Dwell-Time Distribution, Long Pausing and Arrest of Single-Ribosome Translation through the mRNA Duplex
Abstract
:1. Introduction
2. Results
2.1. Comparison between Theoretical and Experimental Data on the Distribution of Dwell Times
| Rate Constants | Values | Reference |
|---|---|---|
| k1 (s–1) | 1 | [14,15,16,17,18] |
| k2 (s–1) | 0 | [17,18,19] |
| k3 (s–1) | 20 | [20] |
| k4 (s–1) | 100 | [20] |
| k5 (s–1) | 20 | [21,22,23,24,25,26] |
| kb (µM–1∙s–1) | 110 | [21,22,23,24,25,26] |
| k–6 (s–1) | 25 | [21,22,23,24,25,26] |
| k7 (s–1) | 100 | [21,22,23,24,25,26] |
| k–7 (s–1) | 0.2 | [21,22,23,24,25,26] |
| k8 (s–1) | 260 | [21,22,23,24,25,26] |
| k9 (s–1) | 60 | [21,22,23,24,25,26] |
| k10 (s–1) | 3 | [21,22,23,24,25,26] |
| k11 (s–1) | 50 | [21,22,23,24,25,26] |
| kr (s–1) | 1 | [21,22,23,24,25,26,27] |


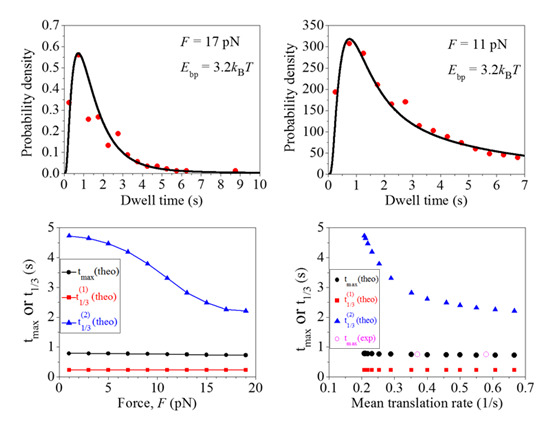
2.2. Effect of the Pulling Force to Unzip the mRNA Duplex on the Distribution of Dwell Times



2.3. Dynamics of Pausing and Arrest Caused by the Interaction of the SD Sequence with Anti-SD Site of the 30S Subunit


3. Discussion
3.1. The Justification of the Occurrence of Futile Translocation and Multiple GTP Hydrolyses


3.2. Three Types of Translational Pausing
4. Methods
4.1. Model of Ribosome Translocation through the mRNA Duplex
4.2. Elongation Cycle of Ribosome Translation through the mRNA Duplex at Saturating Concentration of EF-G∙GTP
4.3. Equations for Probability of Effective Translocation during Ribosome Translation through the Duplex Region of mRNA
4.4. Equations for Distribution of Dwell Times
4.5. Choice of Parameter Values
5. Conclusions
Supplementary Materials
Acknowledgments
Conflicts of Interest
References
- Jou, W.M.; Haegeman, G.; Ysebaert, M.; Fiers, W. Nucleotide sequence of the gene coding for the bacteriophage MS2 coat protein. Nature 1972, 237, 82–88. [Google Scholar] [CrossRef]
- Favre, A.; Morel, C.; Scherrer, K. The secondary structure and poly(A) content of globin messenger RNA as a pure RNA and in polyribosome-derived ribonucleoprotein complexes. Eur. J. Biochem. 1975, 57, 147–157. [Google Scholar] [CrossRef] [PubMed]
- Holder, J.W.; Lingrel, J.B. Determination of secondary structure in rabbit globin messenger RNA by thermal denaturation. Biochemistry 1975, 14, 4209–4215. [Google Scholar] [CrossRef] [PubMed]
- Holbrook, S.R. RNA structure: The long and the short of it. Curr. Opin. Struct. Biol. 2005, 15, 302–308. [Google Scholar] [CrossRef] [PubMed]
- Takyar, S.; Hickerson, R.P.; Noller, H.F. mRNA helicase activity of the ribosome. Cell 2005, 120, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Wen, J.-D.; Lancaster, L.; Hodges, C.; Zeri, A.-C.; Yoshimura, S.H.; Noller, H.F.; Bustamante, C.; Tinoco, I., Jr. Following translation by single ribosomes one codon at a time. Nature 2008, 452, 598–603. [Google Scholar] [CrossRef] [PubMed]
- Qu, X.; Wen, J.-D.; Lancaster, L.; Noller, H.F.; Bustamante, C.; Tinoco, I., Jr. The ribosome uses two active mechanisms to unwind messenger RNA during translation. Nature 2011, 475, 118–121. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Zhang, H.; Broitman, S.L.; Reiche, M.; Farrell, I.; Cooperman, B.S.; Goldman, Y.E. Dynamics of translation by single ribosomes through mRNA secondary structures. Nat. Struct. Mol. Biol. 2013, 20, 582–588. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.-K.; Liu, F.; Fei, J.; Bustamante, C.; Gonzalez, R.L., Jr.; Tinoco, I., Jr. A frameshifting stimulatory stem loop destabilizes the hybrid state and impedes ribosomal translocation. Proc. Natl. Acad. Sci. USA 2014, 111, 5538–5543. [Google Scholar] [CrossRef] [PubMed]
- Tinoco, I., Jr.; Wen, J.D. Simulation and analysis of single-ribosome translation. Phys. Biol. 2009, 6, 025006. [Google Scholar] [CrossRef] [PubMed]
- Garai, A.; Chowdhury, D.; Chowdhury, D.; Ramakrishnan, T.V. Stochastic kinetics of ribosomes: Single motor properties and collective behavior. Phys. Rev. E 2009, 80, 011908. [Google Scholar] [CrossRef]
- Sharma, A.K.; Chowdhury, D. Distribution of dwell times of a ribosome: Effects of infidelity, kinetic proofreading and ribosome crowding. Phys. Biol. 2011, 8, 026005. [Google Scholar] [CrossRef] [PubMed]
- Xie, P. Model of ribosome translation and mRNA unwinding. Eur. Biophys. J. 2013, 42, 347–354. [Google Scholar] [CrossRef] [PubMed]
- Blanchard, S.C.; Kim, H.D.; Gonzalez, R.L., Jr.; Puglisi, J.D.; Chu, S. tRNA dynamics on the ribosome during translation. Proc. Natl. Acad. Sci. USA 2004, 101, 12893–12898. [Google Scholar] [CrossRef] [PubMed]
- Ermolenko, D.N.; Majumdar, Z.K.; Hickerson, R.P.; Spiegel, P.C.; Clegg, R.M.; Noller, H.F. Observation of intersubunit movement of the ribosome in solution using FRET. J. Mol. Biol. 2007, 370, 530–540. [Google Scholar] [CrossRef] [PubMed]
- Munro, J.B.; Altman, R.B.; O’Connor, N.; Blanchard, S.C. Identification of two distinct hybrid state intermediates on the ribosome. Mol. Cell 2007, 25, 505–517. [Google Scholar] [CrossRef] [PubMed]
- Fei, J.; Kosuri, P.; MacDougall, D.D.; Gonzalez, R.L., Jr. Coupling of ribosomal L1stalk and tRNA dynamics during translation elongation. Mol. Cell 2008, 30, 348–359. [Google Scholar] [CrossRef] [PubMed]
- Cornish, P.V.; Ermolenko, D.N.; Noller, H.F.; Ha, T. Spontaneous intersubunit rotation in single ribosomes. Mol. Cell 2008, 30, 578–588. [Google Scholar] [CrossRef] [PubMed]
- Valle, M.; Zavialov, A.; Sengupta, J.; Rawat, U.; Ehrenberg, M.; Frank, J. Locking and unlocking of ribosomal motions. Cell 2003, 114, 123–134. [Google Scholar] [CrossRef]
- Savelsbergh, A.; Katunin, V.I.; Mohr, D.; Peske, F.; Rodnina, M.V.; Wintermeyer, W. An elongation factor G-induced ribosome rearrangement precedes tRNA–mRNA translocation. Mol. Cell 2003, 11, 1517–1523. [Google Scholar] [CrossRef]
- Wintermeyer, W.; Peske, F.; Beringer, M.; Gromadski, K.B.; Savelsbergh, A.; Rodnina, M.V. Mechanisms of elongation on the ribosome: Dynamics of a macromolecular machine. Biochem. Soc. Trans. 2004, 32, 733–737. [Google Scholar] [CrossRef] [PubMed]
- Pape, T.; Wintermeyer, W.; Rodnina, M.V. Complete kinetic mechanism of elongation factor Tu-dependent binding of aminoacyl-tRNA to the A site of the E. coli ribosome. EMBO J. 1998, 17, 7490–7497. [Google Scholar] [CrossRef] [PubMed]
- Pape, T.; Wintermeyer, W.; Rodnina, M.V. Induced fit in initial selection and proofreading of aminocyl-tRNA on the ribosome. EMBO J. 1999, 18, 3800–3807. [Google Scholar] [CrossRef] [PubMed]
- Rodnina, M.V.; Beringer, M.; Wintermeyer, W. How ribosomes make peptide bonds. Trends Biochem. Sci. 2007, 32, 20–26. [Google Scholar] [CrossRef] [PubMed]
- Stark, H.; Rodnina, M.V.; Wieden, H.J.; Zemlin, F.; Wintermeyer, W.; van Heel, M. Ribosome interactions of aminoacyl-tRNA and elongation factor Tu in the codon-recognition complex. Nat. Struct. Biol. 2002, 9, 849–854. [Google Scholar] [CrossRef] [PubMed]
- Wilden, B.; Savelsbergh, A.; Rodnina, M.V.; Wintermeyer, W. Role and timing of GTP binding and hydrolysis during EF-G-dependent tRNA translocation on the ribosome. Proc. Natl. Acad. Sci. USA 2006, 103, 13670–13675. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Petrov, A.; Tsai, A.; O’Leary, S.E.; Puglisi, J.D. Coordinated conformational and compositional dynamics drive ribosome translocation. Nat. Struct. Mol. Biol. 2013, 20, 718–727. [Google Scholar] [CrossRef] [PubMed]
- Li, G.-W.; Oh, E.; Weissman, J.S. The anti-Shine-Dalgarno sequence drives translational pausing and codon choice in bacteria. Nature 2012, 484, 538–541. [Google Scholar] [CrossRef] [PubMed]
- Xie, P. Dynamics of tRNA translocation, mRNA translocation and tRNA dissociation during ribosome translation through mRNA secondary structures. Eur. Biophys. J. 2014, 43, 229–240. [Google Scholar] [CrossRef] [PubMed]
- Xie, P. Origin of multiple intersubunit rotations before EF-G-catalyzed ribosomal translocation through the mRNA with a downstream secondary structure. BMC Biophys. 2014, 7, 12. [Google Scholar] [CrossRef]
- Spiegel, P.C.; Ermolenko, D.N.; Noller, H.F. Elongation factor G stabilizes the hybrid-state conformation of the 70S ribosome. RNA 2007, 13, 1473–1482. [Google Scholar] [CrossRef] [PubMed]
- Xie, P. Dynamics of backtracking long pauses of RNA polymerase. Biochim. Biophys. Acta 2009, 1789, 212–219. [Google Scholar] [CrossRef] [PubMed]
- Misra, R.; Reeves, P. Intermediates in the synthesis of TolC protein include an incomplete peptide stalled at a rare Arg codon. Eur. J. Biochem. 1985, 152, 151–155. [Google Scholar] [CrossRef] [PubMed]
- Sørensen, M.; Pedersen, S. Absolute in vivo translation rates of individual codons in Escherichia coli: The two glutamic acid codons GAA and GAG are translated with a threefold difference in rate. J. Mol. Biol. 1991, 222, 265–280. [Google Scholar] [CrossRef]
- Charneski, C.A.; Hurst, L.D. Positively charged residues are the major determinants of ribosomal velocity. PLoS Biol. 2013, 11, e1001508. [Google Scholar] [CrossRef] [PubMed]
- Goldman, D.H.; Kaiser, C.M.; Milin, A.; Righini, M.; Tinoco, I., Jr.; Bustamante, C. Mechanical force releases nascent chain-mediated ribosome arrest in vitro and in vivo. Science 2015, 348, 457–460. [Google Scholar] [CrossRef] [PubMed]
- Xie, P. Biphasic character of ribosomal translocation and non-Michaelis-Menten kinetics of translation. Phys. Rev. E 2014, 90, 062703. [Google Scholar] [CrossRef]
- Xie, P. Translocation dynamics of tRNA-mRNA in the ribosome. Biophys. Chem. 2013, 180–181, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Gagnon, M.G.; Bulkley, D.; Steitz, T.A. Conformational changes of elongation factor G on the ribosome during tRNA translocation. Cell 2015, 160, 219–227. [Google Scholar] [CrossRef] [PubMed]
- Schuwirth, B.S.; Borovinskaya, M.A.; Hau, C.W.; Zhang, W.; Vila-Sanjurjo, A.; Holton, J.M.; Cate, J.H.D. Structures of the bacterial ribosome at 3.5 Å resolution. Science 2005, 310, 827–834. [Google Scholar] [CrossRef] [PubMed]
- Xie, P. Model of ribosomal translocation coupled with intra- and inter-subunit rotations. Biochem. Biophys. Rep. 2015, 2, 87–93. [Google Scholar] [CrossRef]
- Lill, R.; Robertson, J.M.; Wintermeyer, W. Binding of the 30-terminus of tRNA to 23S rRNA in the ribosomal exit site actively promotes translocation. EMBO J. 1989, 8, 3933–3938. [Google Scholar] [PubMed]
- Feinberg, J.S.; Joseph, S. Identification of molecular interactions between P-site tRNA and the ribosome essential for translocation. Proc. Natl. Acad. Sci. USA 2001, 98, 11120–11125. [Google Scholar] [CrossRef] [PubMed]
- Frank, J.; Agrawal, R.K. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature 2000, 406, 318–322. [Google Scholar] [CrossRef] [PubMed]
- Rodnina, M.V.; Fricke, R.; Kuhn, L.; Wintermeyer, W. Codon dependent conformational change of elongation factor Tu preceding GTP hydrolysis on the ribosome. EMBO J. 1995, 14, 2613–2619. [Google Scholar] [PubMed]
- Dell, V.A.; Miller, D.L.; Johnson, A.E. Effects of nucleotide and aurodox-induced changes in elongation factor Tu conformation upon its interactions with aminoacyl transfer RNA. A fluorescence study. Biochemistry 1990, 29, 1757–1763. [Google Scholar] [CrossRef] [PubMed]
- Abel, K.; Yoder, M.D.; Hilgenfeld, R.; Jurnak, F. An α to β conformational switch in EF-Tu. Structure 1996, 4, 1153–1159. [Google Scholar] [CrossRef]
- Lionnet, T.; Spiering, M.M.; Benkovic, S.J.; Bensimon, D.; Croquette, V. Real-time observation of bacteriophage T4 gp41 helicase reveals an unwinding mechanism. Proc. Natl. Acad. Sci. USA 2007, 104, 19790–19795. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.; Cui, Y.; Bustamante, C. Overstretching B-DNA: The elastic response of individual double-stranded and single-stranded DNA molecules. Science 1996, 271, 795–799. [Google Scholar] [CrossRef] [PubMed]
- Cui, S.; Yu, Y.; Lin, Z. Modeling single chain elasticity of single-stranded DNA: A comparison of three models. Polymer 2009, 50, 930–935. [Google Scholar] [CrossRef]
- Freier, S.M.; Kierzek, R.; Jaeger, J.A.; Sugimoto, N.; Caruthers, M.H.; Neilson, T.; Tuener, D.H. Improved free-energy parameters for predictions of RNA duplex stability. Proc. Natl. Acad. Sci. USA 1986, 83, 9373–9377. [Google Scholar] [CrossRef] [PubMed]
- Buhot, A.; Halperin, A. Effects of stacking on the configurations and elasticity of single-stranded nucleic acids. Phys. Rev. E 2004, 70, 020902. [Google Scholar] [CrossRef]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xie, P. Dwell-Time Distribution, Long Pausing and Arrest of Single-Ribosome Translation through the mRNA Duplex. Int. J. Mol. Sci. 2015, 16, 23723-23744. https://doi.org/10.3390/ijms161023723
Xie P. Dwell-Time Distribution, Long Pausing and Arrest of Single-Ribosome Translation through the mRNA Duplex. International Journal of Molecular Sciences. 2015; 16(10):23723-23744. https://doi.org/10.3390/ijms161023723
Chicago/Turabian StyleXie, Ping. 2015. "Dwell-Time Distribution, Long Pausing and Arrest of Single-Ribosome Translation through the mRNA Duplex" International Journal of Molecular Sciences 16, no. 10: 23723-23744. https://doi.org/10.3390/ijms161023723