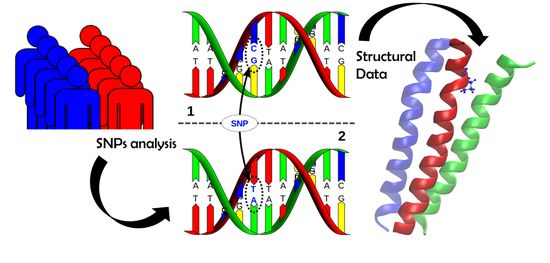
Computational and Experimental Approaches to Reveal the Effects of Single Nucleotide Polymorphisms with Respect to Disease Diagnostics
Abstract
:1. Introduction
2. In Silico Analysis of Pathogenic Mutations
2.1. Sequence and Evolutionary Analyses and Machine Learning Methods
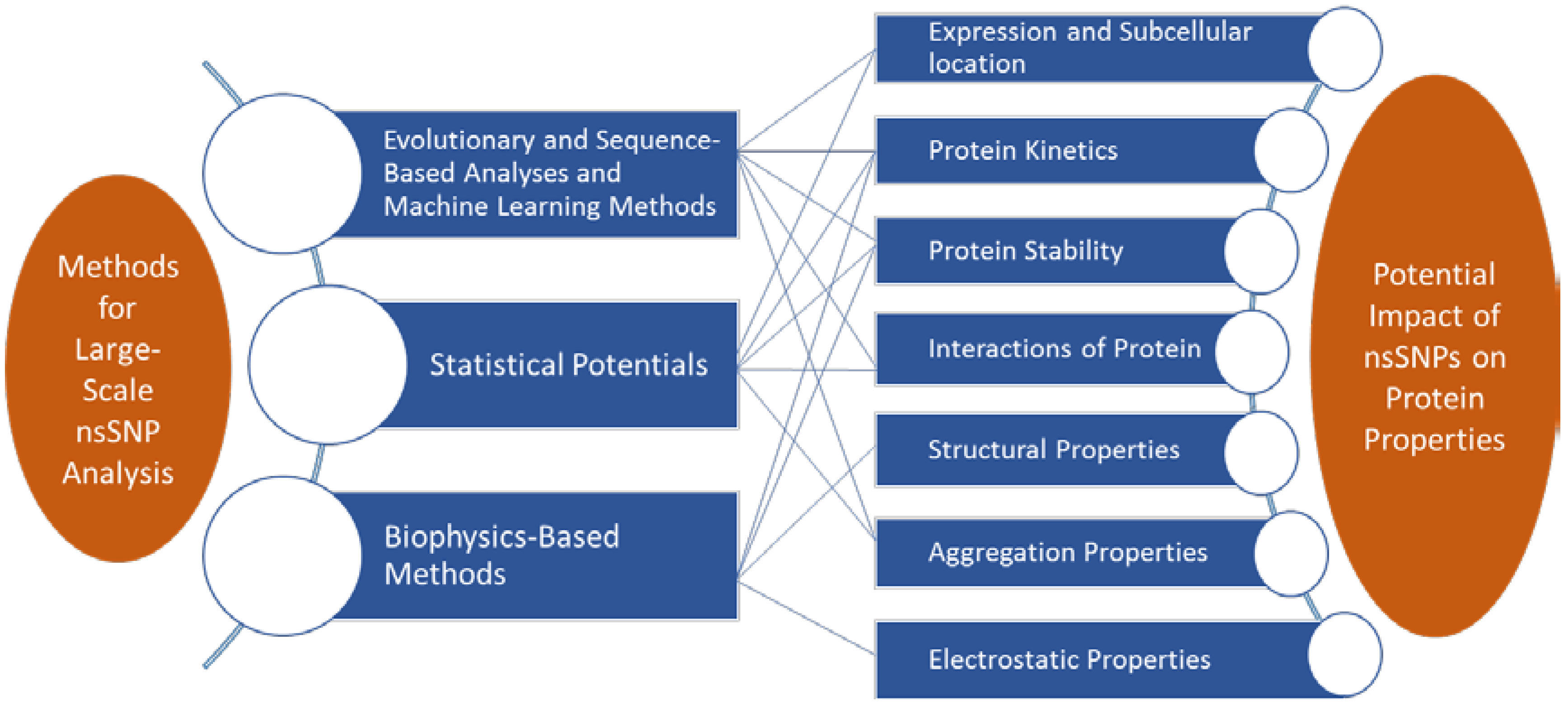
2.2. Statistical Potentials
2.3. Biophysics-Based Methods
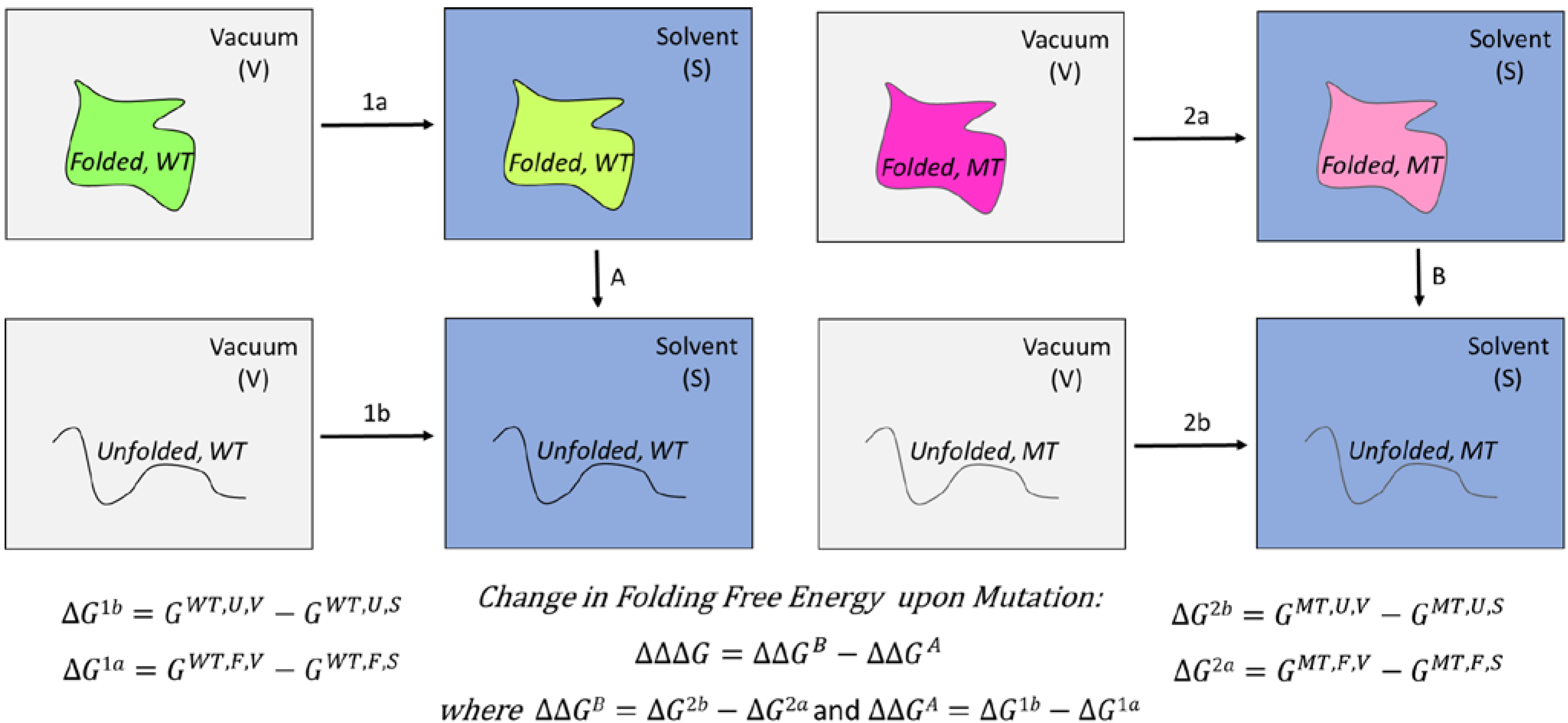
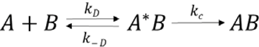
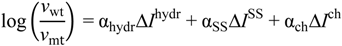
3. In Vitro Analysis of Pathogenic Mutations
3.1. Single Nucleotide Polymorphisms (SNPs)—Detection Methods and Technology
3.1.1. Traditional Methods for SNP Genotyping
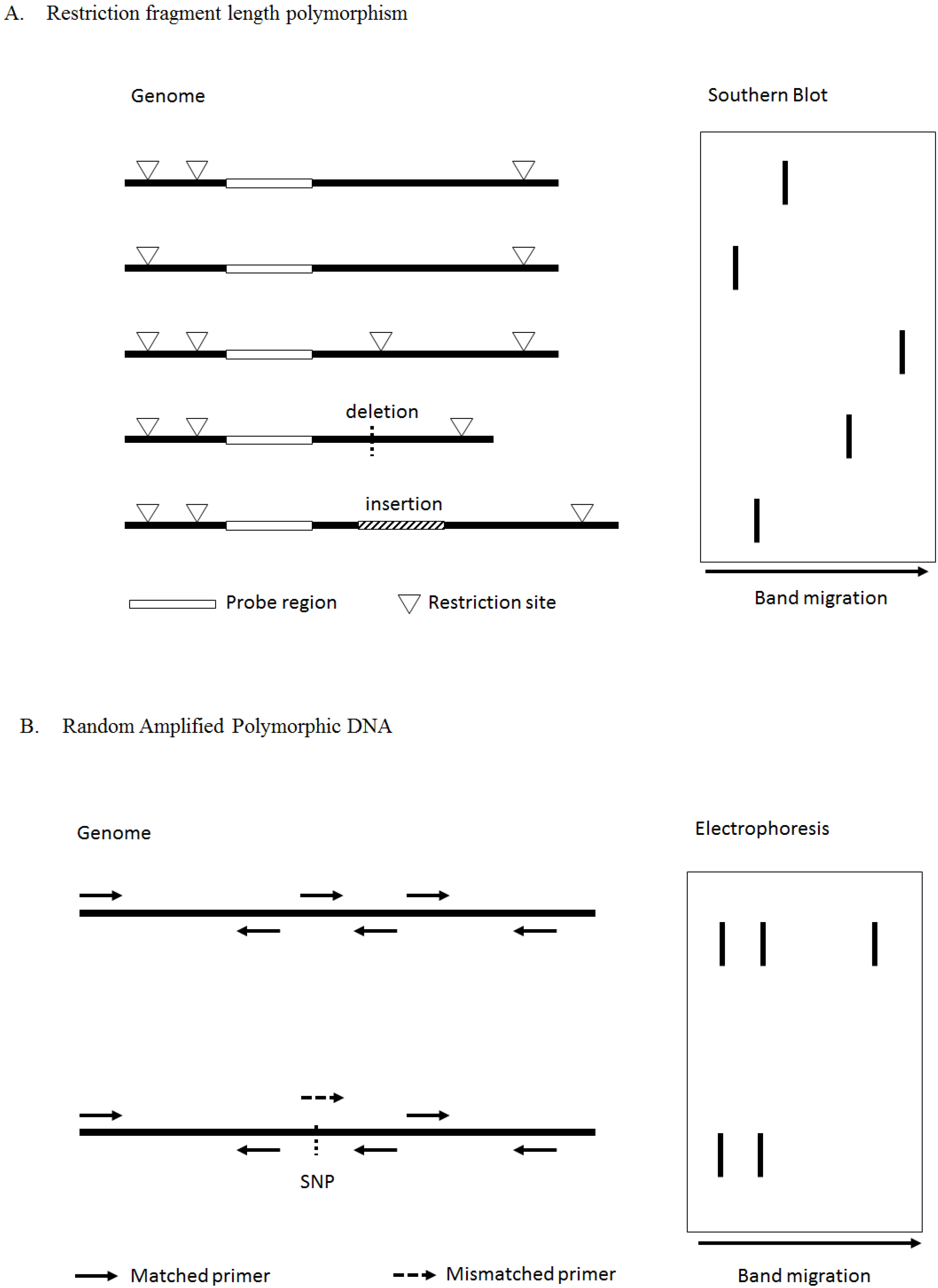
3.1.2. High-Throughput Method for SNP Genotyping
3.2. Functional SNP Screening
| SNP Loci | Subject of Investigation | Methods |
|---|---|---|
| Non-coding region | Gene transcription | Chromatin immunoprecipitation; Electrophoretic Mobility Shift Assay; Dual-luciferase assay; Real Time PCR |
| mRNA degradation | ||
| Gene translation | ||
| Gene expression | Immunoblotting; Enzyme-Linked Immunosorbent Assay; Immunofluorescence | |
| Coding region | Protein structure | Crystallization; Nuclear Magnetic Resonance |
| Conformation Stability | Circular Dichroism Spectroscopy; Intrinsic Fluorescence | |
| Kinetics | Binding assays; Enzyme assays |
| Year | Results | Methods | Reference |
|---|---|---|---|
| 1992 | MECP2 was firstly identified in human X chromosome. MeCP2 belongs to MBD protein family, and binds to methylated DNA | Immunofluorescence; Southwestern Assay | [194] |
| 1997 | MeCP2 acts as a global transcriptional repressor | In vitro Transcription Assay; Western Blot | [195] |
| 1999 | Four mutations of MECP2 gene causing Rett syndrome were identified | Conformation-sensitive gel electrophoresis; DNA sequencing | [196] |
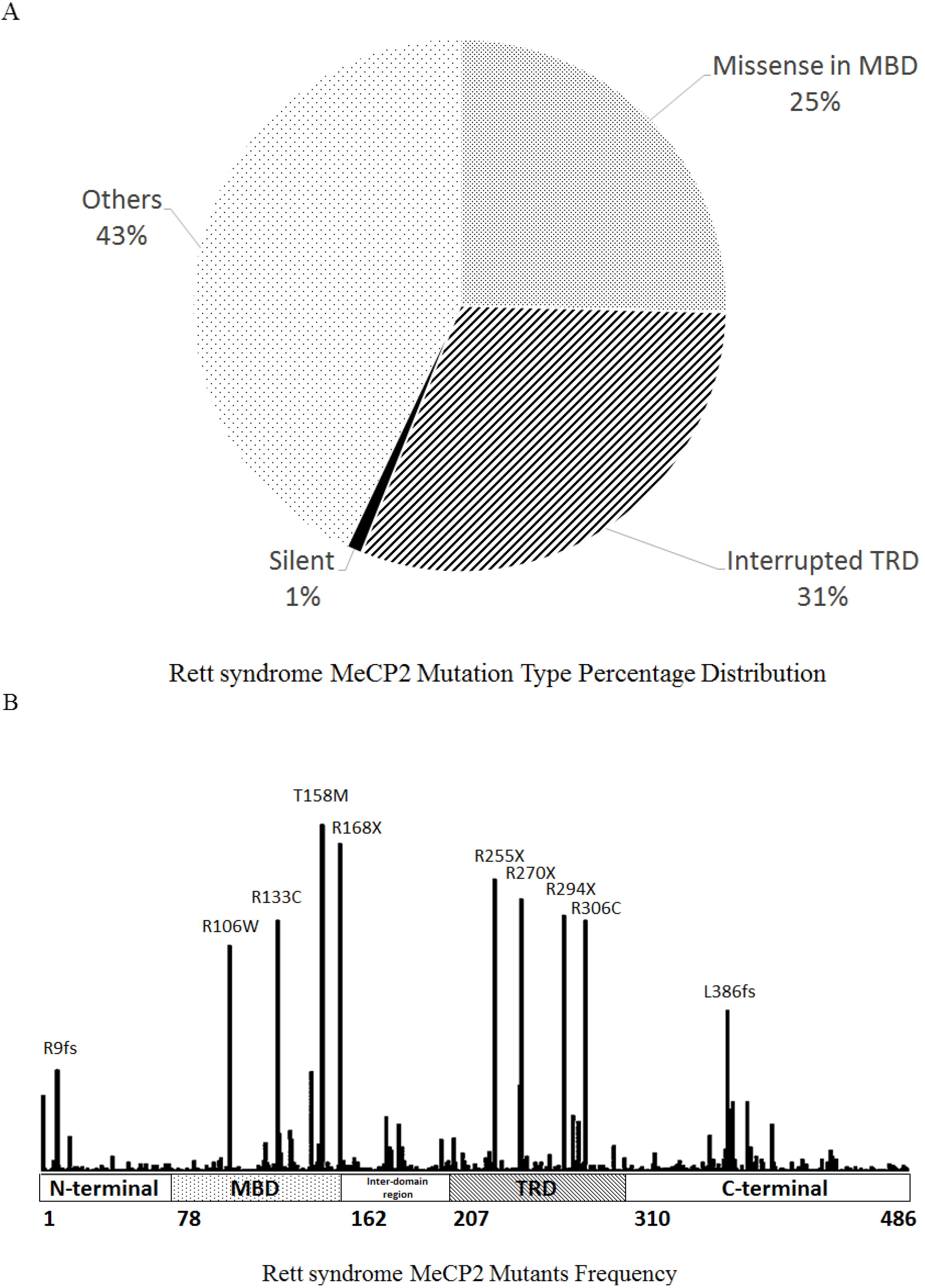
| 2000 | Missense mutations in MBD domain lost 5mC binding specific. Interruption of TRD domain lost repression to various genes | Southwestern assay; In vitro Transcription Assay | [197] |
| 2003 | MeCP2 regulate BDNF gene by binding to promoter IV | EMSA; ChIP; Real-time reverse transcription PCR | [198] |
| 2003 | MeCP2 compact chromatin via its three AT-hook domain | EMSA; Sedimentation Velocity; Electron Microscopy | [199] |
| 2005 | MeCP2 bind to four-way junction DNA in a structure-specific methyl-CpG-independent manner | Gel-retardation assays | [200] |
| 2008 | Mutational hotspots have effects on MeCP2 stability | Fluorescence Spectroscopy; Circular Dichroism | [201] |
| 2012 | MeCP2 bind specifically to 5-hydroxymethylytosine, enriched in the target gene promoter | Substrate Affinity pull-Down; Methyl-DNA-IP Sequencing | [202] |
| 2013 | Mutational hotspot R306C, proximal to the T308 Phosphorylation site, abolishes the binding to the NCoR complex | Phosphotryptic-mapping; Peptide binding assay; Immunofluorescence | [203] |
| 2013 | MeCP2 compact chromatin via its three AT-hook domain | Real-time reverse transcription PCR; Immunofluorescence | [204] |
4. In Vivo Analysis of Pathogenic Mutations
4.1. Zebrafish as a Model for Studying the Downstream Effects of Non-Synonymous Single Nucleotide Polymorphism (nsSNPs)
4.2. Gene Editing to Introduce Individual nsSNPs within the Genome
5. Case Study: KDM5C and MECP2 Genes
| Webservers | Input | Method Summary |
|---|---|---|
| MuStab | Sequence | Support vector machine (SVM), trained on various amino acid features |
| dFIRE/DFIRE2 | Structure | Statistical potentials with orientation-dependent interactions |
| FoldX | Structure | Potential with weighted sum of empirical contributions to free energy |
| Eris | Structure | Molecular mechanics type potential with weighted sum of contributions to free energy |
| MECP2 Mutants | ∆∆G (kcal/mol) | Effect, Confidence | |||||||
|---|---|---|---|---|---|---|---|---|---|
| dDFIRE (1qk9) | DFIRE2 (1qk9) | dDFIRE (3c2i) | DFIRE (3c2i) | Eris (1qk9) | Eris (3c2i) | FoldX (1qk9) | FoldX (3c2i) | MuStab | |
| D97Y | −1.76 | −2.36 | −0.73 | −1.36 | >10 | −2.08 | 5.28 | 1.28 | Stabilizing, 26% |
| L100R | 5.95 | 3.89 | 3.66 | 2.10 | 2.68 | 3.93 | 2.3 | 2.13 | Destabilizing, 90% |
| R106W | −1.47 | −2.24 | −0.63 | −2.13 | >10 | 9.71 | 10.9 | 4.12 | Stabilizing, 27% |
| R111G | 1.73 | 1.01 | 2.15 | 1.51 | 3.22 | 8.57 | 1.56 | 1.71 | Destabilizing, 94% |
| Y120P | 2.69 | 2.30 | 3.66 | 2.45 | −8.67 | 7.48 | 0.53 | 1.95 | Destabilizing, 90% |
| Q128P | 1.59 | 0.52 | 0.65 | 0.57 | >10 | 8 | 1.34 | 0.12 | Destabilizing, 92% |
| E137G | 2.68 | 1.19 | 3.13 | 1.63 | 3.86 | 5.41 | 2.22 | 2.29 | Destabilizing, 88% |
| P152R | 7.54 | 3.74 | 2.05 | 1.13 | >10 | 0.44 | 2.28 | 1.95 | Destabilizing, 81% |
| F155S | 5.16 | 4.38 | 6.62 | 5.17 | 6.8 | 5.04 | 5.73 | 4.03 | Destabilizing, 93% |
| KDM5C Mutant | dDFIRE (2jrz) | DFIRE2 (2jrz) | Eris (2jrz) | FoldX (2jrz) | MuStab | ||||
| D87G | 0.34 | 0.994 | 4.48 | 0.53 | Destabilizing, 94% | ||||
| KDM5C Mutant | dDFIRE (3dxt) | DFIRE2 (3dxt) | dDFIRE (4igo) | DFIRE2 (4igo) | Eris (3dxt) | Eris (4igo) | FoldX (3dxt) | FoldX (4igo) | MuStab |
| D402Y | −0.27 | −1.379 | 0.19 | −0.741 | −4.14 | −1.49 | −1.64 | −0.6 | Stabilizing, 27% |
6. Conclusions
Acknowledgments
Author Contributions
Abbreviations
| MeCP2 | methyl CpG binding protein 2 |
| nsSNP | non-synonymous single nucleotide polymorphism |
| NGS | Next Generation Sequencing |
| RFLP | Restriction Fragment Length Polymorphism |
| SSRs | Simple Sequence Repeats |
| PCR | Polymerase Chain Reaction |
| ASPE | Allele-Specific Primer Extension |
| SBE | Single Base Extension |
| FRET | fluorescence resonance energy transfer |
| L-RAC | Ligation-rolling Circle Amplification |
| MIP | Molecular Inversion Probe |
| UTR | Untranslated Regions |
| EMSA | Electrophoretic Mobility Shift Assay |
| SDS-PAGE | Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis |
| ChIP | Chromatin Immunoprecipitation |
| DLR | Dual-luciferase Reporter Assay System |
| NMR | Magnetic Resonance Spectroscopy |
| CD | Circular Dichroism |
| ATP | Adenosine Triphosphate |
| MBD | Methyl-CpG-binding domain |
| TRD | Transcription Repression Domain |
| ARID | AT-rich interaction domain |
| DNA | Deoxyribonucleic Acid |
| RNA | Ribonucleic acid |
| SVM | Support Vector Machine |
| GOASVM | Gene Ontology Annotation SVM |
| MLSTA | Machine Learning for Protein Stability |
| DFIRE | Distance-Scaled, Finite Ideal Gas Reference |
| PoPMuSiC | Prediction of Protein Mutant Stability Changes |
| dDFIRE | dipolar DFIRE |
| SNPs&GO | SNPs and Gene Ontology |
| CELLO | Subcellular Localization Predictor |
| ngLOC | n-gram-based Bayesian Localization Predictor |
| PPT-DB | Protein Property Prediction and Testing Database |
| PROFcon | Prediction of long-range Contacts |
| ProA-RF | Protein Aggregation Prediction Server-Random Forest |
| ProA-SVM | Protein Aggregation Prediction Server-Support Vector Machine |
| PASTA | Prediction of Amyloid Structure Aggregation |
| SDM | Site Directed Mutator |
| BONGO | Bonds on Graphs |
| MD | Molecular Dynamics |
| LIE | Linear Interaction Energy |
| NCBI | National Center for Biotechnology Information |
| BLAST | Basic Local Alignment Search Tool |
| PDB | Protein Databank |
| VMD | Visual Molecular Dynamics |
| NAMD | Nanoscale Molecular Dynamics |
| MuStab | Predicting Protein Mutant Stability Change |
| SIFT | Sorting Intolerant from Tolerant |
| MAPP | Multivariate Analysis of Protein Polymorphism |
| Align-GVGD | Alignment Grantham-Variation, Grantham-Deviation |
Conflicts of Interest
References and Notes
- Potapov, V.; Cohen, M.; Schreiber, G. Assessing computational methods for predicting protein stability upon mutation: good on average but not in the details. Protein Eng. Des. Sel. 2009, 22, 553–560. [Google Scholar] [CrossRef]
- Thusberg, J.; Vihinen, M. Pathogenic or Not? And if so, then how? Studying the effects of missense mutations using bioinformatics methods. Hum. Mutat. 2009, 30, 703–714. [Google Scholar] [CrossRef]
- Gonzalez-Castejon, M.; Marin, F.; Soler-Rivas, C.; Reglero, G.; Visioli, F.; Rodriguez-Casado, A. Functional non-synonymous polymorphisms prediction methods: Current approaches and future developments. Curr. Med. Chem. 2011, 18, 5095–5103. [Google Scholar] [CrossRef]
- Thiltgen, G.; Goldstein, R.A. Assessing predictors of changes in protein stability upon mutation using self-consistency. PLoS One 2012, 7, e46084. [Google Scholar] [CrossRef]
- Zhang, Z.; Miteva, M.A.; Wang, L.; Alexov, E. Analyzing effects of naturally occurring missense mutations. Comput. Math. Methods Med. 2012, 2012, 805827:1–805827:15. [Google Scholar]
- Stefl, S.; Nishi, H.; Petukh, M.; Panchenko, A.R.; Alexov, E. Molecular mechanisms of disease-causing missense mutations. J. Mol. Biol. 2013, 425, 3919–3936. [Google Scholar] [CrossRef]
- Peterson, T.A.; Doughty, E.; Kann, M.G. Towards Precision Medicine: Advances in computational approaches for the analysis of human variants. J. Mol. Biol. 2013, 425, 4047–4063. [Google Scholar] [CrossRef]
- Chang, C.C.H.; Tey, B.T.; Song, J.; Ramanan, R.N. Towards more accurate prediction of protein folding rates: A review of the existing web-based bioinformatics approaches. Brief. Bioinform. 2014. [Google Scholar] [CrossRef]
- Sander, C.; Schneider, R. Database of homology-derived protein structures and the structural meaning of sequence alignment. Proteins 1991, 9, 56–68. [Google Scholar] [CrossRef]
- Rost, B. Twilight zone of protein sequence alignments. Protein Eng. 1999, 12, 85–94. [Google Scholar] [CrossRef]
- Ng, P.C.; Henikoff, S. Predicting deleterious amino acid substitutions. Genome Res. 2001, 11, 863–874. [Google Scholar] [CrossRef]
- Ng, P.C.; Henikoff, S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003, 31, 3812–3814. [Google Scholar] [CrossRef]
- De Baets, G.; van Durme, J.; Reumers, J.; Maurer-Stroh, S.; Vanhee, P.; Dopazo, J.; Schymkowitz, J.; Rousseau, F. SNPeffect 4.0: On-line prediction of molecular and structural effects of protein-coding variants. Nucleic Acids Res. 2012, 40, D935–D939. [Google Scholar] [CrossRef]
- Yue, P.; Moult, J. Identification and analysis of deleterious human SNPs. J. Mol. Biol. 2006, 356, 1263–1274. [Google Scholar] [CrossRef]
- Tavtigian, S.V.; Deffenbaugh, A.M.; Yin, L.; Judkins, T.; Scholl, T.; Samollow, P.B.; de Silva, D.; Zharkikh, A.; Thomas, A. Comprehensive statistical study of 452 BRCA1 missense substitutions with classification of eight recurrent substitutions as neutral. J. Med. Genet. 2006, 43, 295–305. [Google Scholar]
- Reva, B.; Antipin, Y.; Sander, C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 2011, 39, E118. [Google Scholar] [CrossRef]
- Stone, E.A.; Sidow, A. Physicochemical constraint violation by missense substitutions mediates impairment of protein function and disease severity. Genome Res. 2005, 15, 978–986. [Google Scholar] [CrossRef]
- Adzhubei, I.A.; Schmidt, S.; Peshkin, L.; Ramensky, V.E.; Gerasimova, A.; Bork, P.; Kondrashov, A.S.; Sunyaev, S.R. A method and server for predicting damaging missense mutations. Nat. Methods 2010, 7, 248–249. [Google Scholar]
- Schwarz, J.M.; Roedelsperger, C.; Schuelke, M.; Seelow, D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat. Methods 2010, 7, 575–576. [Google Scholar]
- Ryan, M.; Diekhans, M.; Lien, S.; Liu, Y.; Karchin, R. LS-SNP/PDB: annotated non-synonymous SNPs mapped to Protein Data Bank structures. Bioinformatics 2009, 25, 1431–1432. [Google Scholar]
- Teng, S.; Srivastava, A.K.; Wang, L. Sequence feature-based prediction of protein stability changes upon amino acid substitutions. BMC Genomics 2010, 11, S5. [Google Scholar]
- Teng, S.; Srivastava, A.K.; Wang, L. Biological features for sequence-based prediction of protein stability changes upon amino acid substitutions. In Proceedings of the 2009 International Joint Conference on BioinformaticsSystems Biology and Intelligent Computing; Zhang, J., Li, G.Z., Yang, J.Y., Eds.; IEEE Computer Society: Washington, DC, USA, 2009; pp. 201–206. [Google Scholar]
- Cheng, J.L.; Randall, A.; Baldi, P. Prediction of protein stability changes for single-site mutations using support vector machines. Proteins 2006, 62, 1125–1132. [Google Scholar] [CrossRef]
- Li, B.; Krishnan, V.G.; Mort, M.E.; Xin, F.; Kamati, K.K.; Cooper, D.N.; Mooney, S.D.; Radivojac, P. Automated inference of molecular mechanisms of disease from amino acid substitutions. Bioinformatics 2009, 25, 2744–2750. [Google Scholar] [CrossRef]
- Schaefer, C.; Meier, A.; Rost, B.; Bromberg, Y. SNPdbe: Constructing an nsSNP functional impacts database. Bioinformatics 2012, 28, 601–602. [Google Scholar] [CrossRef]
- Johansen, M.B.; Izarzugaza, J.M.G.; Brunak, S.; Petersen, T.N.; Gupta, R. Prediction of disease causing non-synonymous SNPs by the Artificial Neural Network Predictor NetDiseaseSNP. PLoS One 2013, 8, e68370. [Google Scholar]
- Venselaar, H.; Te Beek, T.A.; Kuipers, R.K.; Hekkelman, M.L.; Vriend, G. Protein structure analysis of mutations causing inheritable diseases. An e-Science approach with life scientist friendly interfaces. BMC Bioinform. 2010, 11, 548. [Google Scholar] [CrossRef]
- Yue, P.; Melamud, E.; Moult, J. SNPs3D: Candidate gene and SNP selection for association studies. BMC Bioinform. 2006, 7, 166. [Google Scholar] [CrossRef]
- Noble, W.S. What is a support vector machine? Nat. Biotechnol. 2006, 24, 1565–1567. [Google Scholar] [CrossRef]
- Joachim, T. Making large-scale SVM learning practical. In Advances in Kernel Methods—Support Vector Learning; Schölkopf, B., Burges, C., Smola, A., Eds.; MIT Press: Cambridge, MA, USA, 1999. [Google Scholar]
- Capriotti, E.; Fariselli, P.; Casadio, R. A neural-network-based method for predicting protein stability changes upon single point mutations. Bioinformatics 2004, 20, 63–68. [Google Scholar] [CrossRef]
- Capriotti, E.; Fariselli, P.; Casadio, R. I-Mutant2.0: Predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res. 2005, 33, W306–W310. [Google Scholar]
- Schymkowitz, J.; Borg, J.; Stricher, F.; Nys, R.; Rousseau, F.; Serrano, L. The FoldX web server: An online force field. Nucleic Acids Res. 2005, 33, W382–W388. [Google Scholar] [CrossRef]
- Schymkowitz, J.W.H.; Rousseau, F.; Martins, I.C.; Ferkinghoff-Borg, J.; Stricher, F.; Serrano, L. Prediction of water and metal binding sites and their affinities by using the Fold-X force field. Proc. Natl. Acad. Sci. USA 2005, 102, 10147–10152. [Google Scholar] [CrossRef]
- Yang, Y.; Zhou, Y. Specific interactions for ab initio folding of protein terminal regions with secondary structures. Proteins 2008, 72, 793–803. [Google Scholar] [CrossRef]
- Yang, Y.; Zhou, Y. Ab initio folding of terminal segments with secondary structures reveals the fine difference between two closely related all-atom statistical energy functions. Protein Sci. 2008, 17, 1212–1219. [Google Scholar] [CrossRef]
- Dehouck, Y.; Grosfils, A.; Folch, B.; Gilis, D.; Bogaerts, P.; Rooman, M. Fast and accurate predictions of protein stability changes upon mutations using statistical potentials and neural networks: PoPMuSiC-2.0. Bioinformatics 2009, 25, 2537–2543. [Google Scholar] [CrossRef]
- Dehouck, Y.; Kwasigroch, J.M.; Gilis, D.; Rooman, M. PoPMuSiC 2.1: A web server for the estimation of protein stability changes upon mutation and sequence optimality. BMC Bioinform. 2011, 12, 151. [Google Scholar] [CrossRef]
- Ozen, A.; Gonen, M.; Alpaydin, E.; Haliloglu, T. Machine learning integration for predicting the effect of single amino acid substitutions on protein stability. BMC Struct. Biol. 2009, 9, 66. [Google Scholar] [CrossRef]
- Folkman, L.; Stantic, B.; Sattar, A. Sequence-only evolutionary and predicted structural features for the prediction of stability changes in protein mutants. BMC Bioinform. 2013, 14, S6. [Google Scholar] [CrossRef]
- Folkman, L.; Stantic, B.; Sattar, A. Towards sequence-based prediction of mutation-induced stability changes in unseen non-homologous proteins. BMC Genomics 2014, 15, S4. [Google Scholar] [CrossRef]
- Bhasin, M.; Raghava, G.P.S. ESLpred: SVM-based method for subcellular localization of eukaryoticproteins using dipeptide composition and PSI-BLAST. Nucleic Acids Res. 2004, 32, W414–W419. [Google Scholar] [CrossRef]
- Cai, C.Z.; Han, L.Y.; Ji, Z.L.; Chen, X.; Chen, Y.Z. SVM-Prot: Web-based support vector machine software for functional classification of a protein from its primary sequence. Nucleic Acids Res. 2003, 31, 3692–3697. [Google Scholar] [CrossRef]
- Rangwala, H.; Kauffman, C.; Karypis, G. svmPRAT: SVM-based protein residue annotation toolkit. BMC Bioinform. 2009, 10, 439. [Google Scholar] [CrossRef]
- Rangwala, H.; Kauffman, C.; Karypis, G. A kernel framework for protein residue annotation. In Advances in Knowledge Discovery and Data Mining, Proceedings; Theeramunkong, T., Kijsirikul, B., Cercone, N., Ho, T.B., Eds.; Springer-Verlag: Berlin/Heidelberg, Germany, 2009; Volume 5476, pp. 439–451. [Google Scholar]
- Masso, M.; Vaisman, I.I. AUTO-MUTE: Web-based tools for predicting stability changes in proteins due to single amino acid replacements. Protein Eng. Des. Sel. 2010, 23, 683–687. [Google Scholar] [CrossRef]
- Masso, M.; Vaisman, I.I. Accurate prediction of stability changes in protein mutants by combining machine learning with structure based computational mutagenesis. Bioinformatics 2008, 24, 2002–2009. [Google Scholar] [CrossRef]
- Tangrot, J.; Wang, L.X.; Kagstrom, B.; Sauer, U.H. FISH—Family identification of sequence homologues using structure floated hidden Markov models. Nucleic Acids Res. 2006, 34, W10–W14. [Google Scholar] [CrossRef]
- Tangrot, J.; Wang, L.; Kagstrom, B.; Sauer, U.H. Design, construction and use of the FISH server. In Applied Parallel Computing: State of the Art in Scientific Computing; Kagstrom, B., Elmroth, E., Dongarra, J., Wasniewski, J., Eds.; Springer-Verlag: Berlin/Heidelberg, Germany, 2007; Volume 4699, pp. 647–657. [Google Scholar]
- Wang, L.X.; Sauer, U.H. OnD-CRF: Predicting order and disorder in proteins conditional random fields. Bioinformatics 2008, 24, 1401–1402. [Google Scholar] [CrossRef]
- Huang, L.T.; Gromiha, M.M.; Ho, S.Y. iPTREE-STAB: Interpretable decision tree based method for predicting protein stability changes upon mutations. Bioinformatics 2007, 23, 1292–1293. [Google Scholar] [CrossRef]
- Dosztanyi, Z.; Fiser, A.; Simon, I. Stabilization centers in proteins: Identification, characterization and predictions. J. Mol. Biol. 1997, 272, 597–612. [Google Scholar] [CrossRef]
- Dosztanyi, Z.; Magyar, C.; Tusnady, G.E.; Simon, I. SCide: Identification of stabilization centers in proteins. Bioinformatics 2003, 19, 899–900. [Google Scholar] [CrossRef]
- Ferrer-Costa, C.; Orozco, M.; de la Cruz, X. Characterization of disease-associated single amino acid polymorphisms in terms of sequence and structure properties. J. Mol. Biol. 2002, 315, 771–786. [Google Scholar] [CrossRef]
- Ferrer-Costa, C.; Orozco, M.; de la Cruz, X. Sequence-based prediction of pathological mutations. Proteins 2004, 57, 811–819. [Google Scholar] [CrossRef]
- Ferrer-Costa, C.; Orozco, M.; de la Cruz, X. Use of bioinformatics tools for the annotation of disease-associated mutations in animal models. Proteins 2005, 61, 878–887. [Google Scholar] [CrossRef]
- Bromberg, Y.; Rost, B. SNAP: Predict effect of non-synonymous polymorphisms on function. Nucleic Acids Res. 2007, 35, 3823–3835. [Google Scholar] [CrossRef]
- Calabrese, R.; Capriotti, E.; Fariselli, P.; Martelli, P.L.; Casadio, R. Functional annotations improvethe predictive score of human disease-related mutations in proteins. Hum. Mutat. 2009, 30, 1237–1244. [Google Scholar] [CrossRef]
- Tian, J.; Wu, N.; Guo, X.; Guo, J.; Zhang, J.; Fan, Y. Predicting the phenotypic effects of non-synonymous single nucleotide polymorphisms based on support vector machines. BMC Bioinform. 2007, 8, 450. [Google Scholar] [CrossRef]
- Kaminker, J.S.; Zhang, Y.; Watanabe, C.; Zhang, Z. CanPredict: A computational tool for predicting cancer-associated missense mutations. Nucleic Acids Res. 2007, 35, W595–W598. [Google Scholar] [CrossRef]
- Saunders, C.T.; Baker, D. Evaluation of structural and evolutionary contributions to deleterious mutation prediction. J. Mol. Biol. 2002, 322, 891–901. [Google Scholar] [CrossRef]
- Bowie, J.U.; Luthy, R.; Eisenberg, D. A method to identify protein sequences that fold into a known 3-dimensional structure. Science 1991, 253, 164–170. [Google Scholar]
- Acharya, V.; Nagarajaram, H.A. Hansa: An automated method for discriminating disease and neutral human nsSNPs. Hum. Mutat. 2012, 33, 332–337. [Google Scholar] [CrossRef]
- Acharya, V.; Nagarajaram, H.A. Response to: Statistical analysis of missense mutation classifiers. Hum. Mutat. 2013, 34, 407. [Google Scholar]
- Dehouck, Y.; Kwasigroch, J.M.; Rooman, M.; Gilis, D. BeAtMuSiC: Prediction of changes in protein–protein binding affinity on mutations. Nucleic Acids Res. 2013, 41, W333–W339. [Google Scholar] [CrossRef]
- Gardy, J.L.; Brinkman, F.S.L. Methods for predicting bacterial protein subcellular localization. Nat. Rev. Microbiol. 2006, 4, 741–751. [Google Scholar] [CrossRef]
- Chou, K.C.; Shen, H.B. Recent progress in protein subcellular location prediction. Anal. Biochem. 2007, 370, 1–16. [Google Scholar] [CrossRef]
- Imai, K.; Nakai, K. Prediction of subcellular locations of proteins: Where to proceed? Proteomics 2010, 10, 3970–3983. [Google Scholar] [CrossRef]
- Nakai, K.; Horton, P. PSORT: A program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 1999, 24, 34–35. [Google Scholar] [CrossRef]
- Nielsen, H.; Engelbrecht, J.; Brunak, S.; vonHeijne, G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997, 10, 1–6. [Google Scholar] [CrossRef]
- Nielsen, H.; Krogh, A. Prediction of signal peptides and signal floats by a hidden Markov model. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1998, 6, 122–130. [Google Scholar]
- Emanuelsson, O.; Brunak, S.; von Heijne, G.; Nielsen, H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2007, 2, 953–971. [Google Scholar] [CrossRef]
- Shen, H.-B.; Chou, K.-C. Predicting protein fold pattern with functional domain and sequential evolution information. J. Theor. Biol. 2009, 256, 441–446. [Google Scholar] [CrossRef]
- Shen, H.-B.; Chou, K.-C. Ensemble classifier for protein fold pattern recognition. Bioinformatics 2006, 22, 1717–1722. [Google Scholar] [CrossRef]
- Chou, K.C. Using amphiphilic pseudo amino acid composition to predict enzyme subfamily classes. Bioinformatics 2005, 21, 10–19. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Chou, K.C. Prediction of protein cellular attributes using pseudo-amino acid composition. Proteins 2001, 43, 246–255. [Google Scholar] [CrossRef]
- Wan, S.B.; Mak, M.W.; Kung, S.Y. GOASVM: A subcellular location predictor by incorporating term-frequency gene ontology into the general form of Chou’s pseudo-amino acid composition. J. Theor. Biol. 2013, 323, 40–48. [Google Scholar] [CrossRef]
- Goldberg, T.; Hamp, T.; Rost, B. LocTree2 predicts localization for all domains of life. Bioinformatics 2012, 28, I458–I465. [Google Scholar] [CrossRef]
- Nair, R.; Rost, B. Mimicking cellular sorting improves prediction of subcellular localization. J. Mol. Biol. 2005, 348, 85–100. [Google Scholar] [CrossRef]
- Mika, S.; Rost, B. UniqueProt: Creating representative protein sequence sets. Nucleic Acids Res. 2003, 31, 3789–3791. [Google Scholar] [CrossRef]
- Yu, C.S.; Lin, C.J.; Hwang, J.K. Predicting subcellular localization of proteins for Gram-negative bacteria by support vector machines based on n-peptide compositions. Protein Sci. 2004, 13, 1402–1406. [Google Scholar] [CrossRef]
- Yu, C.-S.; Chen, Y.-C.; Lu, C.-H.; Hwang, J.-K. Prediction of protein subcellular localization. Proteins 2006, 64, 643–651. [Google Scholar] [CrossRef]
- Horton, P.; Park, K.-J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef]
- King, B.R.; Vural, S.; Pandey, S.; Barteau, A.; Guda, C. ngLOC: Software and web server for predicting protein subcellular localization in prokaryotes and eukaryotes. BMC Res. Notes 2012, 5, 1–7. [Google Scholar]
- Briesemeister, S.; Blum, T.; Brady, S.; Lam, Y.; Kohlbacher, O.; Shatkay, H. SherLoc2: A high-accuracy hybrid method for predicting subcellular localization of proteins. J. Proteome Res. 2009, 8, 5363–5366. [Google Scholar] [CrossRef]
- Chi, S.M.; Nam, D. WegoLoc: Accurate prediction of protein subcellular localization using weighted Gene Ontology terms. Bioinformatics 2012, 28, 1028–1030. [Google Scholar] [CrossRef]
- Guda, C.; Subramaniam, S. pTARGET: A new method for predicting protein subcellular localization in eukaryotes. Bioinformatics 2005, 21, 3963–3969. [Google Scholar] [CrossRef]
- Guda, C. pTARGET: A web server for predicting protein subcellular localization. Nucleic Acids Res. 2006, 34, W210–W213. [Google Scholar] [CrossRef]
- Mer, A.S.; Andrade-Navarro, M.A. A novel approach for protein subcellular location prediction using amino acid exposure. BMC Bioinform. 2013, 14, 342. [Google Scholar] [CrossRef]
- Briesemeister, S.; Rahnenführer, J.; Kohlbacher, O. YLoc—An interpretable web server for predicting subcellular localization. Nucleic Acids Res. 2010, 38, W497–W502. [Google Scholar] [CrossRef]
- Briesemeister, S.; Rahnenführer, J.; Kohlbacher, O. Going from where to why—Interpretable prediction of protein subcellular localization. Bioinformatics 2010, 26, 1232–1238. [Google Scholar] [CrossRef]
- Binder, J.; Pletscher-Frankild, S.; Tsafou, K.; Stolte, C.; O’Donoghue, S.; Schneider, R.; Jensen, L. COMPARTMENTS: Unification and visualization of protein subcellular localization evidence. Database 2014, 2014, bau012. [Google Scholar] [CrossRef]
- Song, J.; Takemoto, K.; Shen, H.; Tan, H.; Gromiha, M.M.; Akutsu, T. Prediction of protein folding rates from structural topology and complex network properties. IPSJ Trans. Bioinform. 2010, 3, 40–53. [Google Scholar] [CrossRef]
- Capriotti, E.; Casadio, R. K-Fold: A tool for the prediction of the protein folding kinetic order and rate. Bioinformatics 2007, 23, 385–386. [Google Scholar] [CrossRef]
- Gromiha, M.M.; Selvaraj, S. Comparison between long-range interactions and contact order in determining the folding rate of two-state proteins: application of long-range order to folding rate prediction. J. Mol. Biol. 2001, 310, 27–32. [Google Scholar] [CrossRef]
- Gromiha, M.M. Importance of native-state topology for determining the folding rate of two-state proteins. J. Chem. Inf. Comput. Sci. 2003, 43, 1481–1485. [Google Scholar] [CrossRef]
- Gromiha, M.M. A statistical model for predicting protein folding rates from amino acid sequence with structural class information. J. Chem. Inf. Model. 2005, 45, 494–501. [Google Scholar] [CrossRef]
- Punta, M.; Rost, B. PROFcon: Novel prediction of long-range contacts. Bioinformatics 2005, 21, 2960–2968. [Google Scholar] [CrossRef]
- Wishart, D.S.; Arndt, D.; Berjanskii, M.; Guo, A.C.; Shi, Y.; Shrivastava, S.; Zhou, J.; Zhou, Y.; Lin, G. PPT-DB: The protein property prediction and testing database. Nucleic Acids Res. 2008, 36, D222–D229. [Google Scholar]
- Fang, Y.; Gao, S.; Tai, D.; Middaugh, C.R.; Fang, J. Identification of properties important to protein aggregation using feature selection. BMC Bioinform. 2013, 14, 314. [Google Scholar] [CrossRef]
- Oliveberg, M. Waltz, an exciting new move in amyloid prediction. Nat. Methods 2010, 7, 187–188. [Google Scholar] [CrossRef]
- Garbuzynskiy, S.O.; Lobanov, M.Y.; Galzitskaya, O.V. FoldAmyloid: A method of prediction of amyloidogenic regions from protein sequence. Bioinformatics 2010, 26, 326–332. [Google Scholar] [CrossRef]
- Tartaglia, G.G.; Vendruscolo, M. The Zyggregator method for predicting protein aggregation propensities. Chem. Soc. Rev. 2008, 37, 1395–1401. [Google Scholar] [CrossRef]
- Thomas, P.D.; Dill, K.A. Statistical potentials extracted from protein structures: How accurate are they? J. Mol. Biol. 1996, 257, 457–469. [Google Scholar] [CrossRef]
- Thomas, P.D.; Dill, K.A. An iterative method for extracting energy-like quantities from protein structures. Proc. Natl. Acad. Sci. USA 1996, 93, 11628–11633. [Google Scholar] [CrossRef]
- BenNaim, A. Statistical potentials extracted from protein structures: Are these meaningful potentials? J. Chem. Phys. 1997, 107, 3698–3706. [Google Scholar] [CrossRef]
- Buchete, N.V.; Straub, J.E.; Thirumalai, D. Development of novel statistical potentials for protein fold recognition. Curr. Opin. Struct. Biol. 2004, 14, 225–232. [Google Scholar] [CrossRef]
- Skolnick, J. In quest of an empirical potential for protein structure prediction. Curr. Opin. Struct. Biol. 2006, 16, 166–171. [Google Scholar] [CrossRef]
- Worth, C.L.; Preissner, R.; Blundell, T.L. SDM—A server for predicting effects of mutations on protein stability and malfunction. Nucleic Acids Res. 2011, 39, W215–W222. [Google Scholar] [CrossRef]
- Parthiban, V.; Gromiha, M.M.; Schomburg, D. CUPSAT: Prediction of protein stability upon point mutations. Nucleic Acids Res. 2006, 34, W239–W242. [Google Scholar] [CrossRef]
- Munson, P.J.; Singh, R.K. Statistical significance of hierarchical multi-body potentials based on Delaunay tessellation and their application in sequence-structure alignment. Protein Sci. 1997, 6, 1467–1481. [Google Scholar] [CrossRef]
- Zhou, H.Y.; Zhou, Y.Q. Distance-scaled, finite ideal-gas reference state improves structure-derived potentials of mean force for structure selection and stability prediction. Protein Sci. 2002, 11, 2714–2726. [Google Scholar] [CrossRef]
- Shen, M.Y.; Sali, A. Statistical potential for assessment and prediction of protein structures. Protein Sci. 2006, 15, 2507–2524. [Google Scholar] [CrossRef]
- Mayewski, S. A multibody, whole-residue potential for protein structures, with testing by Monte Carlo simulated annealing. Proteins 2005, 59, 152–169. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, H.Y.; Zhang, C.; Liu, S. What is a desirable statistical energy function for proteins and how can it be obtained? Cell Biochem. Biophys. 2006, 46, 165–174. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, S.; Zhou, H.Y.; Zhou, Y.Q. The dependence of all-atom statistical potentials on structural training database. Biophys. J. 2004, 86, 3349–3358. [Google Scholar] [CrossRef]
- Liu, T.; Samudrala, R. The effect of experimental resolution on the performance of knowledge-based discriminatory functions for protein structure selection. Protein Eng. Des. Sel. 2006, 19, 431–437. [Google Scholar] [CrossRef]
- Liu, Y.; Kuhlman, B. RosettaDesign server for protein design. Nucleic Acids Res. 2006, 34, W235–W238. [Google Scholar] [CrossRef]
- Kang, S.; Chen, G.; Xiao, G. Robust prediction of mutation-induced protein stability change by property encoding of amino acids. Protein Eng. Des. Sel. 2009, 22, 75–83. [Google Scholar]
- Cohen, M.; Potapov, V.; Schreiber, G. Four distances between pairs of amino acids provide a precise description of their interaction. PLoS Comput. Biol. 2009, 5, e1000470. [Google Scholar] [CrossRef]
- Potapov, V.; Cohen, M.; Inbar, Y.; Schreiber, G. Protein structure modelling and evaluation based on a 4-distance description of side-chain interactions. BMC Bioinform. 2010, 11, 374. [Google Scholar] [CrossRef]
- Pires, D.E.V.; Ascher, D.B.; Blundell, T.L. mCSM: Predicting the effects of mutations in proteins using graph-based signatures. Bioinformatics 2014, 30, 335–342. [Google Scholar] [CrossRef]
- Yin, S.; Ding, F.; Dokholyan, N.V. Eris: An automated estimator of protein stability. Nat. Methods 2007, 4, 466–467. [Google Scholar] [CrossRef]
- Cheng, T.M.K.; Lu, Y.-E.; Vendruscolo, M.; Lio, P.; Blundell, T.L. Prediction by graph theoretic measures of structural effects in proteins arising from non-synonymous single nucleotide polymorphisms. PLoS Comput. Biol. 2008, 4, e1000135. [Google Scholar] [CrossRef]
- Aqvist, J.; Medina, C.; Samuelsson, J.E. New method for predicting binding-affinity in computer-aided drug design. Protein Eng. 1994, 7, 385–391. [Google Scholar] [CrossRef]
- Bueno, M.; Camacho, C.J.; Sancho, J. SIMPLE estimate of the free energy change due to aliphatic mutations: Superior predictions based on first principles. Proteins 2007, 68, 850–862. [Google Scholar] [CrossRef]
- Benedix, A.; Becker, C.M.; de Groot, B.L.; Caflisch, A.; Bockmann, R.A. Predicting free energy changes using structural ensembles. Nat. Methods 2009, 6, 3–4. [Google Scholar]
- Wickstrom, L.; Gallicchio, E.; Levy, R.M. The linear interaction energy method for the prediction of protein stability changes upon mutation. Proteins 2012, 80, 111–125. [Google Scholar] [CrossRef]
- Li, M.; Petukh, M.; Alexov, E.; Panchenko, A.R. Predicting the impact of missense mutations on protein–protein binding affinity. J. Chem. Theor. Comput. 2014, 10, 1770–1780. [Google Scholar] [CrossRef]
- deGroot, B.L.; vanAalten, D.M.F.; Scheek, R.M.; Amadei, A.; Vriend, G.; Berendsen, H.J.C. Prediction of protein conformational freedom from distance constraints. Proteins 1997, 29, 240–251. [Google Scholar] [CrossRef]
- Li, L.; Li, C.; Sarkar, S.; Zhang, J.; Witham, S.; Zhang, Z.; Wang, L.; Smith, N.; Petukh, M.; Alexov, E. DelPhi: A comprehensive suite for DelPhi software and associated resources. BMC Biophys. 2012, 5, 1–11. [Google Scholar] [CrossRef]
- Schlitter, J. Estimation of absolute and relative entropies of macromolecules using the covariance-matrix. Chem. Phys. Lett. 1993, 215, 617–621. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, L.; Gao, Y.; Zhang, J.; Zhenirovskyy, M.; Alexov, E. Predicting folding free energy changes upon single point mutations. Bioinformatics 2012, 28, 664–671. [Google Scholar] [CrossRef]
- Pappu, R.V.; Hart, R.K.; Ponder, J.W. Analysis and application of potential energy smoothing and search methods for global optimization. J. Phys. Chem. B 1998, 102, 9725–9742. [Google Scholar] [CrossRef]
- Guerois, R.; Nielsen, J.E.; Serrano, L. Predicting changes in the stability of proteins and protein complexes: A study of more than 1000 mutations. J. Mol. Biol. 2002, 320, 369–387. [Google Scholar] [CrossRef]
- Pokala, N.; Handel, T.M. Energy functions for protein design: Adjustment with protein-protein complex affinities, models for the unfolded state, and negative design of solubility and specificity. J. Mol. Biol. 2005, 347, 203–227. [Google Scholar] [CrossRef]
- Kumar, A.; Rajendran, V.; Sethumadhavan, R.; Purohit, R. Molecular dynamic simulation reveals damaging impact of RAC1 F28L mutation in the switch I region. PLoS One 2013, 8, e77453. [Google Scholar]
- Beveridge, D.L.; Dicapua, F.M. Free-energy via molecular simulation—Applications to chemical and biomolecular systems. Ann. Rev. Biophys. Biophys. Chem. 1989, 18, 431–492. [Google Scholar] [CrossRef]
- Kirkwood, J.G. Statistical mechanics of fluid mixtures. J. Chem. Phys. 1935, 3, 300–313. [Google Scholar]
- Frenkel, D.; Smit, B. Understanding Molecular Simulation: From Algorithms to Applications, 2nd ed.; Academic Press: New York, NY, USA, 2001. [Google Scholar]
- Chipot, C. Free Energy Calculations: Theory and Applications in Chemistry and Biology; Springer-Verlag: Berlin/Heidelburg, Germany, 2007; volume 86. [Google Scholar]
- Qin, S.; Pang, X.D.; Zhou, H.X. Automated prediction of protein association rate constants. Structure 2011, 19, 1744–1751. [Google Scholar]
- Alsallaq, R.; Zhou, H.X. Electrostatic rate enhancement and transient complex of protein–protein association. Proteins 2008, 71, 320–335. [Google Scholar]
- Bai, H.J.; Yang, K.; Yu, D.Q.; Zhang, C.S.; Chen, F.J.; Lai, L.H. Predicting kinetic constants of protein-protein interactions based on structural properties. Proteins 2011, 79, 720–734. [Google Scholar]
- Hoeting, J.A.; Madigan, D.; Raftery, A.E.; Volinsky, C.T. Bayesian model averaging: A tutorial. Stat. Sci. 1999, 382–401. [Google Scholar]
- Agius, R.; Torchala, M.; Moal, I.H.; Fernandez-Recio, J.; Bates, P.A. Characterizing changes in the rate of protein–protein dissociation upon interface mutation using hotspot energy and organization. PLoS Comput. Biol. 2013, 9, e1003216. [Google Scholar]
- Moretti, R.; Fleishman, S.J.; Agius, R.; Torchala, M.; Bates, P.A.; Kastritis, P.L.; Rodrigues, J.P.; Trellet, M.; Bonvin, A.M.; Cui, M.; et al. Community-wide evaluation of methods for predicting the effect of mutations on protein–protein interactions. Proteins 2013, 81, 1980–1987. [Google Scholar]
- Chiti, F.; Stefani, M.; Taddei, N.; Ramponi, G.; Dobson, C.M. Rationalization of the effects of mutations on peptide and protein aggregation rates. Nature 2003, 424, 805–808. [Google Scholar]
- Tartaglia, G.G.; Cavalli, A.; Pellarin, R.; Caflisch, A. The role of aromaticity, exposed surface, and dipole moment in determining protein aggregation rates. Protein Sci. 2004, 13, 1939–1941. [Google Scholar]
- Fernandez-Escamilla, A.M.; Rousseau, F.; Schymkowitz, J.; Serrano, L. Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat. Biotechnol. 2004, 22, 1302–1306. [Google Scholar]
- Rousseau, F.; Schymkowitz, J.; Serrano, L. Protein aggregation and amyloidosis: confusion of the kinds? Curr. Opin. Struct. Biol. 2006, 16, 118–126. [Google Scholar]
- Linding, R.; Schymkowitz, J.; Rousseau, F.; Diella, F.; Serrano, L. A comparative study of the relationship between protein structure and beta-aggregation in globular and intrinsically disordered proteins. J. Mol. Biol. 2004, 342, 345–353. [Google Scholar]
- Trovato, A.; Chiti, F.; Maritan, A.; Seno, F. Insight into the structure of amyloid fibrils from the analysis of globular proteins. PLoS Comput. Biol. 2006, 2, 1608–1618. [Google Scholar]
- Trovato, A.; Seno, F.; Tosatto, S.C.E. The PASTA server for protein aggregation prediction. Protein Eng. Des. Sel. 2007, 20, 521–523. [Google Scholar]
- Conchillo-Sole, O.; de Groot, N.S.; Aviles, F.X.; Vendrell, J.; Daura, X.; Ventura, S. AGGRESCAN: A server for the prediction and evaluation of “hot spots” of aggregation in polypeptides. BMC Bioinform. 2007, 8, 65. [Google Scholar]
- Lander, E.S.; Schork, N.J. Genetic dissection of complex traits. Science 1994, 265, 2037–2048. [Google Scholar]
- Martin, D.B.; Nelson, P.S. From genomics to proteomics: techniques and applications in cancer research. Trends Cell Biol. 2001, 11, S60–S65. [Google Scholar]
- Landegren, U.; Nilsson, M.; Kwok, P.Y. Reading bits of genetic information: methods for single-nucleotide polymorphism analysis. Genome Res. 1998, 8, 769–776. [Google Scholar]
- Carlson, C.S.; Eberle, M.A.; Kruglyak, L.; Nickerson, D.A. Mapping complex disease loci in whole-genome association studies. Nature 2004, 429, 446–452. [Google Scholar]
- Shendure, J.; Ji, H. Next-generation DNA sequencing. Nat. Biotechnol. 2008, 26, 1135–1145. [Google Scholar]
- Etter, P.D.; Bassham, S.; Hohenlohe, P.A.; Johnson, E.A.; Cresko, W.A. SNP discovery and genotyping for evolutionary genetics using RAD sequencing. Methods Mol. Biol. 2011, 772, 157–178. [Google Scholar]
- Robasky, K.; Lewis, N.E.; Church, G.M. The role of replicates for error mitigation in next-generation sequencing. Nat. Rev. Genet. 2014, 15, 56–62. [Google Scholar]
- Donis-Keller, H.; Green, P.; Helms, C.; Cartinhour, S.; Weiffenbach, B.; Stephens, K.; Keith, T.P.; Bowden, D.W.; Smith, D.R.; Lander, E.S.; et al. A genetic linkage map of the human genome. Cell 1987, 51, 319–337. [Google Scholar]
- Zhang, C.; Liu, S.; Zhu, Q.Q.; Zhou, Y.Q. A knowledge-based energy function for protein–ligand, protein–protein, and protein–DNA complexe. J. Med. Chem. 2005, 48, 2325–2335. [Google Scholar]
- Gurgey, A.; Beksac, S.; Mesci, L.; Cakar, N.; Karakas, U.; Kutlar, A.; Altay, C. Prenatal diagnosis of sickle cell anemia using PCR and restriction enzyme DdeI. Turk. J. Pediatr. 1993, 35, 159–162. [Google Scholar]
- Williams, J.G.; Kubelik, A.R.; Livak, K.J.; Rafalski, J.A.; Tingey, S.V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990, 18, 6531–6535. [Google Scholar]
- Shangkuan, Y.H.; Lin, H.C. Application of random amplified polymorphic DNA analysis to differentiate strains of Salmonella typhi and other Salmonella species. J. Appl. Microbiol. 1998, 85, 693–702. [Google Scholar]
- Quintaes, B.R.; Leal, N.C.; Reis, E.M.; Hofer, E. Optimization of randomly amplified polymorphic DNA-polymerase chain reaction for molecular typing of Salmonella enterica serovar Typhi. Rev. Soc. Bras. Med. Trop. 2004, 37, 143–147. [Google Scholar]
- Konry, T.; Hayman, R.B.; Walt, D.R. Microsphere-based rolling circle amplification microarray for the detection of DNA and proteins in a single assay. Anal. Chem. 2009, 81, 5777–5782. [Google Scholar]
- Epstein, J.R.; Leung, A.P.; Lee, K.H.; Walt, D.R. High-density, microsphere-based fiber optic DNA microarrays. Biosens. Bioelectron. 2003, 18, 541–546. [Google Scholar]
- Shalon, D.; Smith, S.J.; Brown, P.O. A DNA microarray system for analyzing complex DNA samples using two-color fluorescent probe hybridization. Genome Res. 1996, 6, 639–645. [Google Scholar]
- Truco, M.J.; Ashrafi, H.; Kozik, A.; van Leeuwen, H.; Bowers, J.; Reyes Chin, W.O.S.; Stoffel, K.; Xu, H.; Hill, T.; van Deynze, A.; et al. An ultra high-density, transcript-based, genetic map of lettuce. G3 (Bethesda) 2013, 3, 617–631. [Google Scholar]
- Pastinen, T.; Raitio, M.; Lindroos, K.; Tainola, P.; Peltonen, L.; Syvanen, A.C. A system for specific, high-throughput genotyping by allele-specific primer extension on microarrays. Genome Res. 2000, 10, 1031–1042. [Google Scholar]
- Koch, W.H. Technology platforms for pharmacogenomic diagnostic assays. Nat. Rev. Drug Discov. 2004, 3, 749–761. [Google Scholar]
- Baek, I.C.; Jang, J.P.; Choi, H.B.; Choi, E.J.; Ko, W.Y.; Kim, T.G. Microarrays for high-throughput genotyping of MICA alleles using allele-specific primer extension. Tissue Antigens 2013, 82, 259–268. [Google Scholar]
- Shen, R.; Fan, J.B.; Campbell, D.; Chang, W.; Chen, J.; Doucet, D.; Yeakley, J.; Bibikova, M.; Garcia, E.W.; McBride, C.; et al. High-throughput SNP genotyping on universal bead arrays. Mutat. Res. 2005, 573, 70–82. [Google Scholar]
- Van Heek, N.T.; Clayton, S.J.; Sturm, P.D.; Walker, J.; Gouma, D.J.; Noorduyn, L.A.; Offerhaus, G.J.; Fox, J.C. Comparison of the novel quantitative ARMS assay and an enriched PCR-ASO assay for K-ras mutations with conventional cytology on endobiliary brush cytology from 312 consecutive extrahepatic biliary stenoses. J. Clin. Pathol. 2005, 58, 1315–1320. [Google Scholar]
- Macgregor, S.; Zhao, Z.Z.; Henders, A.; Nicholas, M.G.; Montgomery, G.W.; Visscher, P.M. Highly cost-efficient genome-wide association studies using DNA pools and dense SNP arrays. Nucleic Acids Res. 2008, 36, e35. [Google Scholar]
- Goelet, P.; Knapp, M.; Anderson, S.U.S. Methord for Dethermining Nucleotide identity through Primer Extension. U.S. Patent 5,888,819,30, March 1999. [Google Scholar]
- Mandoiu, I.I.; Prajescu, C. High-throughput SNP genotyping by SBE/SBH. IEEE Trans. Nanobiosci. 2007, 6, 28–35. [Google Scholar]
- Hirschhorn, J.N.; Sklar, P.; Lindblad-Toh, K.; Lim, Y.M.; Ruiz-Gutierrez, M.; Bolk, S.; Langhorst, B.; Schaffner, S.; Winchester, E.; Lander, E.S. SBE-TAGS: an array-based method for efficient single-nucleotide polymorphism genotyping. Proc. Natl. Acad. Sci. USA 2000, 97, 12164–12169. [Google Scholar]
- Bell, P.A.; Chaturvedi, S.; Gelfand, C.A.; Huang, C.Y.; Kochersperger, M.; Kopla, R.; Modica, F.; Pohl, M.; Varde, S.; Zhao, R.; et al. NPstream UHT: Ultra-high throughput SNP genotyping for pharmacogenomics and drug discovery. Biotechniques 2002, 74, 76–77. [Google Scholar]
- Liu, H.; Wang, H.; Shi, Z.; Wang, H.; Silke, C.Y.S.; Tan, W.; Lu, Z. TaqMan probe array for quantitative detection of DNA targets. Nucleic Acids Res. 2006, 34, e4. [Google Scholar]
- Shen, G.Q.; Abdullah, K.G.; Wang, Q.K. The TaqMan method for SNP genotyping. Methods Mol. Biol. 2009, 578, 293–306. [Google Scholar]
- Cao, W. Recent developments in ligase-mediated amplification and detection. Trends Biotechnol. 2004, 22, 38–44. [Google Scholar]
- Baner, J.; Isaksson, A.; Waldenstrom, E.; Jarvius, J.; Landegren, U.; Nilsson, M.; et al. Parallel gene analysis with allele-specific padlock probes and tag microarrays. Nucleic Acids Res. 2003, 31, e103. [Google Scholar]
- Hardenbol, P.; Baner, J.; Jain, M.; Nilsson, M.; Namsaraev, E.A.; Karlin-Neumann, G.A.; Fakhrai-Rad, H.; Ronaghi, M.; Willis, T.D.; Landegren, U.; et al. Multiplexed genotyping with sequence-tagged molecular inversion probes. Nat. Biotechnol. 2003, 21, 673–678. [Google Scholar]
- Lamy, P.; Andersen, C.L.; Wikman, F.P.; Wiuf, C. Genotyping and annotation of Affymetrix SNP arrays. Nucleic Acids Res. 2006, 34. [Google Scholar] [CrossRef]
- Orom, U.A.; Nielsen, F.C.; Lund, A.H. MicroRNA-10a binds the 5'UTR of ribosomal protein mRNAs and enhances their translation. Mol. Cell. 2008, 30, 460–471. [Google Scholar]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar]
- Saunders, M.A.; Liang, H.; Li, W.H. Human polymorphism at microRNAs and microRNA target sites. Proc. Natl. Acad. Sci. USA 2007, 104, 3300–3305. [Google Scholar]
- Landi, D.; Gemignani, F.; Naccarati, A.; Pardini, B.; Vodicka, P.; Vodickova, L.; Novotny, J.; Forsti, A.; Hemminki, K.; Canzian, F.; et al. Polymorphisms within micro-RNA-binding sites and risk of sporadic colorectal cancer. Carcinogenesis 2008, 29, 579–584. [Google Scholar]
- Lewis, J.D.; Meehan, R.R.; Henzel, W.J.; Maurer-Fogy, I.; Jeppesen, P.; Klein, F.; Bird, A. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell 1992, 69, 905–914. [Google Scholar]
- Nan, X.; Campoy, F.J.; Bird, A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell 1997, 88, 471–481. [Google Scholar]
- Amir, R.E.; van den, V.E.I.; Wan, M.; Tran, C.Q.; Francke, U.; Zoghbi, H.Y. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat. Genet. 1999, 23, 185–188. [Google Scholar]
- Yusufzai, T.M.; Wolffe, A.P. Functional consequences of Rett syndrome mutations on human MeCP2. Nucleic Acids Res. 2000, 28, 4172–4179. [Google Scholar]
- Chen, W.G.; Chang, Q.; Lin, Y.; Meissner, A.; West, A.E.; Griffith, E.C.; Jaenisch, R.; Greenberg, M.E. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science 2003, 302, 885–889. [Google Scholar]
- Georgel, P.T.; Horowitz-Scherer, R.A.; Adkins, N.; Woodcock, C.L.; Wade, P.A.; Hansen, J.C. Chromatin compaction by human MeCP2. Assembly of novel secondary chromatin structures in the absence of DNA methylation. J. Biol. Chem. 2003, 278, 32181–32188. [Google Scholar]
- Galvao, T.C.; Thomas, J.O. Structure-specific binding of MeCP2 to four-way junction DNA through its methyl CpG-binding domain. Nucleic Acids Res. 2005, 33, 6603–6609. [Google Scholar]
- Ghosh, R.P.; Horowitz-Scherer, R.A.; Nikitina, T.; Gierasch, L.M.; Woodcock, C.L. Rett syndrome-causing mutations in human MeCP2 result in diverse structural changes that impact folding and DNA interactions. J. Biol. Chem. 2008, 283, 20523–20534. [Google Scholar]
- Mellen, M.; Ayata, P.; Dewell, S.; Kriaucionis, S.; Heintz, N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell 2012, 151, 1417–1430. [Google Scholar]
- Lyst, M.J.; Ekiert, R.; Ebert, D.H.; Merusi, C.; Nowak, J.; Selfridge, J.; Guy, J.; Kastan, N.R.; Robinson, N.D.; de Lima Alves, F.; et al. Rett syndrome mutations abolish the interaction of MeCP2 with the NCoR/SMRT co-repressor. Nat. Neurosci. 2013, 16, 898–902. [Google Scholar]
- Baker, S.A.; Chen, L.; Wilkins, A.D.; Yu, P.; Lichtarge, O.; Zoghbi, H.Y. An AT-hook domain in MeCP2 determines the clinical course of Rett syndrome and related disorders. Cell 2013, 152, 984–996. [Google Scholar]
- Erika Hawkins, M.S.; Michael Beck, M.S.; Braeden Butler, B.S.; Keith Wood, P.D. Dual-luciferase reporter assay: An advanced co-reporter technology integrating firefly and renilla luciferase assays. Promega Notes Mag. 1996, 2–8. [Google Scholar]
- McNabb, D.S.; Reed, R.; Marciniak, R.A. Dual luciferase assay system for rapid assessment of gene expression in Saccharomyces cerevisiae. Eukaryot. Cell 2005, 4, 1539–1549. [Google Scholar]
- Nicoloso, M.S.; Sun, H.; Spizzo, R.; Kim, H.; Wickramasinghe, P.; Shimizu, M.; Wojcik, S.E.; Ferdin, J.; Kunej, T.; Xiao, L.; et al. Single-nucleotide polymorphisms inside microRNA target sites influence tumor susceptibility. Cancer Res. 2010, 70, 2789–2798. [Google Scholar]
- Avner, P.; Heard, E. X-chromosome inactivation: counting, choice and initiation. Nat. Rev. Genet. 2001, 2, 59–67. [Google Scholar]
- Tycko, B.; Morison, I.M. Physiological functions of imprinted genes. J. Cell. Physiol. 2002, 192, 245–258. [Google Scholar]
- Kim, J.; Bartel, D.P. Allelic imbalance sequencing reveals that single-nucleotide polymorphisms frequently alter microRNA-directed repression. Nat. Biotechnol. 2009, 27, 472–477. [Google Scholar]
- Bannantine, J.P.; Stabel, J.R.; Lamont, E.A.; Briggs, R.E.; Sreevatsan, S. Monoclonal antibodies bind a SNP-sensitive epitope that is present uniquely in mycobacterium avium subspecies paratuberculosis. Front. Microbiol. 2011, 2, 163. [Google Scholar]
- Wuthrich, K. The way to NMR structures of proteins. Nat. Struct. Biol. 2001, 8, 923–925. [Google Scholar]
- Nakanishi, K.; Berova, N.; Woody, R. Circular Dichroism: Principles and Applications; Jogn Wiley and Sons: New York, NY, USA, 1994; p. 473. [Google Scholar]
- Dubois, A.; Deuve, J.L.; Navarro, P.; Merzouk, S.; Pichard, S.; Commere, P.H.; Louise, A.; Arnaud, D.; Avner, P.; Morey, C. Spontaneous reactivation of clusters of X-linked genes is associated with the plasticity of X-inactivation in mouse trophoblast stem cells. Stem Cells 2014, 32, 377–390. [Google Scholar]
- Yang, Y.; Dou, S.X.; Ren, H.; Wang, P.Y.; Zhang, X.D.; Qian, M.; Pan, B.Y.; Xi, X.G. Evidence for a functional dimeric form of the PcrA helicase in DNA unwinding. Nucleic Acids Res. 2008, 36, 1976–1989. [Google Scholar]
- Ren, H.; Dou, S.X.; Zhang, X.D.; Wang, P.Y.; Kanagaraj, R.; Liu, J.L.; Janscak, P.; Hu, J.S.; Xi, X.G. The zinc-binding motif of human RECQ5beta suppresses the intrinsic strand-annealing activity of its DExH helicase domain and is essential for the helicase activity of the enzyme. Biochem. J. 2008, 412, 425–433. [Google Scholar]
- Sammond, D.W.; Eletr, Z.M.; Purbeck, C.; Kimple, R.J.; Siderovski, D.P.; Kuhlman, B. Structure-based protocol for identifying mutations that enhance protein-protein binding affinities. J. Mol. Biol. 2007, 371, 1392–1404. [Google Scholar]
- Ciucci, A.; Palma, C.; Manzini, S.; Werge, T.M. Point mutation increases a form of the NK1 receptor with high affinity for neurokinin A and B and septide. Br. J. Pharmacol. 1998, 125, 393–401. [Google Scholar]
- Ren, H.; Dou, S.X.; Rigolet, P.; Yang, Y.; Wang, P.Y.; Amor-Gueret, M.; Xi, X.G. The arginine finger of the Bloom syndrome protein: its structural organization and its role in energy coupling. Nucleic Acids Res. 2007, 35, 6029–6041. [Google Scholar]
- Gentile, S.; Martin, N.; Scappini, E.; Williams, J.; Erxleben, C.; Armstrong, D.L. The human ERG1 channel polymorphism, K897T, creates a phosphorylation site that inhibits channel activity. Proc. Natl. Acad. Sci. USA 2008, 105, 14704–14708. [Google Scholar]
- Ebert, D.H.; Gabel, H.W.; Robinson, N.D.; Kastan, N.R.; Hu, L.S.; Cohen, S.; Navarro, A.J.; Lyst, M.J.; Ekiert, R.; Bird, A.P.; et al. Activity-dependent phosphorylation of MeCP2 threonine 308 regulates interaction with NCoR. Nature 2013, 499, 341–345. [Google Scholar]
- Josephy, P.D.; Pan, D.; Ianni, M.D.; Mannervik, B. Functional studies of single-nucleotide polymorphic variants of human glutathione transferase T1–1 involving residues in the dimer interface. Arch. Biochem. Biophys. 2011, 513, 87–93. [Google Scholar]
- Hsu, C.H.; Wen, Z.H.; Lin, C.S.; Chakraborty, C. The zebrafish model: use in studying cellular mechanisms for a spectrum of clinical disease entities. Curr. Neurovasc. Res. 2007, 4, 111–120. [Google Scholar]
- Best, J.D.; Alderton, W.K. Zebrafish: An in vivo model for the study of neurological diseases. Neuropsychiatr. Dis. Treat. 2008, 4, 567–576. [Google Scholar]
- Lieschke, G.J.; Currie, P.D. Animal models of human disease: zebrafish swim into view. Nat. Rev. Genet. 2007, 8, 353–367. [Google Scholar]
- Sager, J.J.; Bai, Q.; Burton, E.A. Transgenic zebrafish models of neurodegenerative diseases. Brain Struct. Funct. 2010, 214, 285–302. [Google Scholar]
- Gupta, A.; Meng, X.; Zhu, L.J.; Lawson, N.D.; Wolfe, S.A. Zinc finger protein-dependent and -independent contributions to the in vivo off-target activity of zinc finger nucleases. Nucleic Acids Res. 2011, 39, 381–392. [Google Scholar]
- Gerety, S.S.; Breau, M.A.; Sasai, N.; Xu, Q.; Briscoe, J.; Wilkinson, D.G. An inducible transgene expression system for zebrafish and chick. Development 2013, 140, 2235–2243. [Google Scholar]
- Kok, F.O.; Gupta, A.; Lawson, N.D.; Wolfe, S.A. Construction and application of site-specific artificial nucleases for targeted gene editing. Methods Mol. Biol. 2014, 1101, 267–303. [Google Scholar]
- Gupta, A.; Hall, V.L.; Kok, F.O.; Shin, M.; McNulty, J.C.; Lawson, N.D.; Wolfe, S.A. Targeted chromosomal deletions and inversions in zebrafish. Genome Res. 2013, 23, 1008–1017. [Google Scholar]
- Sun, N.; Zhao, H. Transcription activator-like effector nucleases (TALENs): A highly efficient and versatile tool for genome editing. Biotechnol. Bioeng. 2013, 110, 1811–1821. [Google Scholar]
- Hwang, W.Y.; Fu, Y.; Reyon, D.; Maeder, M.L.; Kaini, P.; Sander, J.D.; Joung, J.K.; Peterson, R.T.; Yeh, J.R. Heritable and precise zebrafish genome editing using a CRISPR-Cas system. PLoS One 2013, 8, e68708. [Google Scholar]
- Sander, J.D.; Joung, J.K. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 2014, 32, 347–355. [Google Scholar]
- Shalem, O.; Sanjana, N.E.; Hartenian, E.; Shi, X.; Scott, D.A.; Mikkelsen, T.S.; Heckl, D.; Ebert, B.L.; Root, D.E.; Doench, J.G.; et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 2014, 343, 84–87. [Google Scholar]
- Sashital, D.G.; Wiedenheft, B.; Doudna, J.A. Mechanism of foreign DNA selection in a bacterial adaptive immune system. Mol. Cell. 2012, 46, 606–615. [Google Scholar]
- Bhaya, D.; Davison, M.; Barrangou, R. CRISPR-Cas systems in bacteria and archaea: Versatile small RNAs for adaptive defense and regulation. Ann. Rev. Genet. 2011, 45, 273–297. [Google Scholar]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar]
- Blackburn, P.R.; Campbell, J.M.; Clark, K.J.; Ekker, S.C. The CRISPR system—Keeping zebrafish gene targeting fresh. Zebrafish 2013, 10, 116–118. [Google Scholar]
- Huang, P.; Zhu, Z.; Lin, S.; Zhang, B. Reverse genetic approaches in zebrafish. J. Genet. Genomics 2012, 39, 421–433. [Google Scholar]
- Ansai, S.; Inohaya, K.; Yoshiura, Y.; Schartl, M.; Uemura, N.; Takahashi, R.; Kinoshita, M. Design, evaluation, and screening methods for efficient targeted mutagenesis with transcription activator-like effector nucleases in medaka. Dev. Growth Differ. 2014, 56, 98–107. [Google Scholar]
- Edelheit, O.; Hanukoglu, A.; Hanukoglu, I. Simple and efficient site-directed mutagenesis using two single-primer reactions in parallel to generate mutants for protein structure–function studies. BMC Biotechnol. 2009, 9, 61. [Google Scholar]
- Agulnik, A.I.; Mitchell, M.J.; Mattei, M.G.; Borsani, G.; Avner, P.A.; Lerner, J.L.; Bishop, C.E. A novel X gene with a widely transcribed Y-linked homologue escapes X-inactivation in mouse and human. Hum. Mol. Genet. 1994, 3, 879–884. [Google Scholar]
- Takeuchi, T.; Yamazaki, Y.; Katoh-Fukui, Y.; Tsuchiya, R.; Kondo, S.; Motoyama, J.; Higashinakagawa, T. Gene trap capture of a novel mouse gene, jumonji, required for neural tube formation. Genes Dev. 1995, 9, 1211–1222. [Google Scholar]
- Jensen, L.R.; Amende, M.; Gurok, U.; Moser, B.; Gimmel, V.; Tzschach, A.; Janecke, A.R.; Tariverdian, G.; Chelly, J.; Fryns, J.P.; et al. Mutations in the JARID1C gene, which is involved in transcriptional regulation and chromatin remodeling, cause X-linked mental retardation. Am. J. Hum. Genet. 2005, 76, 227–236. [Google Scholar]
- Santos, C.; Rodriguez-Revenga, L.; Madrigal, I.; Badenas, C.; Pineda, M.; Mila, M. A novel mutation in JARID1C gene associated with mental retardation. Eur. J. Hum. Genet. 2006, 14, 583–586. [Google Scholar]
- Harvey, C.G.; Menon, S.D.; Stachowiak, B.; Noor, A.; Proctor, A.; Mensah, A.K.; Mnatzakanian, G.N.; Alfred, S.E.; Guo, R.; Scherer, S.W.; et al. Sequence variants within exon 1 of MECP2 occur in females with mental retardation. Am. J. Med. Genet. B 2007, 144B, 355–360. [Google Scholar]
- Christodoulou, J.; Ho, G. MECP2-Related Disorders. GeneReviews. 1993. Available online: http://www.ncbi.nlm.nih.gov/books/NBK1497/ (accessed on 12 March 2014).
- Chandler, S.P.; Guschin, D.; Landsberger, N.; Wolffe, A.P. The methyl-CpG binding transcriptional repressor MeCP2 stably associates with nucleosomal DNA. Biochemistry 1999, 38, 7008–7018. [Google Scholar]
- Nan, X.; Ng, H.H.; Johnson, C.A.; Laherty, C.D.; Turner, B.M.; Eisenman, R.N.; Bird, A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 1998, 393, 386–389. [Google Scholar]
- Smrt, R.D.; Eaves-Egenes, J.; Barkho, B.Z.; Santistevan, N.J.; Zhao, C.; Aimone, J.B.; Gage, F.H.; Zhao, X. Mecp2 deficiency leads to delayed maturation and altered gene expression in hippocampal neurons. Neurobiol. Dis. 2007, 27, 77–89. [Google Scholar]
- Cohen, S.; Gabel, H.W.; Hemberg, M.; Hutchinson, A.N.; Sadacca, L.A.; Ebert, D.H.; Harmin, D.A.; Greenberg, R.S.; Verdine, V.K.; Zhou, Z.; et al. Genome-wide activity-dependent MeCP2 phosphorylation regulates nervous system development and function. Neuron 2011, 72, 72–85. [Google Scholar]
- Xiang, Z.; Honig, B. Extending the accuracy limits of prediction for side-chain conformations. J. Mol. Biol. 2001, 311, 421–430. [Google Scholar]
- Xiang, Z.; Soto, C.S.; Honig, B. Evaluating conformational free energies: The colony energy and its application to the problem of loop prediction. Proc. Natl. Acad. Sci. USA 2002, 99, 7432–7437. [Google Scholar]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kalé, L.; Schulten, K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar]
- NAMD was developed by the Theoretical and Computational Biophysics Group in the Beckman Institute for Advanced Science and Technology at the University of Illinois at Urbana-Champaign
- Patton, E.E.; Zon, L.I. The art and design of genetic screens: zebrafish. Nat. Rev. Genet. 2001, 2, 956–966. [Google Scholar]
- Pietri, T.; Roman, A.C.; Guyon, N.; Romano, S.A.; Washbourne, P.; Moens, C.B.; de Polavieja, G.G.; Sumbre, G. The first mecp2-null zebrafish model shows altered motor behaviors. Front. Neural. Circuits 2013, 7, 118. [Google Scholar]
- Gibbs, E.M.; Horstick, E.J.; Dowling, J.J. Swimming into prominence: The zebrafish as a valuable tool for studying human myopathies and muscular dystrophies. FEBS J. 2013, 280, 4187–4197. [Google Scholar]
- De Bona, C.; Zappella, M.; Hayek, G.; Meloni, I.; Vitelli, F.; Bruttini, M.; Cusano, R.; Loffredo, P.; Longo, I.; Renieri, A. Preserved speech variant is allelic of classic Rett syndrome. Eur. J. Hum. Genet. 2000, 8, 325–330. [Google Scholar]
- Bebbington, A.; Anderson, A.; Ravine, D.; Fyfe, S.; Pineda, M.; de Klerk, N.; Ben-Zeev, B.; Yatawara, N.; Percy, A.; Kaufmann, W.E.; et al. Investigating genotype-phenotype relationships in Rett syndrome using an international data set. Neurology 2008, 70, 868–875. [Google Scholar]
- Renieri, A.; Mari, F.; Mencarelli, M.A.; Scala, E.; Ariani, F.; Longo, I.; Meloni, I.; Cevenini, G.; Pini, G.; Hayek, G.; et al. Diagnostic criteria for the Zappella variant of Rett syndrome (the preserved speech variant). Brain Dev. 2009, 31, 208–216. [Google Scholar]
- Thisse, B.; Heyer, V.; Lux, A.; Alunni, V.; Degrave, A.; Seiliez, I.; Kirchner, J.; Parkhill, J.P.; Thisse, C. Spatial and temporal expression of the zebrafish genome by large-scale in situ hybridization screening. Methods Cell Biol. 2004, 77, 505–519. [Google Scholar]
- Thisse, C.; Thisse, B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 2008, 3, 59–69. [Google Scholar]
- Clarke, L.; Zheng-Bradley, X.; Smith, R.; Kulesha, E.; Xiao, C.; Toneva, I.; Vaughan, B.; Preuss, D.; Leinonen, R.; Shumway, M.; et al. The 1000 Genomes Project: data management and community access. Nat. Meth. 2012, 9, 459–462. [Google Scholar]
- A map of human genome variation from population-scale sequencing. Nature 2010, 467, 1061–1073.[Green Version]
- Seth, A.; Stemple, D.L.; Barroso, I. The emerging use of zebrafish to model metabolic disease. Dis. Model Mech. 2013, 6, 1080–1088. [Google Scholar]
- Wager, K.; Mahmood, F.; Russell, C. Modelling inborn errors of metabolism in zebrafish. J. Inherit. Metab. Dis. 2014, 1–13. [Google Scholar]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Kucukkal, T.G.; Yang, Y.; Chapman, S.C.; Cao, W.; Alexov, E. Computational and Experimental Approaches to Reveal the Effects of Single Nucleotide Polymorphisms with Respect to Disease Diagnostics. Int. J. Mol. Sci. 2014, 15, 9670-9717. https://doi.org/10.3390/ijms15069670
Kucukkal TG, Yang Y, Chapman SC, Cao W, Alexov E. Computational and Experimental Approaches to Reveal the Effects of Single Nucleotide Polymorphisms with Respect to Disease Diagnostics. International Journal of Molecular Sciences. 2014; 15(6):9670-9717. https://doi.org/10.3390/ijms15069670
Chicago/Turabian StyleKucukkal, Tugba G., Ye Yang, Susan C. Chapman, Weiguo Cao, and Emil Alexov. 2014. "Computational and Experimental Approaches to Reveal the Effects of Single Nucleotide Polymorphisms with Respect to Disease Diagnostics" International Journal of Molecular Sciences 15, no. 6: 9670-9717. https://doi.org/10.3390/ijms15069670