Polar Glycosylated and Lateral Non-Glycosylated Flagella from Aeromonas hydrophila Strain AH-1 (Serotype O11)
Abstract
:1. Introduction
2. Results

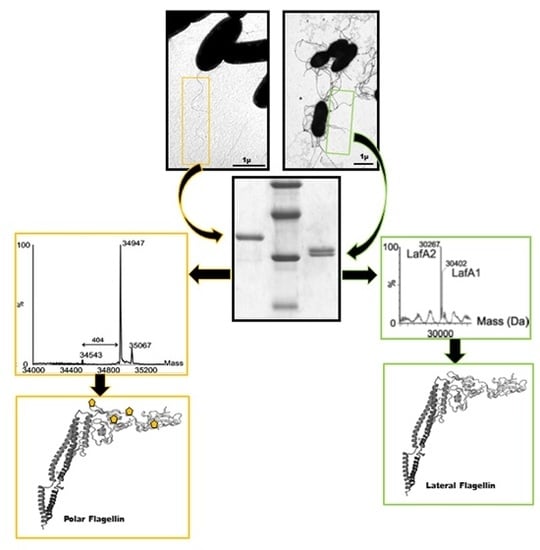
2.1. Mass Spectrometry Analyses of Wild-Type Lateral and Polar Flagellins




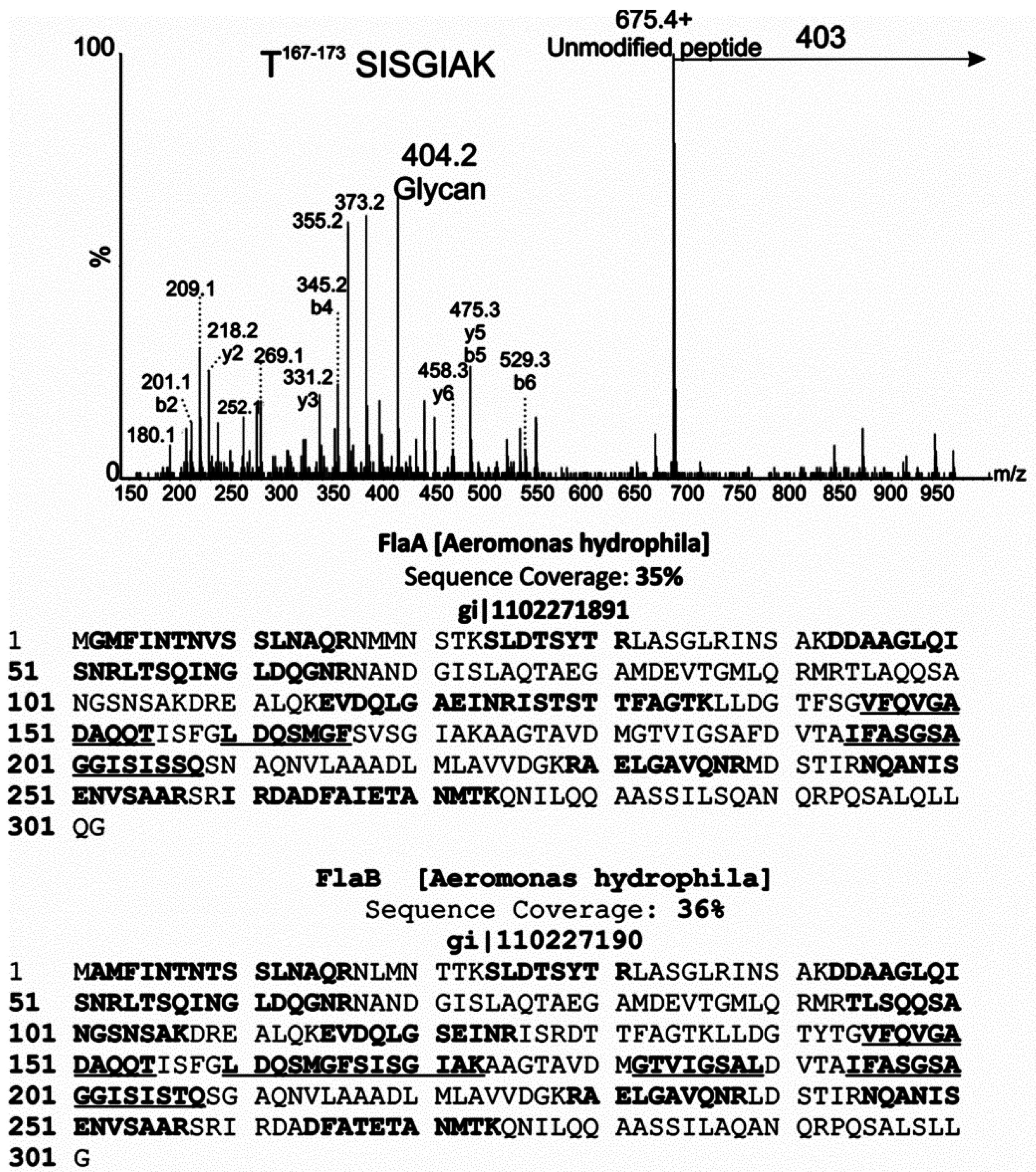
2.2. Tandem Mass Spectrometry Analyses of Proteolytic Digests of Polar Flagellin Proteins
2.3. Putative Pseudaminic Acid Biosynthetic Mutants
2.4. Adhesion to HEp-2 Cells and Biofilm Formation
| Strain and Main Characteristics | Mean No. of Bacteria | % Reduction |
|---|---|---|
| HEp-2 Cell ± SD | in Adhesion a | |
| AH-1; wild-type serotype O11 | 21.4 ± 3.6 | – |
| AH-1ΔpseB (flagella−) (O11+; flagella polar−/lateral−, grown in TSB) | 2.7 ± 0.9 | 87 * |
| AH-1∆pseB (flagella−) (O11+; flagella polar−/lateral+, grown in TSA) | 7.5 ± 1.8 | 65 * |
| AH-1∆pseI (flagella−) (O11+; flagella polar−/lateral−, grown in TSB) | 2.9 ± 0.5 | 86 * |
| flagella polar−/lateral−, grown in TSA | 8.1 ± 1.1 | 63 * |
| AH-1∆pseB + pBAD-pseB (flagella−) (O11+; flagella polar+/lateral−, grown in TSB) | 19.8 ± 2.4 | <8 |
| flagella polar+/lateral−, grown in TSB | 20.7 ± 2.0 | <8 |
| AH-1∆rmlB (O11− mutant) (O11−; flagella+) | 15.4 ± 2.6 | 29 * |
| AH-1∆rmlB + pBAD-rmlB (O11+; flagella+) | 20.4 ± 3.2 | <8 |
| Strain and Characteristics | Value (OD570) |
|---|---|
| AH-1, wild-type serotype O11 | 1.43 ± 0.15 |
| AH-1∆pseB (flagella−) (O11+; flagella polar−/lateral−, grown in TSB) | <0.1 |
| AH-1∆pseB (flagella−) (O11+; flagella polar−/lateral+, grown in TSA) | <0.1 |
| AH-1ΔpseI (flagella−) (O11+; flagella polar−/lateral−, grown in TSB) | <0.1 |
| AH-1∆pseI (flagella−) (O11+; flagella polar−/lateral+, grown in TSA) | <0.1 |
| AH-1∆pseB + pBAD-pseB (flagella−) (O11+; flagella polar+/lateral−, grown in TSB) | 1.37 ± 0.11 |
| AH-1∆pseI + pBAD-pseI (flagella−) (O11+; flagella polar+/lateral−, grown in TSB) | 1.39 ± 0.18 |
| AH-1∆rmlB (O11−; flagella+) | 0.78 ± 0.13 |
| AH-1∆rmlB + pBAD-rmlB | 1.41 ± 0.17 |
2.5. IL-8 Immune Stimulation

3. Discussion
4. Experimental Section
4.1. Bacterial Strains, Plasmids, and Growth Conditions
| Strain or Plasmid | Relevant Characteristics | Reference or Source |
|---|---|---|
| E. coli Strains | ||
| DH5α | F− end A hsdR17 (rK− mK+) supE44 thi-1 recA1 gyr-A96 Ф80lacZM15 | [31] |
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac (F− proABlacIqZ_M15 Tn10) | Stratagene |
| BL21(λD3) | F− ompT hsdSB (rB− mB−) gal dcm(λD3) | Novagen |
| A. hydrophila Strains | ||
| AH-1 | O11, Wild type | [19] |
| AH-Rif R | AH-1, spontaneous rifampicin resistant mutant, Rif R | [19] |
| AH-1ΔflaB-J | AH-1 mutant in frame unable produce polar flagellum but able to produce lateral flagella | This study |
| AH-1ΔrmlB | AH-1 mutant in frame unable produce O11-antigen LPS | [18] |
| AH-1ΔpseB | AH-1 pseB mutant in frame with pDM4 | This study |
| AH-1ΔpseI | AH-1 pseI mutant in frame with pDM4 | This study |
| Plasmids | ||
| pRK2073 | Helper plasmid,,Spc R | [32] |
| pBAD33 | arabinose inducible expression vector, Cm R | [33] |
| pBAD-pseB | pBAD33 with AH-1 pseB | This study |
| pBAD-pseI | pBAD33 with AH-1 pseI | This study |
| pDM4 | pir dependent with sacAB genes, oriR6K, Cm R | [34] |
| pDM4-flaB-J | pDM4 with AH-1 flaB-J fragment, Cm R | This study |
| pDM4-pseB | pDM4 with AH-1 pseB fragment, Cm R | This study |
| pDM4-pseI | pDM4 with AH-1 pseI fragment, Cm R | This study |
| pET-30 Xa/LIC | IPTG inducible expression vector Km R | Novagen |
| pET-30-FlaB-AH1 | pET-30 Xa/LIC with A. hydrophila AH-1 flaB | This study |
4.2. DNA Techniques
4.3. Construction of Defined Mutants
4.4. Plasmid Constructions
4.5. Flagella Purification
4.6. Motility Assays (Swarming and Swimming)
4.7. Transmission Electron Microscopy (TEM)
4.8. Electrospray Liquid Chromatography Mass Spectrometry Analysis of Intact Flagellins
4.9. Solution Enzymatic Digests and Bottom-Up Mass Spectrometry Analysis of Glycopeptides
4.10. Adherence Assay to HEp-2 Cell
4.11. Biofilm Formation
4.12. Purification of A. hydrophila AH-1 His6-FlaB
4.13. Interleukin-8 (IL-8) Assay with Human Embryonic Kidney Cells
4.14. Statistical Analysis
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Janda, J.M.; Kokka, R.P. The pathogenicity of Aeromonas strains relative to genospecies and phenospecies identification. FEMS Microbiol. Lett. 1991, 90, 29–34. [Google Scholar] [CrossRef] [Green Version]
- Janda, J.M.; Brenden, R. Importance of Aeromonas sobria in Aeromonas bacteremia. J. Infect. Dis. 1987, 155, 589–591. [Google Scholar] [CrossRef] [PubMed]
- Janda, J.M.; Guthertz, L.S.; Kokka, R.P.; Shimada, T. Aeromonas species in septicemia: Laboratory characteristics and clinical observations. Clin. Infect. Dis. 1994, 19, 77–83. [Google Scholar] [PubMed]
- Kokka, R.P.; Janda, J.M. Isolation and identification of autoagglutinating serogroup O:11 Aeromonas strains in the clinical laboratory. J. Clin. Microbiol. 1990, 28, 1297–1299. [Google Scholar] [PubMed]
- Shaw, D.H.; Squires, M. O-Antigen structure in a virulent strain of Aeromonas hydrophila. FEMS Microbiol. Lett. 1984, 24, 277–280. [Google Scholar] [CrossRef]
- Murray, R.G.E.; Dooley, J.S.G.; Whippey, P.W.; Trust, T.J. Structure of an S-layer on a pathogenic strain of Aeromonas hydrophila. J. Bacteriol. 1988, 170, 2625–2636. [Google Scholar] [PubMed]
- Frenchel, T. Microbial behavior in a heterogeneous world. Science 2002, 296, 1068–1071. [Google Scholar] [CrossRef] [PubMed]
- Kirov, S.M.; Tassel, B.C.; Semmler, A.B.T.; O’Donovan, L.A.; Rabaan, A.A.; Shaw, J.G. Lateral flagella and swarming motility in Aeromonas species. J. Bacteriol. 2002, 184, 547–555. [Google Scholar] [CrossRef] [PubMed]
- Shimada, T.; Sakazaki, R.; Suzuki, K. Peritrichous flagella in mesophilic strains of Aeromonas. Jpn. J. Med. Sci. Biol. 1985, 38, 141–145. [Google Scholar] [CrossRef] [PubMed]
- Kirov, S.M.; Castrisios, M.; Shaw, J.G. Aeromonas flagella (polar and lateral) are enterocyte adhesins that contribute to biofilm formation on surfaces. Infect. Immun. 2004, 72, 1939–1945. [Google Scholar] [CrossRef] [PubMed]
- Iwashkiw, J.A.; Vozza, N.F.; Kinsella, R.L.; Feldman, M.F. Pour some sugar on it: The expanding world of bacterial protein O-linked glycosylation. Mol. Microbiol. 2013, 89, 14–28. [Google Scholar] [CrossRef] [PubMed]
- Schirm, M.; Soo, E.C.; Aubry, A.J.; Austin, J.; Thibault, P.; Logan, S.M. Structural, genetic and functional characterization of the flagellin glycosylation process in Helicobacter pylori. Mol. Microbiol. 2003, 48, 1579–1592. [Google Scholar] [CrossRef] [PubMed]
- Goon, S.; Kelly, J.F.; Logan, S.M.; Ewing, C.P.; Guerry, P. Pseudaminic acid, the major modification on Campylobacter flagellin, is synthesized via the Cj1293 gene. Mol. Microbiol. 2003, 50, 659–671. [Google Scholar] [CrossRef] [PubMed]
- Faridmoayer, A.; Fentabil, M.A.; Mills, D.C.; Klassen, J.S.; Feldman, M.F. Functional characterization of bacterial oligosaccharyltransferases involved in O-linked protein glycosylation. J. Bacteriol. 2007, 189, 8088–8098. [Google Scholar] [CrossRef] [PubMed]
- Iwashkiw, J.A.; Seper, A.; Weber, B.S.; Scott, N.E.; Vinogradov, E.; Stratilo, C.; Reiz, B.; Cordwell, S.J.; Whittal, R.; Schild, S.; et al. Identification of a general O-linked protein glycosylation system in Acinetobacter baumannii and its role in virulence and biofilm formation. PLoS Pathog. 2012, 8, e1002758. [Google Scholar] [CrossRef] [PubMed]
- Merino, S.; Tomás, J.M. Gram-negative flagella glycosylation. Int. J. Mol. Sci. 2014, 15, 2840–2857. [Google Scholar] [CrossRef] [PubMed]
- Merino, S.; Rubires, X.; Aguilar, A.; Tomás, J.M. The role of flagella and motility on the adherence and invasion to fish cell lines by Aeromonas hydrophila strains serogroup O:34. FEMS Microbiol. Lett. 1997, 151, 213–217. [Google Scholar] [CrossRef] [PubMed]
- Merino, S.; Canals, R.; Knirel, Y.A.; Tomás, J.M. Molecular and chemical analysis of the lipopolysaccharide from Aeromonas hydrophila strain AH-1 (serotype O11). Mar. Drugs 2015, 13, 2233–2249. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.B.; Srinivasa Rao, P.S.; Lee, H.C.; Vilches, S.; Merino, S.; Tomás, J.M.; Leung, K.Y. A Type III secretion system is required for Aeromonas hydrophila AH-1 pathogenesis. Infect. Immun. 2004, 72, 1248–1256. [Google Scholar] [CrossRef] [PubMed]
- Merino, S.; Jiménez, N.; Molero, R.; Bouamama, L.; Regué, M.; Tomás, J.M. A UDP-HexNAc: Polyprenol-P GalNAc-1-P transferase (WecP) representing a new subgroup of this enzyme family. J. Bacteriol. 2011, 193, 1943–1952. [Google Scholar] [CrossRef] [PubMed]
- Canals, R.; Ramirez, S.; Vilches, S.; Horsburgh, G.; Shaw, J.G.; Tomás, J.M.; Merino, S. Polar flagella biogenesis in Aeromonas hydrophila. J. Bacteriol. 2006, 188, 542–555. [Google Scholar] [CrossRef] [PubMed]
- Canals, R.; Altarriba, M.; Vilches, S.; Horsburgh, G.; Shaw, J.G.; Tomás, J.M.; Merino, S. Analysis of the lateral flagella gene system of Aeromonas hydrophila AH-3. J. Bacteriol. 2006, 188, 852–862. [Google Scholar] [CrossRef] [PubMed]
- Wilhelms, M.; Fulton, K.M.; Twine, S.M.; Tomás, J.M.; Merino, S. Differential glycosylation of polar and lateral flagellins in Aeromonas hydrophila AH-3. J. Biol. Chem. 2012, 287, 27851–27862. [Google Scholar] [CrossRef] [PubMed]
- Schirm, M.; Schoenhofen, I.C.; Logan, S.M.; Waldron, K.C.; Thibault, P. Identification of Unusual Bacterial Glycosylation by Tandem Mass Spectrometry Analyses of Intact Proteins. Anal. Chem. 2005, 77, 7774–7782. [Google Scholar] [CrossRef] [PubMed]
- Schoenhofen, I.C.; Vinogradov, E.; Whitfield, D.M.; Brisson, J.R.; Logan, S.M. The CMP-legionaminic acid pathway in Campylobacter: Biosynthesis involving novel GDP-linked precursors. Glycobiology 2009, 19, 715–725. [Google Scholar] [CrossRef] [PubMed]
- Kokka, R.P.; Janda, J.M.; Oshiro, L.S.; Altwegg, M.; Shimada, T.; Sakazaki, R.; Brenner, D.J. Biochemical and genetic characterization of autoagglutinating phenotypes of Aeromonas. species associated with invasive and noninvasive disease. J. Infect. Dis. 1991, 163, 890–894. [Google Scholar] [CrossRef] [PubMed]
- Twine, S.M.; Reid, C.W.; Aubry, A.; McMullin, D.R.; Fulton, K.M.; Austin, J.; Logan, S.M. Motility and flagellar glycosylation in Clostridium difficile. J. Bacteriol. 2009, 191, 7050–7062. [Google Scholar] [CrossRef] [PubMed]
- Miller, W.L.; Matewish, M.J.; McNally, D.J.; Ishiyama, N.; Anderson, E.M.; Brewer, D.; Brisson, J.R.; Berghuis, A.M.; Lam, J.S. Flagellin glycosylation in Pseudomonas aeruginosa PAK requires the O-antigen biosynthesis enzyme WbpO. J. Biol. Chem. 2008, 283, 3507–3518. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.C. Regulation of Salmonella flagellin-induced interleukin-8 in intestinal epithelial cells by muramyl dipeptide. Cell Immunol. 2012, 278, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Basu, M.; Swain, B.; Maiti, N.K.; Routray, P.; Samanta, M. Inductive expression of toll-like receptor 5 (TLR5) and associated downstream signaling molecules following ligand exposure and bacterial infection in the Indian major carp, mrigal (Cirrhinus mrigala). Fish. Shellfish Immunol. 2012, 32, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 1983, 166, 557–580. [Google Scholar] [CrossRef]
- Nogueras, M.M.; Merino, S.; Aguilar, A.; Benedi, V.J.; Tomás, J.M. Cloning, sequencing, and role in serum susceptibility of porin II from mesophilic Aeromonas hydrophila. Infect. Immun. 2000, 68, 1849–1854. [Google Scholar] [CrossRef] [PubMed]
- Guzman, L.M.; Belin, D.; Carson, M.J.; Beckwith, J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose pBAD promoter. J. Bacteriol. 1995, 177, 4121–4130. [Google Scholar] [PubMed]
- Milton, D.L.; O’Toole, R.; Horstedt, P.; Wolf-Watz, H. Flagellin A is essential for the virulence of Vibrio anguillarum. J. Bacteriol. 1996, 178, 1310–1319. [Google Scholar] [PubMed]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 1989. [Google Scholar]
- Sanger, F.; Nicklen, S.; Coulson, A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 1977, 74, 5463–5467. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Studholme, D.J.; Dixon, R. Domain architectures of sigma54-dependent transcriptional activators. J. Bacteriol. 2003, 185, 1757–1767. [Google Scholar] [CrossRef] [PubMed]
- Münch, R.; Hiller, K.; Grote, A.; Scheer, M.; Klein, J.; Schobert, M.; Jahn, D. Virtual Footprint and PRODORIC: An integrative framework for regulon prediction in prokaryotes. Bioinformatics 2005, 21, 4187–4189. [Google Scholar] [CrossRef] [PubMed]
- Merino, S.; Wilhelms, M.; Tomás, J.M. Role of Aeromonas hydrophila flagella glycosylation in adhesion to Hep-2 cells, biofilm formation and immune stimulation. Int. J. Mol. Sci. 2014, 15, 21935–21946. [Google Scholar] [CrossRef] [PubMed]
- Pratt, L.A.; Kolter, R. Genetic analysis of Escherichia coli biofilm formation: Role of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 1998, 30, 285–293. [Google Scholar] [CrossRef] [PubMed]
- Jiménez, N.; Vilches, S.; Lacasta, A.; Regué, M.; Merino, S.; Tomás, J.M. A bifunctional enzyme in a single gene catalyzes the incorporation of GlcN into the Aeromonas core LPS. J. Biol. Chem. 2009, 284, 32995–33005. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fulton, K.M.; Mendoza-Barberá, E.; Twine, S.M.; Tomás, J.M.; Merino, S. Polar Glycosylated and Lateral Non-Glycosylated Flagella from Aeromonas hydrophila Strain AH-1 (Serotype O11). Int. J. Mol. Sci. 2015, 16, 28255-28269. https://doi.org/10.3390/ijms161226097
Fulton KM, Mendoza-Barberá E, Twine SM, Tomás JM, Merino S. Polar Glycosylated and Lateral Non-Glycosylated Flagella from Aeromonas hydrophila Strain AH-1 (Serotype O11). International Journal of Molecular Sciences. 2015; 16(12):28255-28269. https://doi.org/10.3390/ijms161226097
Chicago/Turabian StyleFulton, Kelly M., Elena Mendoza-Barberá, Susan M. Twine, Juan M. Tomás, and Susana Merino. 2015. "Polar Glycosylated and Lateral Non-Glycosylated Flagella from Aeromonas hydrophila Strain AH-1 (Serotype O11)" International Journal of Molecular Sciences 16, no. 12: 28255-28269. https://doi.org/10.3390/ijms161226097
APA StyleFulton, K. M., Mendoza-Barberá, E., Twine, S. M., Tomás, J. M., & Merino, S. (2015). Polar Glycosylated and Lateral Non-Glycosylated Flagella from Aeromonas hydrophila Strain AH-1 (Serotype O11). International Journal of Molecular Sciences, 16(12), 28255-28269. https://doi.org/10.3390/ijms161226097