Curcumin Promotes KLF5 Proteasome Degradation through Downregulating YAP/TAZ in Bladder Cancer Cells
Abstract
:1. Introduction
2. Results
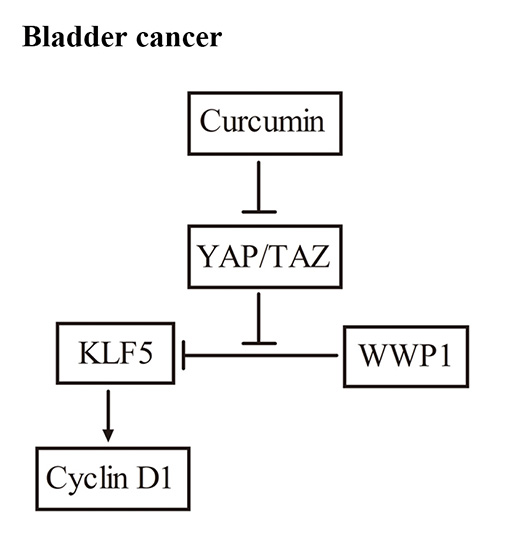
2.1. Curcumin Down-Regulated KLF5 Protein Expression in a Dose- and Time-Dependent Manner in 5637 and WH Bladder Cancer Cells

2.2. Curcumin Promoted Proteasome-Dependent Degradation of KLF5 Protein

2.3. KLF5 Mediated the Anti-Proliferative Effect of Curcumin

2.4. Curcumin Down-Regulated YAP and TAZ Expression

2.5. YAP Played Critical Roles in KLF5 Protein Stability in 5637 Cells

2.6. Curcumin Inhibited Subcutaneous Tumor Growth and the YAP/TAZ/KLF5/Cyclin D1 Axis in Vivo

3. Discussion
4. Experimental Section
4.1. Cell Culture and Reagents
4.2. 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) Assay
4.3. Western Blot Analysis
4.4. Cycloheximide (CHX) Chase Assay
4.5. Plasmids, Lentivirus Preparation, siRNA and Transfection
4.6. Real-Time Quantitative PCR (qPCR)
| Gene | Forward Primer | Reverse Primer |
|---|---|---|
| GAPDH | ATGGGGAAGGTGAAGGTCGG | GACGGTGCCATGGAATTTGC |
| KLF5 | CAGAGGACCTGGTCCAGACAAG | GAGGCCAGTTCTCAGGTGAGTG |
| WWP1 | ATGTTGTGTGGCATGCAGGA | CTGCAATAGTCGCATTCTTACTTCA |
| FBXW7 | CATATGTTGCTCAAAGGTGGCAAG | TGGGACAACAATCCCATATTTGAAG |
| SMURF2 | CAAGATTGCTTCAGTTTGTGACAG | GGGCTTTCGGCAGGTTGTT |
| YAP1 | AATTTGCCCAGTTATACCTCAGTG | CACATCAAGGCTATGATTCAAACTC |
| TAZ | GCTGAGCGTACAGGCAGACA | AGGAACATAAACCATGGGTCCTT |
| CDK6 | TGCACAGTGTCACGAACAGA | ACCTCGGAGAAGCTGAAACA |
| AXL | CGTAACCTCCACCTGGTCTC | TCCCATCGTCTGACAGCA |
| ITGB2 | AAGTGACGCTTTACCTGCGAC | AAGCATGGAGTAGGAGAGGTC |
| CYR61 | GGTCAAAGTTACCGGGCAGT | GGAGGCATCGAATCCCAGC |
4.7. Tumor Xenograft Model
4.8. Immunohistochemistry
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- Siegel, R.; Ma, J.; Zou, Z.; Jemal, A. Cancer statistics. CA Cancer J. Clin. 2014, 64, 9–29. [Google Scholar] [CrossRef]
- Van Rhijn, B.W.; Burger, M.; Lotan, Y.; Solsona, E.; Stief, C.G.; Sylvester, R.J.; Witjes, J.A.; Zlotta, A.R. Recurrence and progression of disease in non-muscle-invasive bladder cancer: From epidemiology to treatment strategy. Eur. Urol. 2009, 56, 430–442. [Google Scholar]
- Dong, J.T.; Chen, C. Essential role of KLF5 transcription factor in cell proliferation and differentiation and its implications for human diseases. Cell. Mol. Life Sci. 2009, 66, 2691–2706. [Google Scholar] [CrossRef]
- Tetreault, M.P.; Yang, Y.; Katz, J.P. Kruppel-like factors in cancer. Nat. Rev. Cancer 2013, 13, 701–713. [Google Scholar] [CrossRef]
- Bell, S.M.; Zhang, L.; Mendell, A.; Xu, Y.; Haitchi, H.M.; Lessard, J.L.; Whitsett, J.A. Kruppel-like factor 5 is required for formation and differentiation of the bladder urothelium. Dev. Biol. 2011, 358, 79–90. [Google Scholar] [CrossRef]
- Chen, C.; Benjamin, M.S.; Sun, X.; Otto, K.B.; Guo, P.; Dong, X.Y.; Bao, Y.; Zhou, Z.; Cheng, X.; Simons, J.W. KLF5 promotes cell proliferation and tumorigenesis through gene regulation and the TSU-Pr1 human bladder cancer cell line. Int. J. Cancer 2006, 118, 1346–1355. [Google Scholar] [CrossRef]
- Zhao, D.; Zheng, H.Q.; Zhou, Z.; Chen, C. The Fbw7 tumor suppressor targets KLF5 for ubiquitin-mediated degradation and suppresses breast cell proliferation. Cancer Res. 2010, 70, 4728–4738. [Google Scholar] [CrossRef]
- Chen, C.; Sun, X.; Guo, P.; Dong, X.Y.; Sethi, P.; Cheng, X.; Zhou, J.; Ling, J.; Simons, J.W.; Lingrel, J.B. Human Kruppel-like factor 5 is a target of the E3 ubiquitin ligase WWP1 for proteolysis in epithelial cells. J. Biol. Chem. 2005, 280, 41553–41561. [Google Scholar] [CrossRef]
- Du, J.X.; Hagos, E.G.; Nandan, M.O.; Bialkowska, A.B.; Yu, B.; Yang, V.W. The E3 ubiquitin ligase SMAD ubiquitination regulatory factor 2 negatively regulates Kruppel-like factor 5 protein. J. Biol. Chem. 2011, 286, 40354–40364. [Google Scholar] [CrossRef]
- Zhao, D.; Zhi, X.; Zhou, Z.; Chen, C. TAZ antagonizes the WWP1-mediated KLF5 degradation and promotes breast cell proliferation and tumorigenesis. Carcinogenesis 2012, 33, 59–67. [Google Scholar] [CrossRef]
- Zhi, X.; Zhao, D.; Zhou, Z.; Liu, R.; Chen, C. YAP promotes breast cell proliferation and survival partially through stabilizing the KLF5 transcription factor. Am. J. Pathol. 2012, 180, 2452–2461. [Google Scholar] [CrossRef]
- Kunnumakkara, A.B.; Anand, P.; Aggarwal, B.B. Curcumin inhibits proliferation, invasion, angiogenesis and metastasis of different cancers through interaction with multiple cell signaling proteins. Cancer Lett. 2008, 269, 199–225. [Google Scholar] [CrossRef]
- Gururaj, A.E.; Belakavadi, M.; Venkatesh, D.A.; Marme, D.; Salimath, B.P. Molecular mechanisms of anti-angiogenic effect of curcumin. Biochem. Biophys. Res. Commun. 2002, 297, 934–942. [Google Scholar] [CrossRef]
- Ye, M.X.; Li, Y.; Yin, H.; Zhang, J. Curcumin: Updated molecular mechanisms and intervention targets in human lung cancer. Int. J. Mol. Sci. 2012, 13, 3959–3978. [Google Scholar] [CrossRef]
- Kamat, A.M.; Tharakan, S.T.; Sung, B.; Aggarwal, B.B. Curcumin potentiates the antitumor effects of Bacillus Calmette-Guerin against bladder cancer through the down-regulation of NF-κB and up-regulation of TRAIL receptors. Cancer Res. 2009, 69, 8958–8966. [Google Scholar] [CrossRef]
- Chadalapaka, G.; Jutooru, I.; Chintharlapalli, S.; Papineni, S.; Smith, R.R.; Li, X.; Safe, S. Curcumin decreases specificity protein expression in bladder cancer cells. Cancer Res. 2008, 68, 5345–5354. [Google Scholar] [CrossRef]
- Chen, C.; Zhou, Z.; Guo, P.; Dong, J.T. Proteasomal degradation of the KLF5 transcription factor through a ubiquitin-independent pathway. FEBS Lett. 2007, 581, 1124–1130. [Google Scholar] [CrossRef]
- Tarapore, R.S.; Yang, Y.; Katz, J.P. Restoring KLF5 in esophageal squamous cell cancer cells activates the JNK pathway leading to apoptosis and reduced cell survival. Neoplasia 2013, 15, 472–480. [Google Scholar]
- Chen, C.; Bhalala, H.V.; Vessella, R.L.; Dong, J.T. KLF5 is frequently deleted and down-regulated but rarely mutated in prostate cancer. Prostate 2003, 55, 81–88. [Google Scholar] [CrossRef]
- Bialkowska, A.; Crisp, M.; Bannister, T.; He, Y.; Chowdhury, S.; Schurer, S.; Chase, P.; Spicer, T.; Madoux, F.; Tian, C. Identification of small-molecule inhibitors of the colorectal cancer oncogene Kruppel-like factor 5 expression by ultrahigh-throughput screening. Mol. Cancer Ther. 2011, 10, 2043–2051. [Google Scholar] [CrossRef]
- Musgrove, E.A.; Caldon, C.E.; Barraclough, J.; Stone, A.; Sutherland, R.L. Cyclin D as a therapeutic target in cancer. Nat. Rev. Cancer 2011, 11, 558–572. [Google Scholar] [CrossRef]
- Cai, X.Z.; Wang, J.; Li, X.D.; Wang, G.L.; Liu, F.N.; Cheng, M.S.; Li, F. Curcumin suppresses proliferation and invasion in human gastric cancer cells by down-regulation of PAK1 activity and cyclin D1 expression. Cancer Biol. Ther. 2009, 8, 1360–1368. [Google Scholar] [CrossRef]
- Suzuki, T.; Sawaki, D.; Aizawa, K.; Munemasa, Y.; Matsumura, T.; Ishida, J.; Nagai, R. Kruppel-like factor 5 shows proliferation-specific roles in vascular remodeling, direct stimulation of cell growth, and inhibition of apoptosis. J. Biol. Chem. 2009, 284, 9549–9557. [Google Scholar] [CrossRef]
- Aggarwal, B.B.; Banerjee, S.; Bharadwaj, U.; Sung, B.; Shishodia, S.; Sethi, G. Curcumin induces the degradation of cyclin E expression through ubiquitin-dependent pathway and up-regulates cyclin-dependent kinase inhibitors p21 and p27 in multiple human tumor cell lines. Biochem. Pharmacol. 2007, 73, 1024–1032. [Google Scholar] [CrossRef]
- Jung, Y.; Xu, W.; Kim, H.; Ha, N.; Neckers, L. Curcumin-induced degradation of ErbB2: A role for the E3 ubiquitin ligase CHIP and the Michael reaction acceptor activity of curcumin. Biochim. Biophys. Acta 2007, 1773, 383–390. [Google Scholar]
- Milacic, V.; Banerjee, S.; Landis-Piwowar, K.R.; Sarkar, F.H.; Majumdar, A.P.; Dou, Q.P. Curcumin inhibits the proteasome activity in human colon cancer cells in vitro and in vivo. Cancer Res. 2008, 68, 7283–7292. [Google Scholar] [CrossRef]
- Jana, N.R.; Dikshit, P.; Goswami, A.; Nukina, N. Inhibition of proteasomal function by curcumin induces apoptosis through mitochondrial pathway. J. Biol. Chem. 2004, 279, 11680–11685. [Google Scholar] [CrossRef]
- Xu, S.; Guo, P.; Gao, Y.; Shi, Q.; He, D.; Gao, Y.; Zhang, H. Acyldepsipeptides inhibit the growth of renal cancer cells through G1 phase cell cycle arrest. Biochem. Biophys. Res. Commun. 2013, 438, 468–472. [Google Scholar] [CrossRef]
- Chen, C.; Sun, X.; Ran, Q.; Wilkinson, K.D.; Murphy, T.J.; Simons, J.W.; Dong, J.T. Ubiquitin-proteasome degradation of KLF5 transcription factor in cancer and untransformed epithelial cells. Oncogene 2005, 24, 3319–3327. [Google Scholar] [CrossRef]
- Guo, P.; Dong, X.Y.; Zhao, K.; Sun, X.; Li, Q.; Dong, J.T. Opposing effects of KLF5 on the transcription of MYC in epithelial proliferation in the context of transforming growth factor beta. J. Biol. Chem. 2009, 284, 28243–28252. [Google Scholar] [CrossRef]
- Zhang, T.; Guo, P.; Zhang, Y.; Xiong, H.; Yu, X.; Xu, S.; Wang, X.; He, D.; Jin, X. The antidiabetic drug metformin inhibits the proliferation of bladder cancer cells in vitro and in vivo. Int. J. Mol. Sci. 2013, 14, 24603–24618. [Google Scholar] [CrossRef]
- Tu, C.T.; Han, B.; Liu, H.C.; Zhang, S.C. Curcumin protects mice against concanavalin A-induced hepatitis by inhibiting intrahepatic intercellular adhesion molecule-1 (ICAM-1) and CXCL10 expression. Mol. Cell. Biochem. 2011, 358, 53–60. [Google Scholar]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Gao, Y.; Shi, Q.; Xu, S.; Du, C.; Liang, L.; Wu, K.; Wang, K.; Wang, X.; Chang, L.S.; He, D.; et al. Curcumin Promotes KLF5 Proteasome Degradation through Downregulating YAP/TAZ in Bladder Cancer Cells. Int. J. Mol. Sci. 2014, 15, 15173-15187. https://doi.org/10.3390/ijms150915173
Gao Y, Shi Q, Xu S, Du C, Liang L, Wu K, Wang K, Wang X, Chang LS, He D, et al. Curcumin Promotes KLF5 Proteasome Degradation through Downregulating YAP/TAZ in Bladder Cancer Cells. International Journal of Molecular Sciences. 2014; 15(9):15173-15187. https://doi.org/10.3390/ijms150915173
Chicago/Turabian StyleGao, Yang, Qi Shi, Shan Xu, Chong Du, Liang Liang, Kaijie Wu, Ke Wang, Xinyang Wang, Luke S. Chang, Dalin He, and et al. 2014. "Curcumin Promotes KLF5 Proteasome Degradation through Downregulating YAP/TAZ in Bladder Cancer Cells" International Journal of Molecular Sciences 15, no. 9: 15173-15187. https://doi.org/10.3390/ijms150915173