The Infection Efficiency and Replication Ability of Circularized HBV DNA Optimized the Linear HBV DNA in Vitro and in Vivo
Abstract
:1. Introduction
2. Results
2.1. Assessment of the Optimal Amount of Circularized HBV DNA and pAAV/HBV1.2 in Vitro and in Vivo


2.2. Transfection and Hydrodynamic Injection of Circularized HBV DNA and pAAV/HBV1.2 Could Lead to Viral Gene Replication and Expression in Vitro and in Vivo
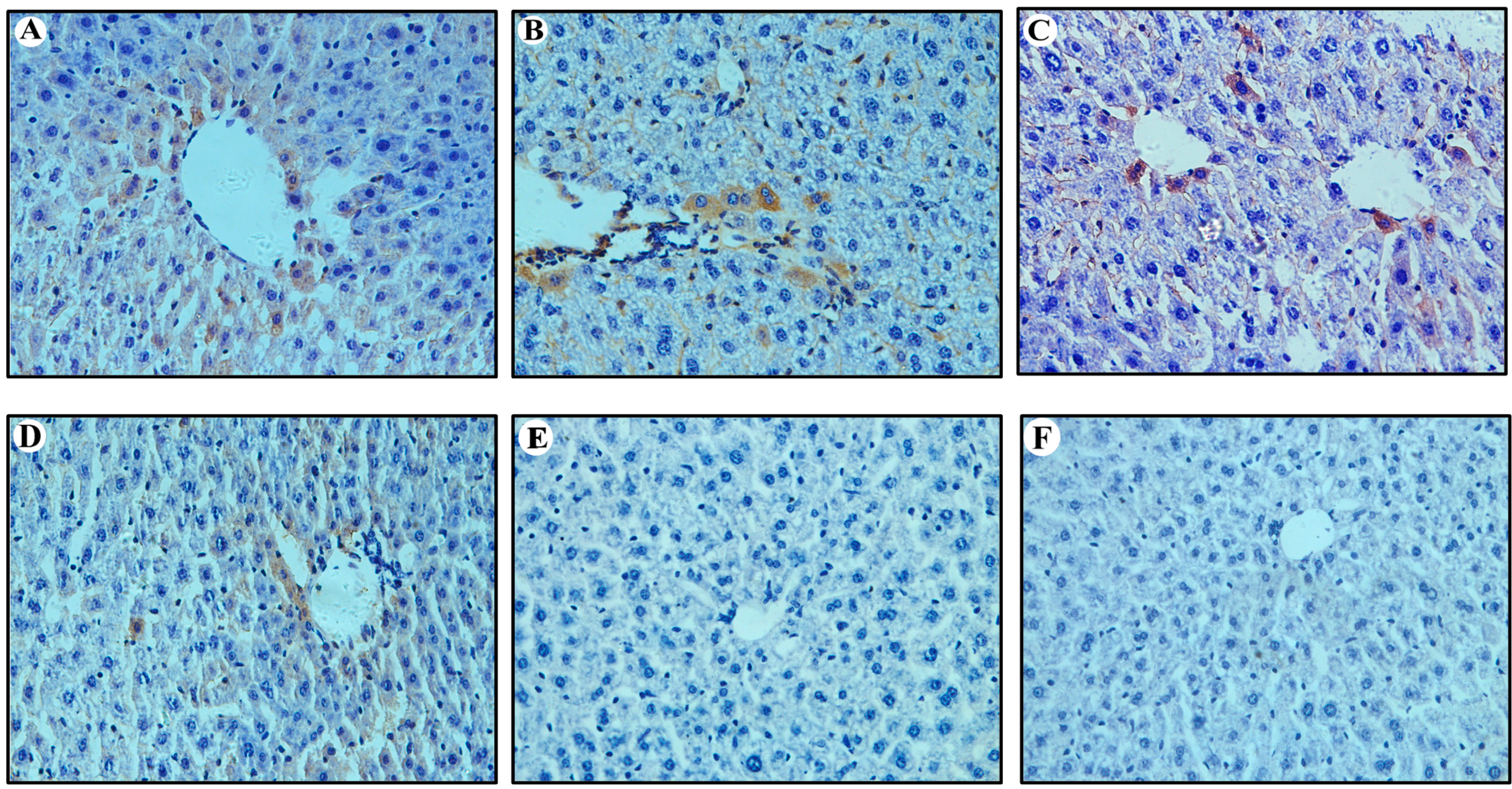
2.3. Expression of HBcAg in Vitro and in Vivo
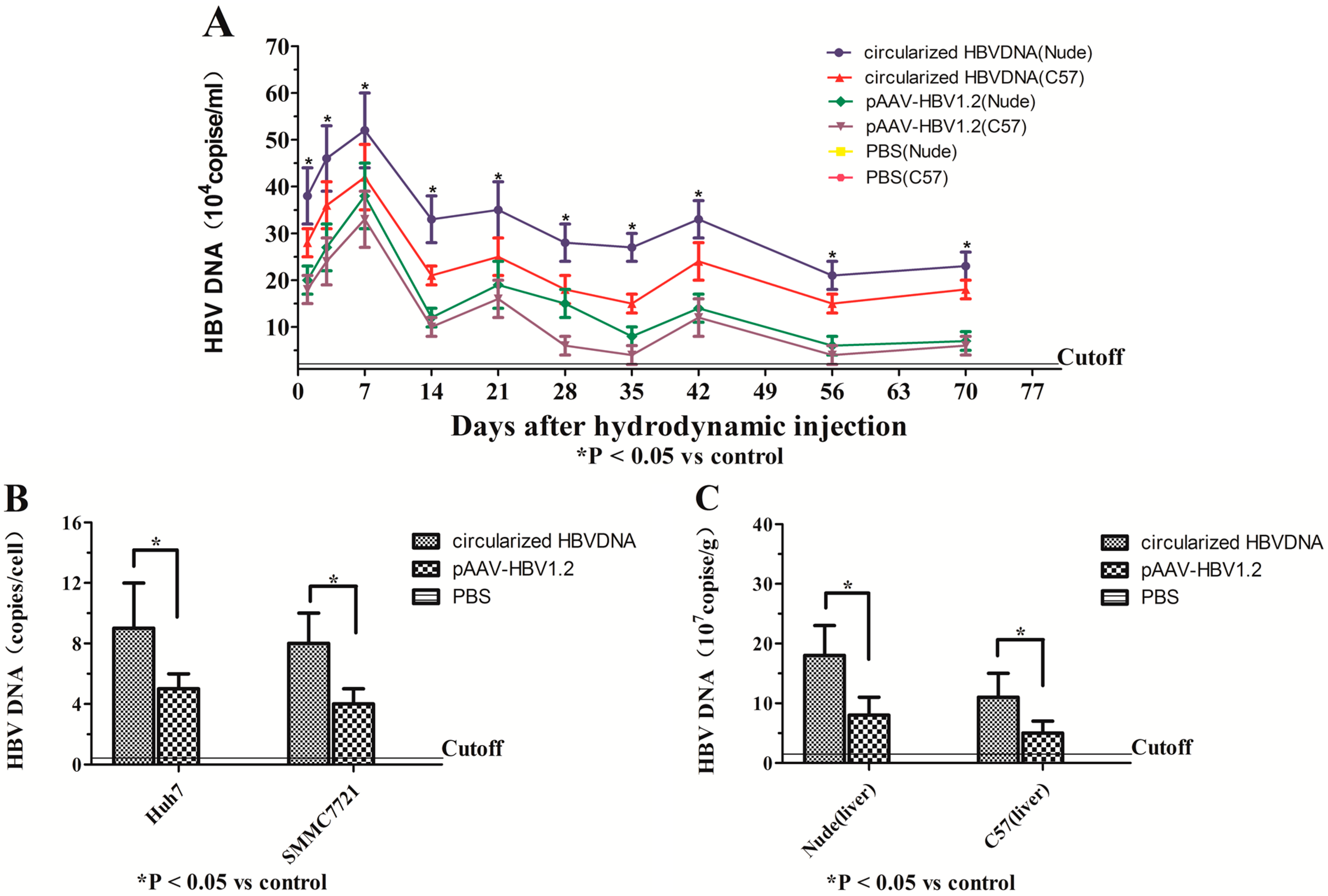



2.4. Expression of ALT, HBsAg and HBeAg in Vitro and in Vivo

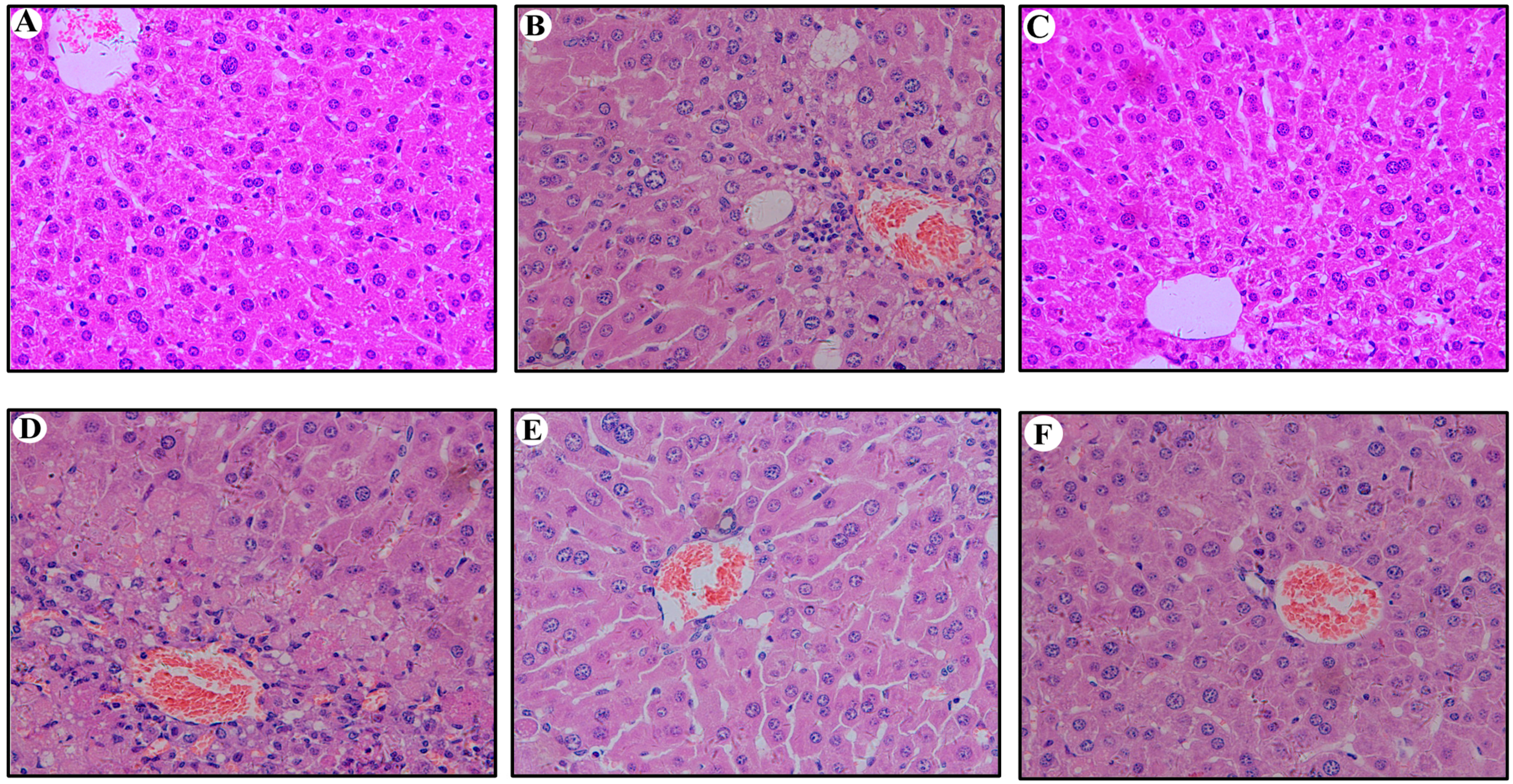
2.5. Hepatic Histopathological Changes in Mouse Liver

3. Discussion
4. Experimental Section
4.1. Primers
4.2. Cells
4.3. Animals
4.4. The Preparation of Circularized HBV DNA and pAAV/HBV1.2

4.5. The Transfection of Circularized HBV DNA and pAAV/HBV1.2 to Cells
4.6. The Hydrodynamic Injection of Circularized HBV DNA and pAAV/HBV1.2 to Mice
4.7. Detection and Quantification of HBV DNA
4.8. Detection and Quantification of HBV pgRNA
4.9. Detection and Quantification of ALT, HBsAg and HBeAg
4.10. Detection and Quantification of TNF-α and IL-6 in Vivo
4.11. Hepatic Histopathological and Immunohistochemical Analysis (IHC)
4.12. Southern Blot Analysis
4.13. Western Blot Analysis
4.14. Statistical Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Barker, L.F.; Maynard, J.E.; Purcell, R.H.; Hoofnagle, J.H.; Berquist, K.R.; London, W.T. Viral hepatitis, type B, in experimental animals. Am. J. Med. Sci. 1975, 270, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Beasley, R.P.; Hwang, L.Y.; Lin, C.C.; Chien, C.S. Hepatocellular carcinoma and hepatitis B virus, A prospective study of 22707 men in Taiwan. Lancet 1981, 2, 1129–1133. [Google Scholar] [CrossRef] [PubMed]
- Ganem, D.; Varmus, H.E. The molecular biology of the hepatitis B viruses. Annu. Rev. Biochem. 1987, 56, 651–693. [Google Scholar] [CrossRef] [PubMed]
- Lupberger, J.; Hildt, E. Hepatitis B virus-induced oncogenesis. World J. Gastroenterol. 2007, 13, 74–81. [Google Scholar] [CrossRef] [PubMed]
- Moriyama, T.; Guilhot, S.; Klopchin, K.; Moss, B.; Pinkert, C.A.; Palmiter, R.D.; Brinster, R.L.; Kanagawa, O.; Chisari, F.V. Immunobiology and pathogenesis of hepatocellular injury in hepatitis B virus transgenic mice. Science 1990, 248, 361–364. [Google Scholar] [CrossRef] [PubMed]
- Rybicka, M.; Stalke, P.; Charmuszko, U.; Bielawski, K.P. The influence of hepatitis B virus polymorphism on the progression of chronic liver disease. Postepy Hig. Med. Dosw. 2011, 21, 244–254. [Google Scholar] [CrossRef]
- Suzuki, T.; Takehara, T.; Ohkawa, K.; Ishida, H.; Jinushi, M.; Miyagi, T.; Sasaki, Y.; Hayashi, N. Intravenous injection of naked plasmid DNA encoding hepatitis B virus (HBV) produces HBV and induces humoral immune response in mice. Biochem. Biophys. Res. Commun. 2003, 300, 784–788. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.L.; Althage, A.; Chung, J.; Chisari, F.V. Hydrodynamic injection of viral DNA: A mouse model of acute hepatitis B virus infection. Proc. Natl. Acad. Sci. USA. 2002, 99, 13825–13830. [Google Scholar] [CrossRef] [PubMed]
- Ketzinel-Gilad, M.; Zauberman, A.; Nussbaum, O.; Shoshany, Y.; Ben-Moshe, O.; Pappo, O.; Felig, Y.; Ilan, E.; Wald, H.; Dagan, S.; et al. The use of the hydrodynamics HBV animal model to study HBV biology and anti-viral therapy. Hepatol. Res. 2006, 34, 228–237. [Google Scholar] [CrossRef] [PubMed]
- Locarnini, S.; Zoulim, F. Molecular genetics of HBV infection. Antivir. Ther. 2010, 15, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Buckwold, V.E.; Xu, Z.; Chen, M.; Yen, T.S.; Ou, J.H. Effects of a naturally occurring mutation in the hepatitis B virus basal core promoter on precore gene expression and viral replication. J. Virol. 1996, 70, 5845–5851. [Google Scholar] [PubMed]
- Baumert, T.F.; Rogers S, A.; Hasegawa, K.; Liang, T.J. Two core promotor mutations identified in a hepatitis B virus strain associated with fulminant hepatitis result in enhanced viral replication. J. Clin. Investig. 1996, 98, 2268–2276. [Google Scholar] [CrossRef] [PubMed]
- Scaglioni, P.P.; Melegari, M.; Wands, J.R. Posttranscriptional regulation of hepatitis B virus replication by the precore protein. J. Virol. 1997, 71, 345–353. [Google Scholar] [PubMed]
- Khan, N.; Guarnieri, M.; Ahn, S.H.; Li, J.; Zhou, Y.; Bang, G.; Kim, K.H.; Wands, J.R.; Tong, S. Modulation of hepatitis B virus secretion by naturally occurring mutations in the S gene. J. Virol. 2004, 78, 3262–3270. [Google Scholar] [CrossRef] [PubMed]
- Durantel, D.; Carrouée-Durantel, S.; Werle-Lapostolle, B.; Brunelle, M.N.; Pichoud, C.; Trépo, C.; Zoulim, F. A new strategy for studying in vitro the drug susceptibility of clinical isolates of human hepatitis B virus. Hepatology 2004, 40, 855–864. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.R.; Wu, H.L.; Chen, P.J.; Chen, D.S. An immunocompetent mouse model for the tolerance of human chronic hepatitis B virus infection. Proc. Natl. Acad. Sci. USA. 2006, 103, 17862–17867. [Google Scholar] [CrossRef] [PubMed]
- Gu¨nther, S.; Li, B.C.; Miska, S.; Krüger, D.H.; Meisel, H.; Will, H. A novel method for efficient amplification of whole hepatitis B virus genomes permits rapid functional analysis and reveals deletion mutants in immunosuppressed patients. J. Virol. 1995, 69, 5437–5444. [Google Scholar] [PubMed]
- Parekh, S.; Zoulim, F.; Ahn, S.H.; Tsai, A.; Li, J.; Kawai, S.; Khan, N.; Trépo, C.; Wands, J.; Tong, S. Genome replication, virion secretion, and E antigen expression of naturally occurring hepatitis B virus core promoter mutants. J. Virol. 2003, 77, 6601–6612. [Google Scholar] [CrossRef] [PubMed]
- Sureau, C.; Romet-Lemonne, J.L.; Mullins, J.I. Production of hepatitis B virus by a differentiated human hepatoma cell line after transfection with cloned circularized HBV DNA. Cell 1986, 47, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Song, Y.; Liu, D. Hydrodynamics-based transfection in animals by systemic administration of plasmid DNA. Gene Ther. 1999, 6, 1258–1266. [Google Scholar] [CrossRef] [PubMed]
- McMahon, B.J. The natural history of chronic hepatitis B virus infection. Semin. Liver Dis. 2004, 24 (Suppl. 1), 17–21. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.S. From hepatitis to hepatoma: Lessons from type B viral hepatitis. Science 1993, 262, 369–370. [Google Scholar] [CrossRef] [PubMed]
- Guidotti, L.G.; Matzke, B.; Schaller, H.; Chisari, F.V. High-level hepatitis B virus replication in transgenic mice. J. Virol. 1995, 69, 6158–6169. [Google Scholar] [PubMed]
- Dienstag, J.L. Hepatitis B virus infection. N. Engl. J. Med. 2008, 359, 1486–1500. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.K.; Seo, Y.B.; Im, S.J.; Bae, S.H.; Song, M.J.; You, C.R.; Jang, J.W.; Yang, S.H.; Suh, Y.S.; Song, J.S.; et al. Safety and immunogenicity of therapeutic DNA vaccine with antiviral drug in chronic HBV patients and its immunogenicity in mice. Liver Int. 2014. [Google Scholar] [CrossRef]
- Seeger, C.; Ganem, D.; Varmus, H.E. The cloned genome of ground squirrel hepatitis virus is infectious in the animal. Proc. Natl. Acad. Sci. USA. 1984, 81, 5849–5852. [Google Scholar] [CrossRef] [PubMed]
- Raney, A.K.; Eggers, C.M.; Kline, E.F.; Guidotti, L.G.; Pontoglio, M.; Yaniv, M.; McLachlan, A. Nuclear covalently closed circularized viral genomic DNA in the liver of hepatocyte nuclear factor 1a-null hepatitis B virus transgenic mice. J. Virol. 2001, 75, 2900–2911. [Google Scholar] [CrossRef] [PubMed]
- Gripon, P.; Rumin, S.; Urban, S.; Le Seyec, J.; Glaise, D.; Cannie, I.; Guyomard, C.; Lucas, J.; Trepo, C.; Guguen-Guillouzo, C. Infection of a human hepatoma cell line by hepatitis B virus. Proc. Natl. Acad. Sci. USA. 2002, 99, 15655–15660. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.J.; Liu, L.; He, F.; Wang, S.; Zhou, T.Y.; Liu, C.; Deng, L.Y.; Tang, H. Establishment and primary application of a mouse model with hepatitis B virus replication. World J. Gastroenterol. 2007, 13, 5324–5330. [Google Scholar] [CrossRef] [PubMed]
- Balsano, C.; Alisi, A. Viral hepatitis B: Established and emerging therapies. Curr. Med. Chem. 2008, 15, 930–939. [Google Scholar] [CrossRef] [PubMed]
- Carman, W.F.; Waters, J.; Manzillo, G.; Tanzi, E.; Zuckerman, A.J.; Thomas, H.C. Vaccine-induced escape mutant of hepatitis B virus. Lancet 1990, 336, 325–329. [Google Scholar] [CrossRef] [PubMed]
- Cooreman, M.P.; van Roosmalen, M.H.; te Morsche, R.; Sünnen, C.M.; de Ven, E.M.; Jansen, J.B.; Tytgat, G.N.; de Wit, P.L.; Paulij, W.P. Characterization of the reactivity pattern of murine monoclonal antibodies against wild-type hepatitis B surface antigen to G145R and other naturally occurring “A” loop escape mutations. Hepatology 1999, 30, 1287–1292. [Google Scholar] [CrossRef] [PubMed]
- Waters, J.A.; Kennedy, M.; Voet, P.; Hauser, P.; Petre, J.; Carman, W.; Thomas, H.C. Loss of the common “A” determinant of hepatitis B surface antigen by a vaccine-induced escape mutant. J. Clin. Investig. 1992, 90, 2543–2547. [Google Scholar] [CrossRef] [PubMed]
- El Chaar, M.; Candotti, D.; Crowther, R.A.; Allain, J.P. Impact of hepatitis B virus surface protein mutations on the diagnosis of occult hepatitis B virus infection. Hepatology 2010, 52, 1600–1610. [Google Scholar] [CrossRef] [PubMed]
- Jeantet, D.; Chemin, I.; Mandrand, B.; Tran, A.; Zoulim, F.; Merle, P.; Trepo, C.; Kay, A. Cloning and expression of surface antigens from occult chronic hepatitis B virus infections and their recognition by commercial detection assays. J. Med. Virol. 2004, 73, 508–515. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.J.; Huang, L.R.; Yang, H.C.; Tzeng, H.T.; Hsu, P.N.; Wu, H.L.; Chen, P.J.; Chen, D.S. Hepatitis B virus core antigen determines viral persistence in a C57BL/6 mouse model. Proc. Natl. Acad. Sci. USA 2010, 107, 9340–9345. [Google Scholar] [CrossRef] [PubMed]
- Belloni, L.; Allweiss, L.; Guerrieri, F.; Pediconi, N.; Volz, T.; Pollicino, T.; Petersen, J.; Raimondo, G.; Dandri, M.; Levrero, M. IFN-α inhibits HBV transcription and replication in cell culture and in humanized mice by targeting the epigenetic regulation of the nuclear cccDNA minichromosome. J. Clin. Investig. 2012, 122, 529–537. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Zhang, J.; Garcia, T.; Ito, K.; Gutelius, D.; Li, J.; Wands, J.; Tong, S. Improved method for rapid and efficient determination of genome replication and protein expression of clinical hepatitis B virus isolates. J. Clin. Microbiol. 2011, 49, 1226–1233. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Tang, X.; Garcia, T.; Hussain, M.; Zhang, J.; Lok, A.; Wands, J.; Li, J.; Tong, S. Hepatitis B virus genotype C isolates with wild-type core promoter sequence replicate less efficiently than genotype B isolates but possess higher virion secretion capacity. J. Virol. 2011, 85, 10167–10177. [Google Scholar] [CrossRef] [PubMed]
- Liang, T.J.; Hasegawa, K.; Rimon, N.; Wands, J.R.; Ben-Porath, E. A hepatitis B virus mutant associated with an epidemic of fulminant hepatitis. N. Engl. J. Med. 1991, 324, 1705–1709. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Huang, L. Improving plasmid DNA-mediated liver gene transfer by prolonging its retention in the hepatic vasculature. J. Gene Med. 2001, 3, 569–576. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.R.; Gäbel, Y.A.; Graf, S.; Arzberger, S.; Kurts, C.; Heikenwalder, M.; Knolle, P.A.; Protzer, U. Transfer of HBV genomes using low doses of adenovirus vectors leads to persistent infection in immune competent mice. Gastroenterology 2012, 142, 1447–1450. [Google Scholar] [CrossRef] [PubMed]
- Garcia, T.; Li, J.; Sureau, C.; Ito, K.; Qin, Y.; Wands, J.; Tong, S. Drastic reduction in the production of subviral particles does not impair hepatitis B virus virion secretion. J. Virol. 2009, 83, 11152–11165. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.L.; Chen, P.J.; Lin, M.H.; Chen, D.S. Temporal aspects of major viral transcript expression in Hep G2 cells transfected with cloned hepatitis B virus DNA: With emphasis on the X transcript. Virology 1991, 185, 644–651. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Budker, V.; Wolff, J.A. High levels of foreign gene expression in hepatocytes after tail vein injections of naked plasmid DNA. Hum. Gene Ther. 1999, 10, 1735–1737. [Google Scholar] [CrossRef] [PubMed]
- Shintani, M.; Urano, M.; Takakuwa, Y.; Kuroda, M.; Kamoshida, S. Immunohistochemical characterization of pyrimidine synthetic enzymes, thymidine kinase-1 and thymidylate synthase, in various types of cancer. Oncol. Rep. 2010, 23, 1345–1350. [Google Scholar] [PubMed]
- Shimizu, S.; Nomura, F.; Tomonaga, T.; Sunaga, M.; Noda, M.; Ebara, M.; Saisho, H. Expression of poly(ADP-ribose) polymerase in human hepatocellular carcinoma and analysis of biopsy specimens obtained under sonographic guidance. Oncol. Rep. 2004, 12, 821–825. [Google Scholar] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Zhu, J.; Lai, G.; Yan, L.; Hu, J.; Chen, J.; Tang, N.; Huang, A. The Infection Efficiency and Replication Ability of Circularized HBV DNA Optimized the Linear HBV DNA in Vitro and in Vivo. Int. J. Mol. Sci. 2015, 16, 5141-5160. https://doi.org/10.3390/ijms16035141
Li X, Zhu J, Lai G, Yan L, Hu J, Chen J, Tang N, Huang A. The Infection Efficiency and Replication Ability of Circularized HBV DNA Optimized the Linear HBV DNA in Vitro and in Vivo. International Journal of Molecular Sciences. 2015; 16(3):5141-5160. https://doi.org/10.3390/ijms16035141
Chicago/Turabian StyleLi, Xiaosong, Junke Zhu, Guoqi Lai, Lei Yan, Jieli Hu, Juan Chen, Ni Tang, and Ailong Huang. 2015. "The Infection Efficiency and Replication Ability of Circularized HBV DNA Optimized the Linear HBV DNA in Vitro and in Vivo" International Journal of Molecular Sciences 16, no. 3: 5141-5160. https://doi.org/10.3390/ijms16035141





