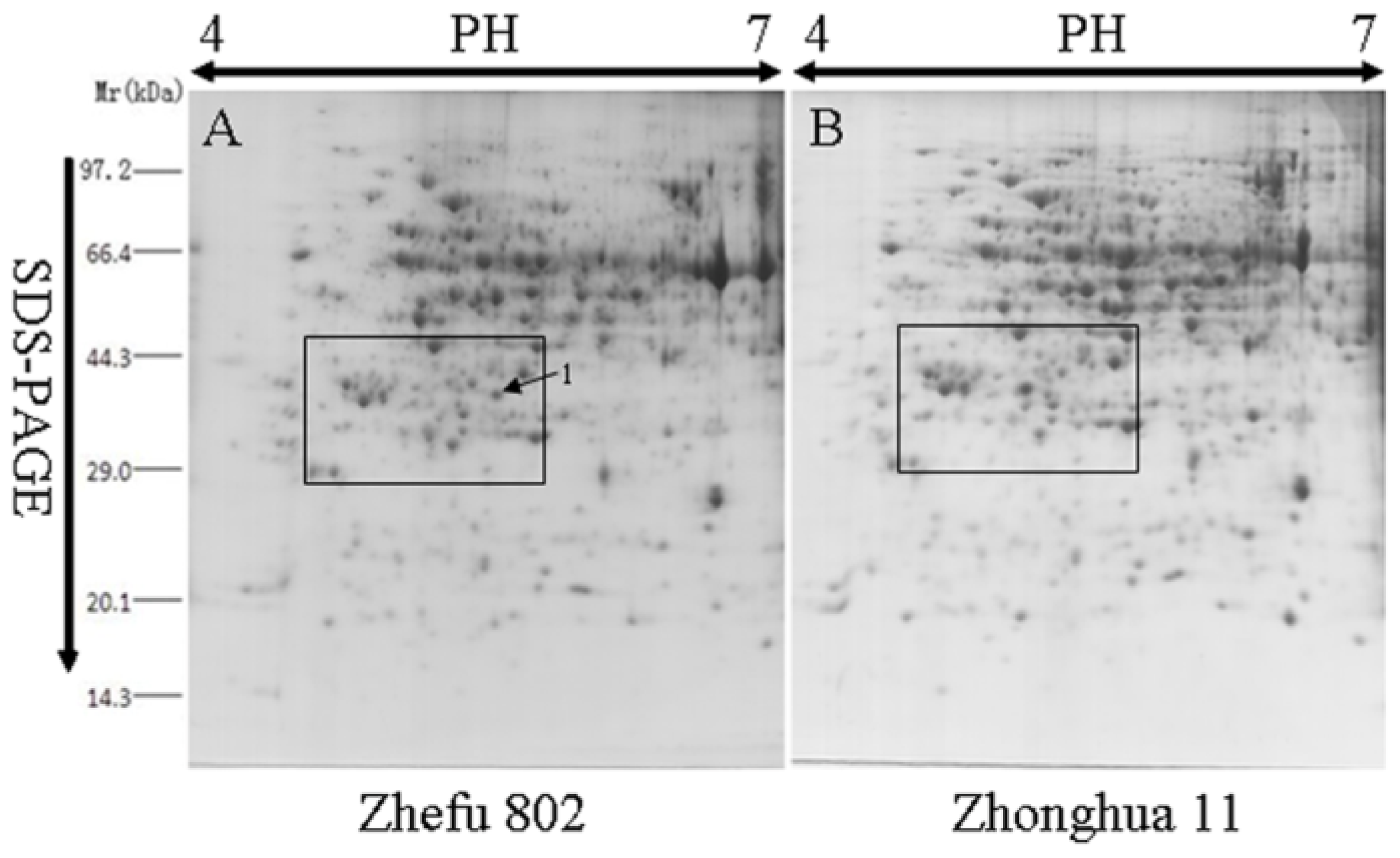

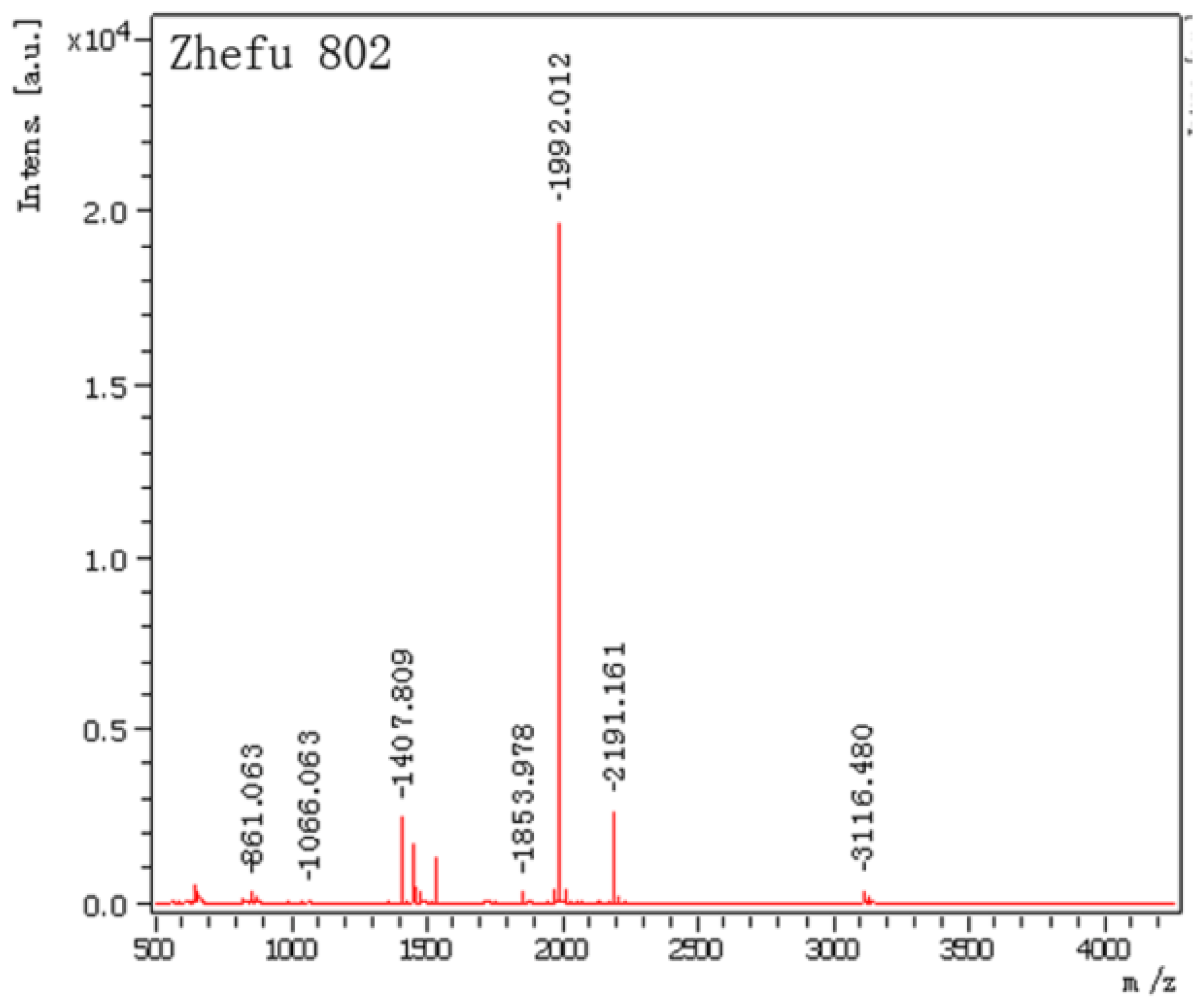


| Spot No. | Protein name | Monoisotopic masses | Matched peptides |
|---|---|---|---|
| 1 | salt-induced protein (salT) (score: 100 protein sequence coverage: 66%) | 1535.9189 | K.KLLGVTIYSSDAIR.S |
| 1407.8090 | K.LLGVTIYSSDAIR.S | ||
| 3116.4797 | R.SIAFNYIGVDGQEYAIGPWGGGEGTSTEIK.L | ||
| 2191.1609 | K.EISGTHGPVYDLADIVTYLK.I | ||
| 1992.0117 | K.EFSIPLQDSGHVVGFFGR.S | ||
| 1455.8126 | R.SGTLIDAIGIYVHP. |
© 2014 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Zhu, K.; Min, C.; Xia, H.; Yang, Y.; Wang, B.; Chen, K. Characterisation of Indica Special Protein (ISP), a Marker Protein for the Differentiation of Oryza sativa Subspecies indica and japonica. Int. J. Mol. Sci. 2014, 15, 7332-7343. https://doi.org/10.3390/ijms15057332
Zhu K, Min C, Xia H, Yang Y, Wang B, Chen K. Characterisation of Indica Special Protein (ISP), a Marker Protein for the Differentiation of Oryza sativa Subspecies indica and japonica. International Journal of Molecular Sciences. 2014; 15(5):7332-7343. https://doi.org/10.3390/ijms15057332
Chicago/Turabian StyleZhu, Keming, Chao Min, Hengchuan Xia, Yanhua Yang, Bin Wang, and Keping Chen. 2014. "Characterisation of Indica Special Protein (ISP), a Marker Protein for the Differentiation of Oryza sativa Subspecies indica and japonica" International Journal of Molecular Sciences 15, no. 5: 7332-7343. https://doi.org/10.3390/ijms15057332
APA StyleZhu, K., Min, C., Xia, H., Yang, Y., Wang, B., & Chen, K. (2014). Characterisation of Indica Special Protein (ISP), a Marker Protein for the Differentiation of Oryza sativa Subspecies indica and japonica. International Journal of Molecular Sciences, 15(5), 7332-7343. https://doi.org/10.3390/ijms15057332





