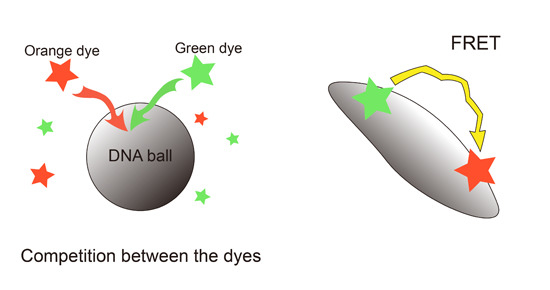
Investigation of Förster Resonance Energy Transfer (FRET) and Competition of Fluorescent Dyes on DNA Microparticles
Abstract
:1. Introduction
2. Results and Discussion
2.1. Multicolored DNA Balls



2.2. Förster Resonance Energy Transfer (FRET) Efficiency Difference between ssDNA Balls and dsDNA Ball

2.3. Interference between the Dyes Dependent on Staining Method

3. Experimental Section
3.1. Preparation of Circular DNA
3.2. Synthesis of DNA Balls
3.3. Multicolor Staining of the DNA Balls
3.4. Measurement of Fluorescence Intensity
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Winfree, E.; Liu, F.; Wenzler, L.A.; Seeman, N.C. Design and self-assembly of two-dimensional DNA crystals. Nature 1998, 394, 539–544. [Google Scholar] [CrossRef] [PubMed]
- Chelyapov, N.; Brun, Y.; Gopalkrishnan, M.; Reishus, D.; Shaw, B.; Adleman, L. DNA triangles and self-assembled hexagonal tilings. J. Am. Chem. Soc. 2004, 126, 13924–13925. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Seeman, N.C. Synthesis from DNA of a molecule with the connectivity of a cube. Nature 1991, 350, 631–633. [Google Scholar] [CrossRef] [PubMed]
- Russell, P. The single-step synthesis of a DNA tetrahedron. Chem. Commun. 2004, 12, 1372–1373. [Google Scholar]
- Li, Y.; Tseng, Y.D.; Kwon, S.Y.; d'Espaux, L.; Bunch, J.S.; McEuen, P.L.; Luo, D. Controlled assembly of dendrimer-like DNA. Nat. Mater. 2003, 3, 38–42. [Google Scholar] [CrossRef] [PubMed]
- Um, S.H.; Lee, J.B.; Park, N.; Kwon, S.Y.; Umbach, C.C.; Luo, D. Enzyme-catalysed assembly of DNA hydrogel. Nat. Mater. 2006, 5, 797–801. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.; Hu, R.; Zhao, Z.; Chen, Z.; Zhang, X.; Tan, W. Noncanonical self-assembly of multifunctional DNA nanoflowers for biomedical applications. J. Am. Chem. Soc. 2013, 135, 16438–16445. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.-X.; Shaw, A.; Zeng, X.; Benson, E.; Nyström, A.M.; Högberg, B.R. DNA origami delivery system for cancer therapy with tunable release properties. ACS Nano 2012, 6, 8684–8691. [Google Scholar] [CrossRef] [PubMed]
- Roh, Y.H.; Lee, J.B.; Shopsowitz, K.E.; Dreaden, E.C.; Morton, S.W.; Poon, Z.; Hong, J.; Yamin, I.; Bonner, D.K.; Hammond, P.T. Layer-by-layer assembled antisense DNA microsponge particles for efficient delivery of cancer therapeutics. ACS Nano 2014, 8, 9767–9780. [Google Scholar] [CrossRef] [PubMed]
- Douglas, S.M.; Bachelet, I.; Church, G.M. A logic-gated nanorobot for targeted transport of molecular payloads. Science 2012, 335, 831–834. [Google Scholar] [CrossRef] [PubMed]
- List, J.; Weber, M.; Simmel, F.C. Hydrophobic actuation of a DNA origami bilayer structure. Angew. Chem. Int. Ed. 2014, 53, 4236–4239. [Google Scholar] [CrossRef]
- Pei, H.; Lu, N.; Wen, Y.; Song, S.; Liu, Y.; Yan, H.; Fan, C. A DNA nanostructure-based biomolecular probe carrier platform for electrochemical biosensing. Adv. Mater. 2010, 22, 4754–4758. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.B.; Roh, Y.H.; Um, S.H.; Funabashi, H.; Cheng, W.; Cha, J.J.; Kiatwuthinon, P.; Muller, D.A.; Luo, D. Multifunctional nanoarchitectures from DNA-based ABC monomers. Nat. Nanotechnol. 2009, 4, 430–436. [Google Scholar] [CrossRef] [PubMed]
- Schotter, J.; Kamp, P.-B.; Becker, A.; Pühler, A.; Reiss, G.; Brückl, H. Comparison of a prototype magnetoresistive biosensor to standard fluorescent DNA detection. Biosens. Bioelectron. 2004, 19, 1149–1156. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Wieckowska, A.; Willner, I. Optical analysis of Hg2+ ions by oligonucleotide–gold-nanoparticle hybrids and DNA-based machines. Angew. Chem. 2008, 120, 3991–3995. [Google Scholar] [CrossRef]
- Li, J.; Lu, Y. A highly sensitive and selective catalytic DNA biosensor for lead ions. J. Am. Chem. Soc. 2000, 122, 10466–10467. [Google Scholar] [CrossRef]
- Song, Y.; Xu, X.; MacRenaris, K.W.; Zhang, X.Q.; Mirkin, C.A.; Meade, T.J. Multimodal gadolinium-enriched DNA–gold nanoparticle conjugates for cellular imaging. Angew. Chem. Int. Ed. 2009, 48, 9143–9147. [Google Scholar] [CrossRef]
- Hu, R.; Zhang, X.; Zhao, Z.; Zhu, G.; Chen, T.; Fu, T.; Tan, W. DNA nanoflowers for multiplexed cellular imaging and traceable targeted drug delivery. Angew. Chem. 2014, 126, 5931–5936. [Google Scholar] [CrossRef]
- Ho, Y.-P.; Chen, H.H.; Leong, K.W.; Wang, T.-H. Evaluating the intracellular stability and unpacking of DNA nanocomplexes by quantum dots-fret. J. Control. Release 2006, 116, 83–89. [Google Scholar] [CrossRef] [PubMed]
- Derfus, A.M.; Chan, W.C.; Bhatia, S.N. Probing the cytotoxicity of semiconductor quantum dots. Nano Lett. 2004, 4, 11–18. [Google Scholar] [CrossRef]
- Gerion, D.; Pinaud, F.; Williams, S.C.; Parak, W.J.; Zanchet, D.; Weiss, S.; Alivisatos, A.P. Synthesis and properties of biocompatible water-soluble silica-coated CdSe/ZnS semiconductor quantum dots. J. Phys. Chem. B 2001, 105, 8861–8871. [Google Scholar] [CrossRef]
- Chehab, F.F.; Kan, Y. Detection of specific DNA sequences by fluorescence amplification: A color complementation assay. Proc. Natl. Acad. Sci. USA 1989, 86, 9178–9182. [Google Scholar] [CrossRef] [PubMed]
- Zipper, H.; Brunner, H.; Bernhagen, J.; Vitzthum, F. Investigations on DNA intercalation and surface binding by SYBR Green I, its structure determination and methodological implications. Nucleic Acids Res. 2004, 32, e103. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Habbersett, R.C.; Cordek, J.M.; Nolan, J.P.; Yoshida, T.M.; Jett, J.H.; Marrone, B.L. Development of a mechanism-based, DNA staining protocol using SYTOX orange nucleic acid stain and DNA fragment sizing flow cytometry. Anal. Biochem. 2000, 286, 138–148. [Google Scholar] [CrossRef] [PubMed]
- Valeur, B.; Berberan-Santos, M.N. Molecular Fluorescence: Principles and Applications; John Wiley & Sons: Hoboken, NJ, USA, 2012. [Google Scholar]
- Singer, V.L.; Lawlor, T.E.; Yue, S. Comparison of SYBR® green I nucleic acid gel stain mutagenicity and ethidium bromide mutagenicity in the salmonella/mammalian microsome reverse mutation assay (Ames test). Mutation Res. Genet. Toxicol. Environ. Mutagen. 1999, 439, 37–47. [Google Scholar] [CrossRef]
- Johnson, I.; Spence, M. The Molecular Probes Handbook; Life Technologies Corporation: Carlsbad, CA, USA, 2010. [Google Scholar]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, J.; Lee, J.S.; Lee, J.B. Investigation of Förster Resonance Energy Transfer (FRET) and Competition of Fluorescent Dyes on DNA Microparticles. Int. J. Mol. Sci. 2015, 16, 7738-7747. https://doi.org/10.3390/ijms16047738
Kim J, Lee JS, Lee JB. Investigation of Förster Resonance Energy Transfer (FRET) and Competition of Fluorescent Dyes on DNA Microparticles. International Journal of Molecular Sciences. 2015; 16(4):7738-7747. https://doi.org/10.3390/ijms16047738
Chicago/Turabian StyleKim, Jieun, Jae Sung Lee, and Jong Bum Lee. 2015. "Investigation of Förster Resonance Energy Transfer (FRET) and Competition of Fluorescent Dyes on DNA Microparticles" International Journal of Molecular Sciences 16, no. 4: 7738-7747. https://doi.org/10.3390/ijms16047738
APA StyleKim, J., Lee, J. S., & Lee, J. B. (2015). Investigation of Förster Resonance Energy Transfer (FRET) and Competition of Fluorescent Dyes on DNA Microparticles. International Journal of Molecular Sciences, 16(4), 7738-7747. https://doi.org/10.3390/ijms16047738