MicroRNAs Regulate Bone Development and Regeneration
Abstract
:1. Introduction: Functions and Canonical Biogenesis of MiRNAs
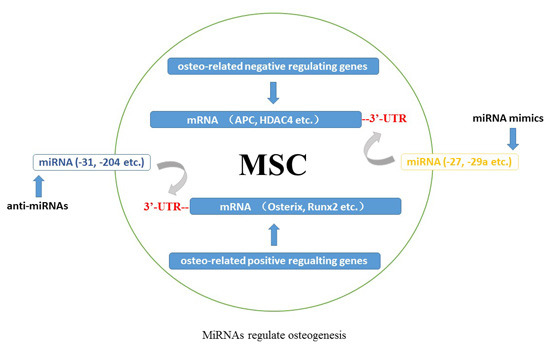
2. Evidence Supporting MiRNAs Participating in Osteogenesis
3. In Vitro Profiling Analysis of Osteogenesis-Related MiRNAs
| MiRNA | Variation Trend | Cell Line | Reference |
|---|---|---|---|
| pre-miR-15 | + | hBMSC | [39] |
| miR-29b | + | MC3T3 primary rat osteoblasts | [29] |
| miR-34a | + | hBMSC | [33] |
| miR-93 | − | primary mouse osteoblast | [41] |
| miR-138 | − | MC3T3 hBMSC | [17] |
| miR-140-5-p | − | hBMSC hADCS hUCSC | [22] |
| miR-181a | + | C2C12 MC3T3 | [34] |
| miR-196a | + | hADSC | [37] |
| miR-210 | + | ST2 | [35] |
| miR-218 | + | hADSC | [36] |
| miR-335-5p | + | MC3T3 MLO-A5 C3H10T1/2 | [19] |
| miR-338-3p | − | mouse BMSC | [42] |
| miR-346 | + | hBMSC | [38] |
| miR-433 | − | C3H10T1/2 | [44] |
| miR-2861 | + | ST2 | [20] |
| miR-3960 | + | ST2 | [21] |
4. MiRNAs and Bone Development
4.1. MiRNAs and Osteo-Related Transcription Factors in Bone Formation
4.2. Other Target Molecules of MiRNAs in Skeletogenesis
4.3. The Interplay between MiRNAs and Signaling Pathways in Bone Homeostasis
4.4. Coordinate Regulation by Feedback Loops of MiRNAs in Osteoblasts
5. MiRNAs and Bone Regeneration

| MiRNA | Target Gene | Osteogenesis | Cell Line | Reference |
|---|---|---|---|---|
| let-7f | Axin2 | + | hBMSC | [104] |
| miR-10a | KLF4 | + | hBMSC | [31] |
| miR-15b | Smurf1 | + | hBMSC | [39] |
| miR-20a | PPARγ Bambi Crim1 | + | hBMSC | [116] |
| miR-23a | Runx2 | − | MC3T3 | [50] |
| miR-23a~27a~24-2 | SATB2 | − | MC3T3 | [107] |
| miR-26a | Smad-1 | − | hADSC | [90] |
| miR-27 | APC | + | hFOB1.19 | [100] |
| SFRP1 | hFOB1.19 | [101] | ||
| miR-29a | osteonectin | + | MC3T3 | [75] |
| DKK1 Kremen2 SFRP2 | hFOB1.19 | [102] | ||
| HDAC4 | MC3T3 | [99] | ||
| miR-29b | AcvR2a CTNNBIP1 DUSP2 TGF-β3 HDAC4 | + | MC3T3 | [29] |
| miR-29c | osteonectin | + | MC3T3 | [75] |
| miR-30c | Runx2 | − | MC3T3 | [50] |
| miR-31 | Osx | − | hBMSC | [32] |
| SATB2 | hADSC | [72] | ||
| rat BMSC | [109] | |||
| rat ADSC | [126] | |||
| miR-34a | JAG1 | − | hBMSC | [33] |
| miR-34b | SATB2 | − | primary mouse osteoblasts | [115] |
| miR-34c | SATB2 | − | primary mouse osteoblasts | [115] |
| Runx2 | MC3T3 | [50] | ||
| miR-93 | Osx | − | primary mouse osteoblasts | [41] |
| miR-103a | Runx2 | − | hFOB1.19 | [50] |
| miR-125b | Osx | − | hCASMC | [64] |
| hBMSC | [65] | |||
| Smad-4 | hADSC | [91] | ||
| Cbfβ | C3H10T1/2 | [68] | ||
| miR-133 | Runx2 | − | C2C12 | [28] |
| MC3T3 | [50] | |||
| miR-135a | Smad-5 | − | C2C12 | [28] |
| Runx2 | MC3T3 | [50] | ||
| miR-137 | Runx2 | − | MC3T3 | [50] |
| miR-138 | FAK | − | hBMSC | [17] |
| miR-140-5p | BMP-2 | − | hBMSC hADCS hUCSC | [22] |
| miR-141 | Dlx5 | − | MC3T3 | [69] |
| miR-143 | Osx | − | MC3T3 | [58] |
| miR-145 | Osx | − | C2C12 MC3T3 | [62] |
| miR-146a | JMJD3 | − | hUCSC | [74] |
| Smad-2 Smad-3 | + | hSSC | [92] | |
| miR-181a | TgfbI TβR-I Rgs4 Gata6 | + | MC3T3 | [34] |
| miR-196a | HOXC8 | + | hADSC | [37] |
| miR-199a | Smad-1 | − | C3H10T1/2 | [88] |
| miR-200a | Dlx5 | ‑ | MC3T3 | [69] |
| miR-206 | Connexin43 | − | C2C12 | [27] |
| miR-204 | Runx2 | − | ST2 hMSC | [18] |
| MC3T3 | [50] | |||
| miR-205 | Runx2 | − | MC3T3 | [50] |
| miR-210 | AcvR1b | + | ST2 | [35] |
| miR-211 | Runx2 | − | ST2 hMSC | [18] |
| miR-214 | Osx | − | C2C12 | [63] |
| ATF4 | C2C12 | [70] | ||
| miR-217 | Runx2 | − | MC3T3 | [50] |
| miR-218 | SOST DKK2 SFRP2 | + | MC3T3 | [103] |
| DKK2 SFRP2 | hADSC | [36] | ||
| Runx2 | − | hDSC | [53] | |
| miR-335-5p | DKK1 | + | C3H10T1/2 | [19] |
| miR-338 | Runx2 | − | MC3T3 | [50] |
| miR-338-3p | Runx2 Fgfr2 | − | mouse BMSC | [42] |
| miR-346 | GSK-3β | + | hBMSC | [38] |
| miR-378 | + | C2C12 | [76] | |
| miR-433 | Runx2 | − | C3H10T1/2 | [44] |
| miR-542-3p | BMP-7 | − | primary mouse osteoblasts | [89] |
| miR-548d-5p | PPARγ | + | hBMSC | [117] |
| miR-637 | Osx | − | hBMSC | [66] |
| miR-654-5p | BMP-2 | − | hBMSC | [93] |
| miR-2861 | HDAC5 | + | ST2 | [20] |
| miR-3960 | HOXA2 | + | ST2 | [21] |
| MiRNA | Target Component | Regulation on Signaling | Reference |
|---|---|---|---|
| BMP signaling | |||
| miR-26a | Smad-1 | − | [90] |
| miR-29b | TGF-β3 AcvR2a | + | [29] |
| miR-125b | Smad-4 | − | [91] |
| miR-135a | Smad-5 | − | [28] |
| miR-140-5p | BMP-2 | − | [22] |
| miR-146a | Smad-2 Smad-3 | + | [92] |
| miR-181a | TgfbI TβR-I | + | [34] |
| miR-199a | Smad-1 | − | [88] |
| miR-210 | AcvR1b | + | [35] |
| miR-542-3p | BMP-7 | − | [89] |
| miR-654-5p | BMP-2 | − | [93] |
| Wnt signaling | |||
| let-7f | Axin2 | + | [104] |
| miR-27 | APC SFRP1 | + | [100] |
| miR-29a | HDAC4 DKK1 Kremen2 SFRP2 | + | [99,102] |
| miR-29b | HDAC4 CTNNBIP1 | + | [29] |
| miR-218 | SOST DKK2 SFRP2 | + | [36,103] |
| miR-335-5p | DKK1 | + | [19] |
| miR-346 | GSK-3β | + | [38] |
| Notch signaling | |||
| miR-34c | Notch1 Notch2 Jag1 | − | [106] |
| MAPK signaling | |||
| miR-29b | DUSP2 | + | [29] |
6. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [PubMed]
- Van Rooij, E.; Kauppinen, S. Development of microRNA therapeutics is coming of age. EMBO Mol. Med. 2014, 6, 851–864. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Paroo, Z. Biochemical principles of small RNA pathways. Annu. Rev. Biochem. 2010, 79, 295–319. [Google Scholar] [CrossRef] [PubMed]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef] [PubMed]
- Xu, S. microRNA expression in the eyes and their significance in relation to functions. Prog. Retin. Eye Res. 2009, 28, 87–116. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Xie, H.; Xiong, W.; Xu, D.; Cao, K.; Liu, R.; Zhou, J.; Luo, C. MicroRNA and metabolism regulation. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2013, 38, 318–322. [Google Scholar] [PubMed]
- Kim, V.N.; Han, J.; Siomi, M.C. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009, 10, 126–139. [Google Scholar] [CrossRef] [PubMed]
- Roberts, T.C. The microRNA biology of the mammalian nucleus. Mol. Ther. Nucleic Acids. 2014, 3, e188. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Fortin, K.; Mourelatos, Z. MicroRNAs: Biogenesis and molecular functions. Brain Pathol. 2008, 18, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Lian, J.B.; Stein, G.S.; van Wijnen, A.J.; Stein, J.L.; Hassan, M.Q.; Gaur, T.; Zhang, Y. MicroRNA control of bone formation and homeostasis. Nat. Rev. Endocrinol. 2012, 8, 212–227. [Google Scholar] [CrossRef] [PubMed]
- Sinha, K.M.; Zhou, X. Genetic and molecular control of osterix in skeletal formation. J. Cell. Biochem. 2013, 114, 975–984. [Google Scholar] [CrossRef] [PubMed]
- Katagiri, T.; Takahashi, N. Regulatory mechanisms of osteoblast and osteoclast differentiation. Oral Dis. 2002, 8, 147–159. [Google Scholar] [CrossRef] [PubMed]
- Van Wijnen, A.J.; van de Peppel, J.; van Leeuwen, J.P.; Lian, J.B.; Stein, G.S.; Westendorf, J.J.; Oursler, M.J.; Im, H.J.; Taipaleenmäki, H.; Hesse, E.; et al. MicroRNA functions in osteogenesis and dysfunctions in osteoporosis. Curr. Osteoporos. Rep. 2013, 11, 72–82. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.; Yang, B.; Guo, H.; Kang, F. MicroRNAs regulate osteogenesis and chondrogenesis. Biochem. Biophys. Res. Commun. 2012, 418, 587–591. [Google Scholar] [CrossRef] [PubMed]
- Komori, T. Regulation of bone development and extracellular matrix protein genes by RUNX2. Cell Tissue Res. 2010, 339, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Lian, J.B.; Javed, A.; Zaidi, S.K.; Lengner, C.; Montecino, M.; van Wijnen, A.J.; Stein, J.L.; Stein, G.S. Regulatory controls for osteoblast growth and differentiation: Role of Runx/Cbfa/AML factors. Crit. Rev. Eukaryot. Gene Expr. 2004, 14, 1–41. [Google Scholar] [CrossRef] [PubMed]
- Eskildsen, T.; Taipaleenmäki, H.; Stenvang, J.; Abdallah, B.M.; Ditzel, N.; Nossent, A.Y.; Bak, M.; Kauppinen, S.; Kassem, M. MicroRNA-138 regulates osteogenic differentiation of human stromal (mesenchymal) stem cells in vivo. Proc. Natl. Acad. Sci. USA 2011, 108, 6139–6144. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Zhao, L.; Xing, L.; Chen, D. MicroRNA-204 regulates Runx2 protein expression and mesenchymal progenitor cell differentiation. Stem Cells 2010, 28, 357–364. [Google Scholar] [PubMed]
- Zhang, J.; Tu, Q.; Bonewald, L.F.; He, X.; Stein, G.; Lian, J.; Chen, J. Effects of miR-335-5p in modulating osteogenic differentiation by specifically downregulating Wnt antagonist DKK1. J. Bone Miner. Res. 2011, 26, 1953–1963. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Xie, H.; Liu, W.; Hu, R.; Huang, B.; Tan, Y.F.; Xu, K.; Sheng, Z.F.; Zhou, H.D.; Wu, X.P.; et al. A novel microRNA targeting HDAC5 regulates osteoblast differentiation in mice and contributes to primary osteoporosis in humans. J. Clin. Investig. 2009, 119, 3666–3677. [Google Scholar] [CrossRef] [PubMed]
- Hu, R.; Liu, W.; Li, H.; Yang, L.; Chen, C.; Xia, Z.Y.; Guo, L.J.; Xie, H.; Zhou, H.D.; Wu, X.P.; et al. A Runx2/miR-3960/miR-2861 regulatory feedback loop during mouse osteoblast differentiation. J. Biol. Chem. 2011, 286, 12328–12339. [Google Scholar] [CrossRef] [PubMed]
- Hwang, S.; Park, S.K.; Lee, H.Y.; Kim, S.W.; Lee, J.S.; Choi, E.K.; You, D.; Kim, C.S.; Suh, N. miR-140-5p suppresses BMP2-mediated osteogenesis in undifferentiated human mesenchymal stem cells. FEBS Lett. 2014, 588, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Lu, J.; Cobb, B.S.; Rodda, S.J.; McMahon, A.P.; Schipani, E.; Merkenschlager, M.; Kronenberg, H.M. Dicer-dependent pathways regulate chondrocyte proliferation and differentiation. Proc. Natl. Acad. Sci. USA 2008, 105, 1949–1954. [Google Scholar] [CrossRef] [PubMed]
- Gaur, T.; Hussain, S.; Mudhasani, R.; Parulkar, I.; Colby, J.L.; Frederick, D.; Kream, B.E.; van Wijnen, A.J.; Stein, J.L.; Stein, G.S.; et al. Dicer inactivation in osteoprogenitor cells compromises fetal survival and bone formation, while excision in differentiated osteoblasts increases bone mass in the adult mouse. Dev. Biol. 2010, 340, 10–21. [Google Scholar] [CrossRef] [PubMed]
- Raaijmakers, M.H.; Mukherjee, S.; Guo, S.; Zhang, S.; Kobayashi, T.; Schoonmaker, J.A.; Ebert, B.L.; Al-Shahrour, F.; Hasserjian, R.P.; Scadden, E.O.; et al. Bone progenitor dysfunction induces myelodysplasia and secondary leukaemia. Nature 2010, 464, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Hu, R.; Li, H.; Liu, W.; Yang, L.; Tan, Y.F.; Luo, X.H. Targeting miRNAs in osteoblast differentiation and bone formation. Expert Opin. Ther. Targets 2010, 14, 1109–1120. [Google Scholar] [CrossRef] [PubMed]
- Inose, H.; Ochi, H.; Kimura, A.; Fujita, K.; Xu, R.; Sato, S.; Iwasaki, M.; Sunamura, S.; Takeuchi, Y.; Fukumoto, S.; et al. A microRNA regulatory mechanism of osteoblast differentiation. Proc. Natl. Acad. Sci. USA 2009, 106, 20794–20799. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Hassan, M.Q.; Volinia, S.; van Wijnen, A.J.; Stein, J.L.; Croce, C.M.; Lian, J.B.; Stein, G.S. A microRNA signature for a BMP2-induced osteoblast lineage commitment program. Proc. Natl. Acad. Sci. USA 2008, 105, 13906–13911. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Hassan, M.Q.; Jafferji, M.; Aqeilan, R.I.; Garzon, R.; Croce, C.M.; van Wijnen, A.J.; Stein, J.L.; Stein, G.S.; Lian, J.B. Biological functions of miR-29b contribute to positive regulation of osteoblast differentiation. J. Biol. Chem. 2009, 284, 15676–15684. [Google Scholar] [CrossRef] [PubMed]
- Trompeter, H.I.; Dreesen, J.; Hermann, E.; Iwaniuk, K.M.; Hafner, M.; Renwick, N.; Tuschl, T.; Wernet, P. MicroRNAs miR-26a, miR-26b, and miR-29b accelerate osteogenic differentiation of unrestricted somatic stem cells from human cord blood. BMC Genomics 2013, 14, 111. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Dong, J.; Zhang, Z.H.; Zhang, D.C.; You, X.Y.; Zhong, Y.; Chen, M.S.; Liu, S.M. miR-10a restores human mesenchymal stem cell differentiation by repressing KLF4. J. Cell. Physiol. 2013, 228, 2324–2336. [Google Scholar] [PubMed]
- Baglìo, S.R.; Devescovi, V.; Granchi, D.; Baldini, N. MicroRNA expression profiling of human bone marrow mesenchymal stem cells during osteogenic differentiation reveals Osterix regulation by miR-31. Gene 2013, 527, 321–331. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Holmstrøm, K.; Qiu, W.; Ditzel, N.; Shi, K.; Hokland, L.; Kassem, M. MicroRNA-34a inhibits osteoblast differentiation and in vivo bone formation of human stromal stem cells. Stem Cells 2014, 32, 902–912. [Google Scholar] [CrossRef] [PubMed]
- Bhushan, R.; Grünhagen, J.; Becker, J.; Robinson, P.N.; Ott, C.E.; Knaus, P. miR-181a promotes osteoblastic differentiation through repression of TGF-β signaling molecules. Int. J. Biochem. Cell Biol. 2013, 45, 696–705. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, Y.; Tokuzawa, Y.; Ninomiya, Y.; Yagi, K.; Yatsuka-Kanesaki, Y.; Suda, T.; Fukuda, T.; Katagiri, T.; Kondoh, Y.; Amemiya, T.; et al. miR-210 promotes osteoblastic differentiation through inhibition of AcvR1b. FEBS Lett. 2009, 583, 2263–2268. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.B.; Zhong, W.J.; Wang, L. A signal-amplification circuit between miR-218 and Wnt/β-catenin signal promotes human adipose tissue-derived stem cells osteogenic differentiation. Bone 2014, 58, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.J.; Bae, S.W.; Yu, S.S.; Bae, Y.C.; Jung, J.S. miR-196a regulates proliferation and osteogenic differentiation in mesenchymal stem cells derived from human adipose tissue. J. Bone Miner. Res. 2009, 24, 816–825. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Cai, J.; Cai, X.H.; Chen, L. miR-346 regulates osteogenic differentiation of human bone marrow-derived mesenchymal stem cells by targeting the Wnt/β-catenin pathway. PLoS ONE 2013, 8, e72266. [Google Scholar] [CrossRef] [PubMed]
- Vimalraj, S.; Partridge, N.C.; Selvamurugan, N. A positive role of microRNA-15b on regulation of osteoblast differentiation. J. Cell. Physiol. 2014, 229, 1236–1244. [Google Scholar] [CrossRef] [PubMed]
- O’Hara, A.J.; Vahrson, W.; Dittmer, D.P. Gene alteration and precursor and mature microRNA transcription changes contribute to the miRNA signature of primary effusion lymphoma. Blood 2008, 111, 2347–2353. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Cheng, P.; Chen, C.; He, H.B.; Xie, G.Q.; Zhou, H.D.; Xie, H.; Wu, X.P.; Luo, X.H. MiR-93/Sp7 function loop mediates osteoblast mineralization. J. Bone Miner. Res. 2012, 27, 1598–1606. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Sun, Q.; Wan, C.; Li, L.; Zhang, L.; Chen, Z. MicroRNA-338-3p regulates osteogenic differentiation of mouse bone marrow stromal stem cells by targeting Runx2 and Fgfr2. J. Cell. Physiol. 2014, 229, 1494–1502. [Google Scholar] [CrossRef] [PubMed]
- Kahai, S.; Lee, S.C.; Lee, D.Y.; Yang, J.; Li, M.; Wang, C.H.; Jiang, Z.; Zhang, Y.; Peng, C.; Yang, B.B. MicroRNA miR-378 regulates nephronectin expression modulating osteoblast differentiation by targeting GalNT-7. PLoS ONE 2009, 4, e7535. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.J.; Kang, I.H.; Lee, J.W.; Jang, W.G.; Koh, J.T. MiR-433 mediates ERRγ-suppressed osteoblast differentiation via direct targeting to Runx2 mRNA in C3H10T1/2 cells. Life Sci. 2013, 92, 562–568. [Google Scholar] [CrossRef] [PubMed]
- Vimalraj, S.; Selvamurugan, N. MicroRNAs: Synthesis, gene regulation and osteoblast differentiation. Curr. Issues Mol. Biol. 2012, 15, 7–18. [Google Scholar] [PubMed]
- Tou, L.; Quibria, N.; Alexander, J.M. Transcriptional regulation of the human Runx2/Cbfa1 gene promoter by bone morphogenetic protein-7. Mol. Cell. Endocrinol. 2003, 205, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Kundu, M.; Javed, A.; Jeon, J.P.; Horner, A.; Shum, L.; Eckhaus, M.; Muenke, M.; Lian, J.B.; Yang, Y.; Nuckolls, G.H.; et al. Cbfbeta interacts with Runx2 and has a critical role in bone development. Nat. Genet. 2002, 32, 639–644. [Google Scholar] [CrossRef] [PubMed]
- Komori, T. A fundamental transcription factor for bone and cartilage. Biochem. Biophys. Res. Commun. 2000, 276, 813–816. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.M.; Lee, E.H. Transcriptional regulatory cascades in Runx2-dependent bone development. Tissue Eng. 2013, 19, 254–263. [Google Scholar] [CrossRef]
- Zhang, Y.; Xie, R.L.; Croce, C.M.; Stein, J.L.; Lian, J.B.; van Wijnen, A.J.; Stein, G.S. A program of microRNAs controls osteogenic lineage progression by targeting transcription factor Runx2. Proc. Natl. Acad. Sci. USA 2011, 108, 9863–9868. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xie, R.L.; Gordon, J.; LeBlanc, K.; Stein, J.L.; Lian, J.B.; van Wijnen, A.J.; Stein, G.S. Control of mesenchymal lineage progression by microRNAs targeting skeletal gene regulators Trps1 and Runx2. J. Biol. Chem. 2012, 287, 21926–21935. [Google Scholar] [CrossRef] [PubMed]
- Zuo, B.; Zhu, J.F.; Li, J.; Wang, C.D.; Zhao, X.Y.; Cai, G.Q.; Li, Z.; Peng, J.; Wang, P.; Shen, C.; et al. MicroRNA-103a functions as a mechno-sensitive microRNA to inhibit bone formation through targeting Runx2. J. Bone Miner. Res. 2015, 30, 330–345. [Google Scholar] [CrossRef] [PubMed]
- Gay, I.; Cavender, A.; Peto, D.; Sun, Z.; Speer, A.; Cao, H.; Amendt, B.A. Differentiation of human dental stem cells reveals a role for microRNA-218. J. Periodontal Res. 2014, 49, 110–120. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.M.; Lim, S.K. Role of miRNAs in bone and their potential as therapeutic targets. Curr. Opin. Pharmacol. 2014, 16, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Nishio, Y.; Dong, Y.; Paris, M.; O’Keefe, R.J.; Schwarz, E.M.; Drissi, H. Runx2-mediated regulation of the zinc finger Osterix/Sp7 gene. Gene 2006, 372, 62–70. [Google Scholar] [CrossRef] [PubMed]
- Barbuto, R.; Mitchell, J. Regulation of the osterix (Osx, Sp7) promoter by osterix and its inhibition by parathyroid hormone. J. Mol. Endocrinol. 2013, 51, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.H.; Lu, X.; Nanes, M.S.; Mitchell, J. Regulation of osterix (Osx, Sp7) and the Osx promoter by parathyroid hormone in osteoblasts. J. Mol. Endocrinol. 2009, 43, 197–207. [Google Scholar] [CrossRef] [PubMed]
- Li, E.; Zhang, J.; Yuan, T.; Ma, B. miR-143 suppresses osteogenic differentiation by targeting Osterix. Mol. Cell. Biochem. 2014, 390, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Xia, H.; Sun, S.; Wang, B.; Wang, T.; Liang, C.; Li, G.; Huang, C.; Qi, D.; Chu, X. miR-143 inhibits NSCLC cell growth and metastasis by targeting Limk1. Int. J. Mol. Sci. 2014, 15, 11973–11983. [Google Scholar] [CrossRef] [PubMed]
- Pham, H.; Rodriguez, C.E.; Donald, G.W.; Hertzer, K.M.; Jung, X.S.; Chang, H.H.; Moro, A.; Reber, H.A.; Hines, O.J.; Eibl, G. miR-143 decreases COX-2 mRNA stability and expression in pancreatic cancer cells. Biochem. Biophys. Res. Commun. 2013, 439, 6–11. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Chen, X.; Liang, H.; Deng, T.; Chen, W.; Zhang, S.; Liu, M.; Gao, X.; Liu, Y.; Zhao, C.; et al. miR-143 and miR-145 synergistically regulate ERBB3 to suppress cell proliferation and invasion in breast cancer. Mol. Cancer 2014, 13, 220. [Google Scholar] [CrossRef] [PubMed]
- Jia, J.; Tian, Q.; Ling, S.; Liu, Y.; Yang, S.; Shao, Z. miR-145 suppresses osteogenic differentiation by targeting Sp7. FEBS Lett. 2013, 587, 3027–3031. [Google Scholar] [CrossRef] [PubMed]
- Shi, K.; Lu, J.; Zhao, Y.; Wang, L.; Li, J.; Qi, B.; Li, H.; Ma, C. MicroRNA-214 suppresses osteogenic differentiation of C2C12 myoblast cells by targeting Osterix. Bone 2013, 55, 487–494. [Google Scholar] [CrossRef] [PubMed]
- Goettsch, C.; Rauner, M.; Pacyna, N.; Hempel, U.; Bornstein, S.R.; Hofbauer, L.C. miR-125b regulates calcification of vascular smooth muscle cells. Am. J. Pathol. 2011, 179, 1594–1600. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Yang, L.; Jie, Q.; Lin, Y.S.; Meng, G.L.; Fan, J.Z.; Zhang, J.K.; Fan, J.; Luo, Z.J.; Liu, J. MicroRNA-125b suppresses the proliferation and osteogenic differentiation of human bone marrow-derived mesenchymal stem cells. Mol. Med. Rep. 2014, 9, 1820–1826. [Google Scholar] [PubMed]
- Zhang, J.F.; Fu, W.M.; He, M.L.; Wang, H.; Wang, W.M.; Yu, S.C.; Bian, X.W.; Zhou, J.; Lin, M.C.; Lu, G.; et al. miR-637 maintains the balance between adipocytes and osteoblasts by directly targeting Osterix. Mol. Biol. Cell 2011, 22, 3955–3961. [Google Scholar] [CrossRef] [PubMed]
- Gámez, B.; Rodriguez-Carballo, E.; Ventura, F. MicroRNAs and post-transcriptional regulation of skeletal development. J. Mol. Endocrinol. 2014, 52, R179–R197. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.; Fu, J.; Zhou, W.; Li, W.; Dong, S.; Yu, S.; Hu, Z.; Wang, H.; Xie, Z. MicroRNA-125b regulates osteogenic differentiation of mesenchymal stem cells by targeting Cbfβ in vitro. Biochimie 2014, 102, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Itoh, T.; Nozawa, Y.; Akao, Y. MicroRNA-141 and -200a are involved in bone morphogenetic protein-2-induced mouse pre-osteoblast differentiation by targeting distal-less homeobox 5. J. Biol. Chem. 2009, 284, 19272–19279. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Guo, B.; Li, Q.; Peng, J.; Yang, Z.; Wang, A.; Li, D.; Hou, Z.; Lv, K.; Kan, G.; et al. miR-214 targets ATF4 to inhibit bone formation. Nat. Med. 2013, 19, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Andrianakos, R.; Yang, Y.; Liu, C.; Lu, W. Kruppel-like factor 4 (Klf4) prevents embryonic stem (ES) cell differentiation by regulating Nanog gene expression. J. Biol. Chem. 2010, 285, 9180–9189. [Google Scholar] [CrossRef] [PubMed]
- Xie, Q.; Wang, Z.; Bi, X.; Zhou, H.; Wang, Y.; Gu, P.; Fan, X. Effects of miR-31 on the osteogenesis of human mesenchymal stem cells. Biochem. Biophys. Res. Commun. 2014, 446, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Dobreva, G.; Chahrour, M.; Dautzenberg, M.; Chirivella, L.; Kanzler, B.; Fariñas, I.; Karsenty, G.; Grosschedl, R. SATB2 is a multifunctional determinant of craniofacial patterning and osteoblast differentiation. Cell 2006, 125, 971–986. [Google Scholar] [CrossRef] [PubMed]
- Huszar, J.M.; Payne, C.J. MIR146A inhibits JMJD3 expression and osteogenic differentiation in human mesenchymal stem cells. FEBS Lett. 2014, 588, 1850–1856. [Google Scholar] [CrossRef] [PubMed]
- Kapinas, K.; Kessler, C.B.; Delany, A.M. miR-29 suppression of osteonectin in osteoblasts: Regulation during differentiation and by canonical Wnt signaling. J. Cell. Biochem. 2009, 108, 216–224. [Google Scholar] [CrossRef] [PubMed]
- Hupkes, M.; Sotoca, A.M.; Hendriks, J.M.; van Zoelen, E.J.; Dechering, K.J. MicroRNA miR-378 promotes BMP2-induced osteogenic differentiation of mesenchymal progenitor cells. BMC Mol. Biol. 2014, 15, 1. [Google Scholar] [CrossRef] [PubMed]
- Pulikkan, J.A.; Peramangalam, P.S.; Dengler, V.; Ho, P.A.; Preudhomme, C.; Meshinchi, S.; Christopeit, M.; Nibourel, O.; Müller-Tidow, C.; Bohlander, S.K.; et al. C/EBPα regulated microRNA-34a targets E2F3 during granulopoiesis and is down-regulated in AML with CEBPA mutations. Blood 2010, 116, 5638. [Google Scholar] [CrossRef] [PubMed]
- Fujita, Y.; Kojima, K.; Hamada, N.; Ohhashi, R.; Akao, Y.; Nozawa, Y.; Deguchi, T.; Ito, M. Effects of miR-34a on cell growth and chemoresistance in prostate cancer PC3 cells. Biochem. Biophys. Res. Commun. 2008, 377, 114–119. [Google Scholar] [CrossRef] [PubMed]
- Hermeking, H. The miR-34 family in cancer and apoptosis. Cell Death Differ. 2010, 17, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Jurkiewicz, D.; Gliwicz, D.; Ciara, E.; Gerfen, J.; Pelc, M.; Piekutowska-Abramczuk, D.; Kugaudo, M.; Chrzanowska, K.; Spinner, N.B.; Krajewska-Walasek, M. Spectrum of JAG1 gene mutations in Polish patients with Alagille syndrome. J. Appl. Genet. 2014, 55, 329–336. [Google Scholar] [CrossRef] [PubMed]
- Vajro, P.; Ferrante, L.; Paolella, G. Alagille syndrome: an overview. Clin. Res. Hepatol. Gastroenterol. 2012, 36, 275–277. [Google Scholar] [CrossRef] [PubMed]
- Celil, A.B.; Campbell, P.G. BMP-2 and insulin-like growth factor-I mediate Osterix (Osx) expression in human mesenchymal stem cells via the MAPK and protein kinase D signaling pathways. J. Biol. Chem. 2005, 280, 31353–31359. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Zhao, M.; Mundy, G.R. Bone morphogenetic proteins. Growth Factors 2004, 22, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.D.; Zhang, J.C.; Zhang, Q.; Wang, S.X.; Yang, M.S. TGF-β/BMP signaling pathway is involved in cerium-promoted osteogenic differentiation of mesenchymal stem cells. J. Cell. Biochem. 2013, 114, 1105–1114. [Google Scholar] [CrossRef] [PubMed]
- Phimphilai, M.; Zhao, Z.; Boules, H.; Roca, H.; Franceschi, R.T. BMP signaling is required for RUNX2-dependent induction of the osteoblast phenotype. J. Bone Miner. Res. 2006, 21, 637–646. [Google Scholar] [CrossRef] [PubMed]
- Javed, A.; Bae, J.S.; Afzal, F.; Gutierrez, S.; Pratap, J.; Zaidi, S.K.; Lou, Y.; van Wijnen, A.J.; Stein, J.L.; Stein, G.S.; et al. Structural coupling of Smad and Runx2 for execution of the BMP2 osteogenic signal. J. Biol. Chem. 2008, 283, 8412–8422. [Google Scholar] [CrossRef] [PubMed]
- Hendy, G.N.; Kaji, H.; Sowa, H.; Lebrun, J.J.; Canaff, L. Menin and TGF-β superfamily member signaling via the Smad pathway in pituitary, parathyroid and osteoblast. Horm. Metab. Res. 2005, 37, 375–379. [Google Scholar] [CrossRef] [PubMed]
- Lin, E.A.; Kong, L.; Bai, X.H.; Luan, Y.; Liu, C.J. miR-199a, a bone morphogenic protein 2-responsive microRNA, regulates chondrogenesis via direct targeting to Smad1. J. Biol. Chem. 2009, 284, 11326–11335. [Google Scholar] [CrossRef] [PubMed]
- Kureel, J.; Dixit, M.; Tyagi, A.M.; Mansoori, M.N.; Srivastava, K.; Raghuvanshi, A.; Maurya, R.; Trivedi, R.; Goel, A.; Singh, D. miR-542-3p suppresses osteoblast cell proliferation and differentiation, targets BMP-7 signaling and inhibits bone formation. Cell Death Dis. 2014, 5, e1050. [Google Scholar] [CrossRef] [PubMed]
- Luzi, E.; Marini, F.; Sala, S.C.; Tognarini, I.; Galli, G.; Brandi, M.L. Osteogenic differentiation of human adipose tissue-derived stem cells is modulated by the miR-26a targeting of the SMAD1 transcription factor. J. Bone Miner. Res. 2008, 23, 287–295. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Deng, M.; He, H.; Zeng, D.; Zhang, W. miR-125b regulates osteogenic differentiation of human bone marrow mesenchymal stem cells by targeting Smad4. Zhong Nan Da Xue Xue Bao Yi Xue Ban 2013, 38, 341–346. [Google Scholar] [PubMed]
- Cheung, K.S.; Sposito, N.; Stumpf, P.S.; Wilson, D.I.; Sanchez-Elsner, T.; Oreffo, R.O. MicroRNA-146a regulates human foetal femur derived skeletal stem cell differentiation by down-regulating SMAD2 and SMAD3. PLoS ONE 2014, 9, e98063. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.Q.; Chen, H.; Zheng, X.F.; Zhang, B.X.; Wang, Y.; Tang, P.F.; She, F.; Song, Q.; Li, T.S. Hsa-miR-654-5p regulates osteogenic differentiation of human bone marrow mesenchymal stem cells by repressing bone morphogenetic protein 2. Nan Fang Yi Ke Da Xue Xue Bao 2012, 32, 291–295. [Google Scholar] [PubMed]
- Taipaleenmäki, H.; Bjerre Hokland, L.; Chen, L.; Kauppinen, S.; Kassem, M. Mechanisms in endocrinology: Micro-RNAs: Targets for enhancing osteoblast differentiation and bone formation. Eur. J. Endocrinol. 2012, 166, 359–371. [Google Scholar] [CrossRef] [PubMed]
- Rosenbluh, J.; Wang, X.; Hahn, W.C. Genomic insights into Wnt/β-catenin signaling. Trends Pharmacol. Sci. 2014, 35, 103–109. [Google Scholar] [PubMed]
- Wang, Y.; Li, Y.P.; Paulson, C.; Shao, J.Z.; Zhang, X.; Wu, M.; Chen, W. Wnt and the Wnt signaling pathway in bone development and disease. Front. Biosci. (Landmark Ed.) 2014, 19, 379–407. [Google Scholar] [CrossRef]
- Lian, J.B.; Stein, G.S.; Javed, A.; van Wijnen, A.J.; Stein, J.L.; Montecino, M.; Hassan, M.Q.; Gaur, T.; Lengner, C.J.; Young, D.W. Networks and hubs for the transcriptional control of osteoblastogenesis. Rev. Endocr. Metab. Disord. 2006, 7, 1–16. [Google Scholar] [CrossRef] [PubMed]
- James, A.W. Review of signaling pathways governing MSC osteogenic and adipogenic differentiation. Scientifica (Cairo) 2013, 2013, 684736. [Google Scholar]
- Ko, J.Y.; Chuang, P.C.; Chen, M.W.; Ke, H.C.; Wu, S.L.; Chang, Y.H.; Chen, Y.S.; Wang, F.S. MicroRNA-29a ameliorates glucocorticoid-induced suppression of osteoblast differentiation by regulating β-catenin acetylation. Bone 2013, 57, 468–475. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Xu, Z. miR-27 promotes osteoblast differentiation by modulating Wnt signaling. Biochem. Biophys. Res. Commun. 2010, 402, 186–189. [Google Scholar] [CrossRef] [PubMed]
- Guo, D.; Li, Q.; Lv, Q.; Wei, Q.; Cao, S.; Gu, J. miR-27a targets sFRP1 in hFOB cells to regulate proliferation, apoptosis and differentiation. PLoS ONE 2014, 9, e91354. [Google Scholar] [CrossRef] [PubMed]
- Kapinas, K.; Kessler, C.; Ricks, T.; Gronowicz, G.; Delany, A.M. miR-29 modulates Wnt signaling in human osteoblasts through a positive feedback loop. J. Biol. Chem. 2010, 285, 25221–25231. [Google Scholar] [CrossRef] [PubMed]
- Hassan, M.Q.; Maeda, Y.; Taipaleenmaki, H.; Zhang, W.; Jafferji, M.; Gordon, J.A.; Li, Z.; Croce, C.M.; van Wijnen, A.J.; Stein, J.L.; et al. miR-218 directs a Wnt signaling circuit to promote differentiation of osteoblasts and osteomimicry of metastatic cancer cells. J. Biol. Chem. 2012, 287, 42084–42092. [Google Scholar] [CrossRef] [PubMed]
- Egea, V.; Zahler, S.; Rieth, N.; Neth, P.; Popp, T.; Kehe, K.; Jochum, M.; Ries, C. Tissue inhibitor of metalloproteinase-1 (TIMP-1) regulates mesenchymal stem cells through let-7f microRNA and Wnt/β-catenin signaling. Proc. Natl. Acad. Sci. USA 2012, 109, E309–E316. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Xu, D.; Li, Y.; Zhang, J.; Liu, T.; Ji, Y.; Wang, J.; Zhou, G.; Xie, X. MicroRNAs regulate bone metabolism. J. Bone Miner. Metab. 2014, 32, 221–231. [Google Scholar] [CrossRef] [PubMed]
- Bae, Y.; Yang, T.; Zeng, H.C.; Campeau, P.M.; Chen, Y.; Bertin, T.; Dawson, B.C.; Munivez, E.; Tao, J.; Lee, B.H. miRNA-34c regulates Notch signaling during bone development. Hum. Mol. Genet. 2012, 21, 2991–3000. [Google Scholar] [CrossRef] [PubMed]
- Hassan, M.Q.; Gordon, J.A.; Beloti, M.M.; Croce, C.M.; van Wijnen, A.J.; Stein, J.L.; Stein, G.S.; Lian, J.B. A network connecting Runx2, SATB2, and the miR-23a~27a~24-2 cluster regulates the osteoblast differentiation program. Proc. Natl. Acad. Sci. USA 2010, 107, 19879–19884. [Google Scholar] [CrossRef] [PubMed]
- Kalajzic, I.; Staal, A.; Yang, W.P.; Wu, Y.; Johnson, S.E.; Feyen, J.H.; Krueger, W.; Maye, P.; Yu, F.; Zhao, Y.; et al. Expression profile of osteoblast lineage at defined stages of differentiation. J. Biol. Chem. 2005, 280, 24618–24626. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Wu, S.; Zhou, H.; Bi, X.; Wang, Y.; Hu, Y.; Gu, P.; Fan, X. Effects of a miR-31, Runx2, and Satb2 regulatory loop on the osteogenic differentiation of bone mesenchymal stem cells. Stem Cells Dev. 2013, 22, 2278–2286. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Liu, W.; Sinha, K.M.; Yasuda, H.; de Crombrugghe, B. Identification and characterization of microRNAs controlled by the osteoblast-specific transcription factor Osterix. PLoS ONE 2013, 8, e58104. [Google Scholar] [CrossRef] [PubMed]
- Pittenger, M.F.; Mackay, A.M.; Beck, S.C.; Jaiswal, R.K.; Douglas, R.; Mosca, J.D.; Moorman, M.A.; Simonetti, D.W.; Craig, S.; Marshak, D.R. Multilineage potential of adult human mesenchymal stem cells. Science 1999, 284, 143–147. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Jahagirdar, B.N.; Reinhardt, R.L.; Schwartz, R.E.; Keene, C.D.; Ortiz-Gonzalez, X.R.; Reyes, M.; Lenvik, T.; Lund, T.; Blackstad, M.; et al. Pluripotency of mesenchymal stem cells derived from adult marrow. Nature 2002, 418, 41–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laine, S.K.; Alm, J.J.; Virtanen, S.P.; Aro, H.T.; Laitala-Leinonen, T.K. MicroRNAs miR-96, miR-124, and miR-199a regulate gene expression in human bone marrow-derived mesenchymal stem cells. J. Cell. Biochem. 2012, 113, 2687–2695. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Yang, T.; Han, J.; Yan, K.; Qiu, X.; Zhou, Y.; Fan, Q.; Ma, B. MicroRNA expression during osteogenic differentiation of human multipotent mesenchymal stromal cells from bone marrow. J. Cell. Biochem. 2011, 112, 1844–1856. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Shi, Y.; Zheng, L.; Zhou, B.; Inose, H.; Wang, J.; Guo, X.E.; Grosschedl, R.; Karsenty, G. miR-34s inhibit osteoblast proliferation and differentiation in the mouse by targeting SATB2. J. Cell. Biol. 2012, 197, 509–521. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.F.; Fu, W.M.; He, M.L.; Xie, W.D.; Lv, Q.; Wan, G.; Li, G.; Wang, H.; Lu, G.; Hu, X.; et al. MiRNA-20a promotes osteogenic differentiation of human mesenchymal stem cells by co-regulating BMP signaling. RNA Biol. 2011, 8, 829–838. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Wang, Y.; Li, Y.; Zhao, G. Downregulation of PPARγ by miR-548d-5p suppresses the adipogenic differentiation of human bone marrow mesenchymal stem cells and enhances their osteogenic potential. J. Transl. Med. 2014, 12, 168. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Zhang, R.; Xu, J.; Wu, C.; Lu, X. Functional conservation of both CDS- and 3'-UTR-located microRNA binding sites between species. Mol. Biol. Evol. 2015, 32, 623–628. [Google Scholar] [CrossRef] [PubMed]
- Tay, Y.; Zhang, J.; Thomson, A.M.; Lim, B.; Rigoutsos, I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature 2008, 455, 1124–1128. [Google Scholar] [CrossRef] [PubMed]
- Ebert, M.S.; Neilson, J.R.; Sharp, P.A. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat. Methods. 2007, 4, 721–726. [Google Scholar] [CrossRef] [PubMed]
- Frith, J.E.; Porrello, E.R.; Cooper-White, J.J. Concise review: New frontiers in microRNA-based tissue regeneration. Stem Cells Transl. Med. 2014, 3, 969–976. [Google Scholar] [CrossRef] [PubMed]
- Qureshi, A.T.; Monroe, W.T.; Dasa, V.; Gimble, J.M.; Hayes, D.J. miR-148b-nanoparticle conjugates for light mediated osteogenesis of human adipose stromal/stem cells. Biomaterials 2013, 34, 7799–7810. [Google Scholar] [CrossRef] [PubMed]
- Suh, J.S.; Lee, J.Y.; Choi, Y.S.; Chung, C.P.; Park, Y.J. Peptide-mediated intracellular delivery of miRNA-29b for osteogenic stem cell differentiation. Biomaterials 2013, 34, 4347–4359. [Google Scholar] [CrossRef] [PubMed]
- Effendy, N.M.; Khamis, M.F.; Shuid, A.N. Micro-CT assessments of potential anti-osteoporotic agents. Curr. Drug Targets 2013, 14, 1542–1551. [Google Scholar] [CrossRef] [PubMed]
- Wise, L.D.; Winkelmann, C.T.; Dogdas, B.; Bagchi, A. Micro-computed tomography imaging and analysis in developmental biology and toxicology. Birth Defects Res. 2013, 99, 71–82. [Google Scholar] [CrossRef]
- Deng, Y.; Zhou, H.; Zou, D.; Xie, Q.; Bi, X.; Gu, P.; Fan, X. The role of miR-31-modified adipose tissue-derived stem cells in repairing rat critical-sized calvarial defects. Biomaterials 2013, 34, 6717–6728. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Zhou, H.; Gu, P.; Fan, X. Repair of canine medial orbital bone defects with miR-31-modified bone marrow mesenchymal stem cells. Investig. Ophthalmol. Vis. Sci. 2014, 55, 6016–6023. [Google Scholar] [CrossRef]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fang, S.; Deng, Y.; Gu, P.; Fan, X. MicroRNAs Regulate Bone Development and Regeneration. Int. J. Mol. Sci. 2015, 16, 8227-8253. https://doi.org/10.3390/ijms16048227
Fang S, Deng Y, Gu P, Fan X. MicroRNAs Regulate Bone Development and Regeneration. International Journal of Molecular Sciences. 2015; 16(4):8227-8253. https://doi.org/10.3390/ijms16048227
Chicago/Turabian StyleFang, Sijie, Yuan Deng, Ping Gu, and Xianqun Fan. 2015. "MicroRNAs Regulate Bone Development and Regeneration" International Journal of Molecular Sciences 16, no. 4: 8227-8253. https://doi.org/10.3390/ijms16048227
APA StyleFang, S., Deng, Y., Gu, P., & Fan, X. (2015). MicroRNAs Regulate Bone Development and Regeneration. International Journal of Molecular Sciences, 16(4), 8227-8253. https://doi.org/10.3390/ijms16048227