Overexpressing Ferredoxins in Chlamydomonas reinhardtii Increase Starch and Oil Yields and Enhance Electric Power Production in a Photo Microbial Fuel Cell
Abstract
:1. Introduction
2. Results and Discussion
2.1. Result
2.1.1. Generation and Characterization of Transgenic Lines Overexpressing Ferredoxin Genes
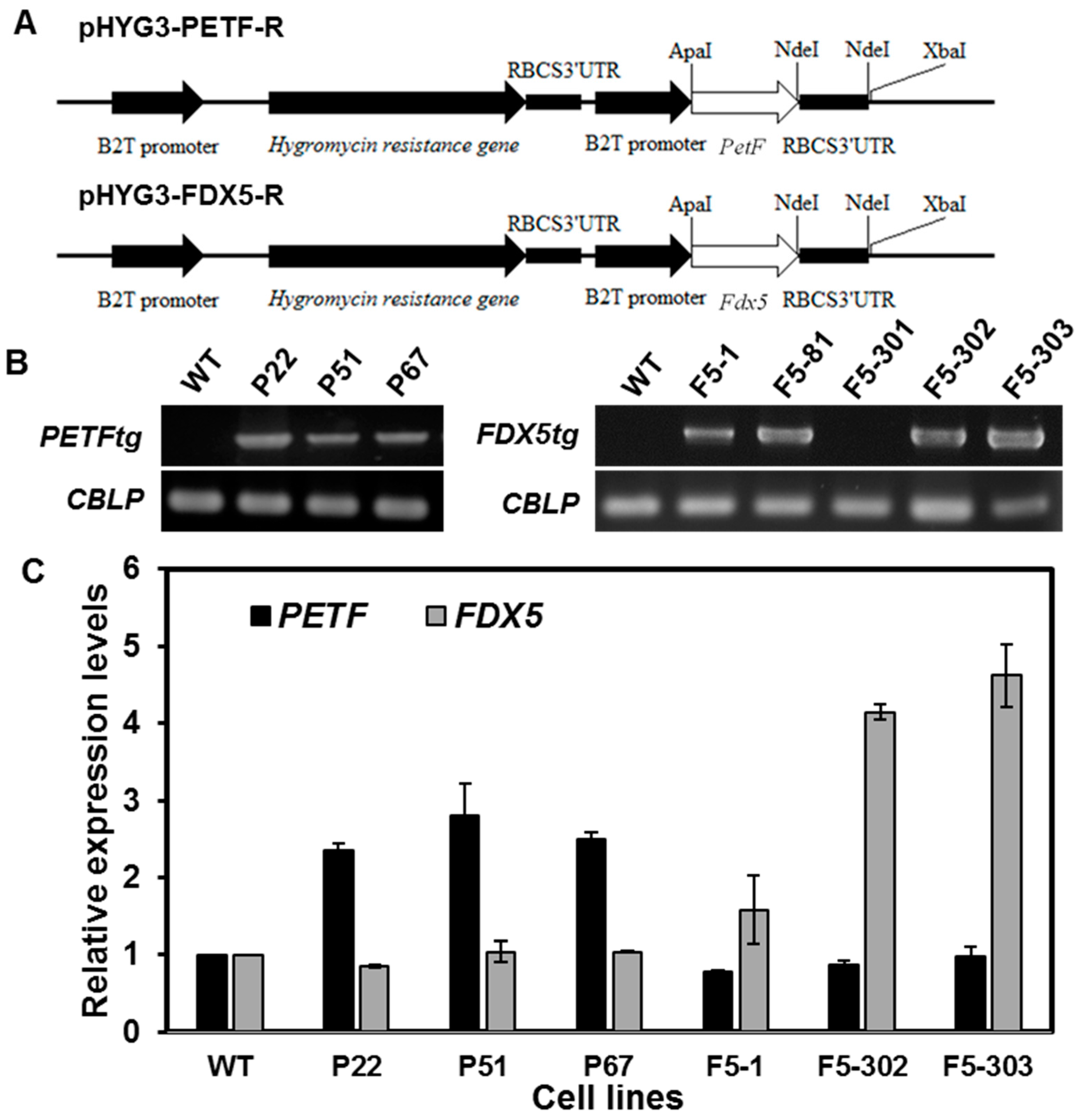
2.1.2. Overexpression of Ferredoxins Promotes Tolerance to Heat and Salt Stresses in Chlamydomonas


2.1.3. Overexpression of Ferredoxins Promotes Tolerance to Heat and Salt Stresses in Chlamydomonas
| Line | +N | −N1 | −N2 | −N3 | −N4 | −N5 | −N6 | −N7 |
|---|---|---|---|---|---|---|---|---|
| WT | 13.05 ± 0.06 | 44.13 ± 0.05 | 42.50 ± 0.05 | 41.33 ± 0.03 | 43.33 ± 0.09 | 40.53 ± 0.04 | 40.53 ± 0.06 | 38.77 ± 0.13 |
| PETF overexpression lines | ||||||||
| P22 | 19.40 ± 0.15 | 63.40 ± 0.30 | 73.91 ± 0.12 | 77.96 ± 1.06 | 80.00 ± 0.24 | 85.08 ± 0.34 | 80.82 ± 0.03 | 83.69 ± 0.11 |
| P51 | 19.13 ± 0.03 | 66.46 ± 0.07 | 70.82 ± 0.08 | 71.74 ± 0.12 | 69.69 ± 0.16 | 65.14 ± 0.30 | 63.84 ± 0.10 | 61.43 ± 0.24 |
| P67 | 18.86 ± 0.08 | 56.08 ± 0.11 | 87.19 ± 0.38 | 75.71 ± 0.12 | 73.07 ± 0.24 | 88.26 ± 0.21 | 84.34 ± 0.02 | 84.90 ± 0.06 |
| FDX5 overexpression lines | ||||||||
| F5-1 | 15.04 ± 0.05 | 56.85 ± 0.01 | 49.59 ± 0.07 | 47.14 ± 0.09 | 62.89 ± 0.15 | 65.25 ± 0.10 | 65.25 ± 0.16 | 57.11 ± 0.08 |
| F5-302 | 18.60 ± 0.02 | 52.97 ± 0.18 | 89.13 ± 0.18 | 101.79 ± 0.11 | 103.30 ± 0.10 | 101.31 ± 0.62 | 103.21 ± 0.18 | 106.88 ± 0.10 |
| F5-303 | 18.11 ± 0.01 | 49.22 ± 0.10 | 69.77 ± 0.19 | 69.52 ± 0.36 | 70.31 ± 0.07 | 87.54 ± 0.01 | 82.25 ± 0.12 | 83.69 ± 0.21 |
| Line | +N | −N1 | −N2 | −N3 | −N4 | −N5 | −N6 | −N7 |
|---|---|---|---|---|---|---|---|---|
| WT | 1.22 ± 0.12 | 1.72 ± 0.17 | 2.12 ± 0.08 | 2.31 ± 0.14 | 2.89 ± 0.06 | 3.10 ± 0.06 | 3.00 ± 0.15 | 3.21 ± 0.10 |
| PETF overexpression lines | ||||||||
| P22 | 1.56 ± 0.16 | 1.77 ± 0.07 | 2.57 ± 0.13 | 2.65 ± 0.08 | 3.63 ± 0.22 | 3.80 ± 0.19 | 4.12 ± 0.16 | 4.05 ± 0.20 |
| P51 | 1.60 ± 0.16 | 1.75 ± 0.17 | 2.65 ± 0.21 | 3.29 ± 0.13 | 3.98 ± 0.16 | 4.12 ± 0.16 | 4.20 ± 0.25 | 4.10 ± 0.12 |
| P67 | 1.61 ± 0.16 | 1.88 ± 0.15 | 2.68 ± 0.11 | 3.43 ± 0.17 | 3.71 ± 0.19 | 4.0 ± 0.20 | 4.13 ± 0.17 | 4.11 ± 0.16 |
| FDX5 overexpression lines | ||||||||
| F5-1 | 1.38 ± 0.14 | 1.80 ± 0.09 | 2.30 ± 0.14 | 2.82 ± 0.11 | 3.31 ± 0.13 | 3.52 ± 0.14 | 3.52 ± 0.21 | 3.60 ± 0.14 |
| F5-302 | 1.90 ± 0.19 | 2.26 ± 0.09 | 2.71 ± 0.19 | 3.67 ± 0.22 | 4.01 ± 0.12 | 4.10 ± 0.21 | 4.15 ± 0.21 | 4.16 ± 0.25 |
| F5-303 | 1.91 ± 0.19 | 2.12 ± 0.17 | 2.72 ± 0.14 | 3.13 ± 0.16 | 3.72 ± 0.19 | 3.99 ± 0.12 | 4.01 ± 0.24 | 4.11 ± 0.25 |

2.1.4. Application of Transgenic Chlamydomonas Ferredoxin Lines in a Photo Microbial Fuel Cell


2.2. Discussion
3. Experimental Section
3.1. Cultivation of Chlamydomonas reinhardtii
3.2. Construction of Plasmids
3.3. Transformation of Chlamydomonas reinhardtii
3.4. Heat and Salt Treatment
3.5. Quantitative RT-PCR
3.6. Detection of H2O2 and Reactive Oxygen Species (ROS)
3.7. Starch Quantitation
3.8. Lipid Quantitation
3.9. Photo Microbial Fuel Cell System (PMFC)
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Hariskos, I.; Posten, C. Biorefinery of microalgae—Opportunities and constraints for different production scenarios. Biotechnol. J. 2014, 9, 739–752. [Google Scholar] [CrossRef] [PubMed]
- Lopez Barreiro, D.; Zamalloa, C.; Boon, N.; Vyverman, W.; Ronsse, F.; Brilman, W.; Prins, W. Influence of strain-specific parameters on hydrothermal liquefaction of microalgae. Bioresour. Technol. 2013, 146, 463–471. [Google Scholar] [CrossRef] [PubMed]
- Atta, M.; Idris, A.; Bukhari, A.; Wahidin, S. Intensity of blue led light: A potential stimulus for biomass and lipid content in fresh water microalgae chlorella vulgaris. Bioresour. Technol. 2013, 148, 373–378. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.H.; Chang, J.S.; Wang, H.Y. Current developments in high-throughput analysis for microalgae cellular contents. Biotechnol. J. 2013, 8, 1301–1314. [Google Scholar] [CrossRef] [PubMed]
- Posten, C.; Schaub, G. Microalgae and terrestrial biomass as source for fuels—A process view. J. Biotechnol. 2009, 142, 64–69. [Google Scholar] [CrossRef] [PubMed]
- Raja, R.; Hemaiswarya, S.; Kumar, N.A.; Sridhar, S.; Rengasamy, R. A perspective on the biotechnological potential of microalgae. Crit. Rev. Microbiol. 2008, 34, 77–88. [Google Scholar] [CrossRef] [PubMed]
- Rochaix, J.D. Chlamydomonas, a model system for studying the assembly and dynamics of photosynthetic complexes. FEBS Lett. 2002, 529, 34–38. [Google Scholar] [CrossRef]
- Hema, R.; Senthil-Kumar, M.; Shivakumar, S.; Chandrasekhara Reddy, P.; Udayakumar, M. Chlamydomonas reinhardtii, a model system for functional validation of abiotic stress responsive genes. Planta 2007, 226, 655–670. [Google Scholar] [CrossRef] [PubMed]
- Siaut, M.; Cuine, S.; Cagnon, C.; Fessler, B.; Nguyen, M.; Carrier, P.; Beyly, A.; Beisson, F.; Triantaphylides, C.; Li-Beisson, Y.; et al. Oil accumulation in the model green alga Chlamydomonas reinhardtii: Characterization, variability between common laboratory strains and relationship with starch reserves. BMC Biotechnol. 2011, 11, 7. [Google Scholar] [CrossRef] [PubMed]
- Anderson, A.; Laohavisit, A.; Blaby, I.K.; Bombelli, P.; Howe, C.J.; Merchant, S.S.; Davies, J.M.; Smith, A.G. Exploiting algal NADPH oxidase for biophotovoltaic energy. Plant Biotechnol. J. 2015. [Google Scholar] [CrossRef] [PubMed]
- Rosenbaum, M.; He, Z.; Angenent, L.T. Light energy to bioelectricity: Photosynthetic microbial fuel cells. Curr. Opin. Biotechnol. 2010, 21, 259–264. [Google Scholar] [CrossRef] [PubMed]
- Hanke, G.; Mulo, P. Plant type ferredoxins and ferredoxin-dependent metabolism. Plant Cell Environ. 2013, 36, 1071–1084. [Google Scholar] [CrossRef] [PubMed]
- Arnon, D.I. The discovery of ferredoxin: The photosynthetic path. Trends Biochem. Sci. 1988, 13, 30–33. [Google Scholar] [CrossRef]
- Schurmann, P.; Buchanan, B.B. The ferredoxin/thioredoxin system of oxygenic photosynthesis. Antioxid. Redox Signal. 2008, 10, 1235–1274. [Google Scholar] [CrossRef] [PubMed]
- Nigro, D.; Blanco, A.; Anderson, O.D.; Gadaleta, A. Characterization of ferredoxin-dependent glutamine-oxoglutarate amidotransferase (Fd-GOGAT) genes and their relationship with grain protein content QTL in wheat. PLoS ONE 2014, 9, e103869. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, J.; Pudollek, S.; Hemschemeier, A.; Happe, T. A novel, anaerobically induced ferredoxin in Chlamydomonas reinhardtii. FEBS Lett. 2009, 583, 325–329. [Google Scholar] [CrossRef] [PubMed]
- Buchanan, B.B.; Arnon, D.I. Ferredoxins: Chemistry and function in photosynthesis, nitrogen fixation, and fermentative metabolism. Adv. Enzymol. Relat. Areas Mol. Biol. 1970, 33, 119–176. [Google Scholar] [PubMed]
- Rizhsky, L.; Liang, H.; Mittler, R. The water-water cycle is essential for chloroplast protection in the absence of stress. J. Biol. Chem. 2003, 278, 38921–38925. [Google Scholar] [CrossRef] [PubMed]
- Asada, K. The water–water cycle in chloroplasts: Scavenging of active oxygens and dissipation of excess photons. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 601–639. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, S.; Murata, N. How do environmental stresses accelerate photoinhibition? Trends Plant Sci. 2008, 13, 178–182. [Google Scholar] [CrossRef] [PubMed]
- Noctor, G.; Foyer, C.H. Ascorbate and glutathione: Keeping active oxygen under control. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1998, 49, 249–279. [Google Scholar] [CrossRef] [PubMed]
- Tognetti, V.B.; Palatnik, J.F.; Fillat, M.F.; Melzer, M.; Hajirezaei, M.R.; Valle, E.M.; Carrillo, N. Functional replacement of ferredoxin by a cyanobacterial flavodoxin in tobacco confers broad-range stress tolerance. Plant Cell 2006, 18, 2035–2050. [Google Scholar] [CrossRef] [PubMed]
- Ceccoli, R.D.; Blanco, N.E.; Medina, M.; Carrillo, N. Stress response of transgenic tobacco plants expressing a cyanobacterial ferredoxin in chloroplasts. Plant Mol. Biol. 2011, 76, 535–544. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.H.; Pan, K.Y.; Hung, C.H.; Huang, H.E.; Chen, C.L.; Feng, T.Y.; Huang, L.F. Overexpression of ferredoxin, PETF, enhances tolerance to heat stress in Chlamydomonas reinhardtii. Int. J. Mol. Sci. 2013, 14, 20913–20929. [Google Scholar] [CrossRef] [PubMed]
- Terauchi, A.M.; Lu, S.F.; Zaffagnini, M.; Tappa, S.; Hirasawa, M.; Tripathy, J.N.; Knaff, D.B.; Farmer, P.J.; Lemaire, S.D.; Hase, T.; et al. Pattern of expression and substrate specificity of chloroplast ferredoxins from Chlamydomonas reinhardtii. J. Biol. Chem. 2009, 284, 25867–25878. [Google Scholar] [CrossRef] [PubMed]
- Winkler, M.; Hemschemeier, A.; Jacobs, J.; Stripp, S.; Happe, T. Multiple ferredoxin isoforms in Chlamydomonas reinhardtii—Their role under stress conditions and biotechnological implications. Eur. J. Cell Biol. 2010, 89, 998–1004. [Google Scholar] [CrossRef] [PubMed]
- Lambertz, C.; Hemschemeier, A.; Happe, T. Anaerobic expression of the ferredoxin-encoding FDX5 gene of Chlamydomonas reinhardtii is regulated by the crr1 transcription factor. Eukaryot. Cell 2010, 9, 1747–1754. [Google Scholar] [CrossRef] [PubMed]
- Zuo, Z.; Chen, Z.; Zhu, Y.; Bai, Y.; Wang, Y. Effects of NaCl and Na2CO3 stresses on photosynthetic ability of Chlamydomonas reinhardtii. Biologia 2014, 69, 1314–1322. [Google Scholar] [CrossRef]
- Zurbriggen, M.D.; Tognetti, V.B.; Fillat, M.F.; Hajirezaei, M.R.; Valle, E.M.; Carrillo, N. Combating stress with flavodoxin: A promising route for crop improvement. Trends Biotechnol. 2008, 26, 531–537. [Google Scholar] [CrossRef] [PubMed]
- Ball, S.G.; Dirick, L.; Decq, A.; Martiat, J.C.; Matagne, R.F. Physiology of starch storage in the monocellular alga Chlamydomonas reinhardtii. Plant Sci. 1990, 66, 1–9. [Google Scholar] [CrossRef]
- Jing, S.; Björn, P.; Michael, M. Removal of nitrogen and phosphorus from wastewater using microalgae immobilized on twin layers: An experimental study. J. Appl. Phycol. 2007, 19, 417–423. [Google Scholar]
- Chinnasamy, S.; Bhatnagar, A.; Hunt, R.W.; Das, K.C. Microalgae cultivation in a wastewater dominated by carpet mill effluents for biofuel applications. Bioresour. Technol. 2010, 101, 3097–3105. [Google Scholar] [CrossRef] [PubMed]
- Kumar, D.; Yusuf, M.A.; Singh, P.; Sardar, M.; Sarin, N.B. Modulation of antioxidant machinery in α-tocopherol-enriched transgenic brassica juncea plants tolerant to abiotic stress conditions. Protoplasma 2013, 250, 1079–1089. [Google Scholar] [CrossRef] [PubMed]
- Choe, Y.H.; Kim, Y.S.; Kim, I.S.; Bae, M.J.; Lee, E.J.; Kim, Y.H.; Park, H.M.; Yoon, H.S. Homologous expression of γ-glutamylcysteine synthetase increases grain yield and tolerance of transgenic rice plants to environmental stresses. J. Plant Physiol. 2013, 170, 610–618. [Google Scholar] [CrossRef] [PubMed]
- Kavitha, K.; George, S.; Venkataraman, G.; Parida, A. A salt-inducible chloroplastic monodehydroascorbate reductase from halophyte avicennia marina confers salt stress tolerance on transgenic plants. Biochimie 2010, 92, 1321–1329. [Google Scholar] [CrossRef] [PubMed]
- Mano, J.; Ohno, C.; Domae, Y.; Asada, K. Chloroplastic ascorbate peroxidase is the primary target of methylviologen-induced photooxidative stress in spinach leaves: Its relevance to monodehydroascorbate radical detected with in vivo ESR. Biochim. Biophys. Acta. 2001, 1504, 275–287. [Google Scholar] [CrossRef]
- Giacomelli, L.; Masi, A.; Ripoll, D.R.; Lee, M.J.; van Wijk, K.J. Arabidopsis thaliana deficient in two chloroplast ascorbate peroxidases shows accelerated light-induced necrosis when levels of cellular ascorbate are low. Plant Mol. Biol. 2007, 65, 627–644. [Google Scholar] [CrossRef] [PubMed]
- Peden, E.A.; Boehm, M.; Mulder, D.W.; Davis, R.; Old, W.M.; King, P.W.; Ghirardi, M.L.; Dubini, A. Identification of global ferredoxin interaction networks in Chlamydomonas reinhardtii. J. Biol. Chem. 2013, 288, 35192–35209. [Google Scholar] [CrossRef] [PubMed]
- Allahverdiyeva, Y.; Ermakova, M.; Eisenhut, M.; Zhang, P.; Richaud, P.; Hagemann, M.; Cournac, L.; Aro, E.M. Interplay between flavodiiron proteins and photorespiration in Synechocystis sp. PCC 6803. J. Biol. Chem. 2011, 286, 24007–24014. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Nigam, P.S.; Murphy, J.D. Renewable fuels from algae: An answer to debatable land based fuels. Bioresour. Technol. 2011, 102, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Mussgnug, J.H.; Thomas-Hall, S.; Rupprecht, J.; Foo, A.; Klassen, V.; McDowall, A.; Schenk, P.M.; Kruse, O.; Hankamer, B. Engineering photosynthetic light capture: Impacts on improved solar energy to biomass conversion. Plant Biotechnol. J. 2007, 5, 802–814. [Google Scholar] [CrossRef] [PubMed]
- Aliverti, A.; Pandini, V.; Pennati, A.; de Rosa, M.; Zanetti, G. Structural and functional diversity of ferredoxin-NADP+ reductases. Arch. Biochem. Biophys. 2008, 474, 283–291. [Google Scholar] [CrossRef] [PubMed]
- Hendriks, J.H.; Kolbe, A.; Gibon, Y.; Stitt, M.; Geigenberger, P. ADP-glucose pyrophosphorylase is activated by posttranslational redox-modification in response to light and to sugars in leaves of arabidopsis and other plant species. Plant Physiol. 2003, 133, 838–849. [Google Scholar] [CrossRef] [PubMed]
- Geigenberger, P. Regulation of starch biosynthesis in response to a fluctuating environment. Plant Physiol. 2011, 155, 1566–1577. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, H.; Heinz, E. Involvement of ferredoxin in desaturation of lipid-bound oleate in chloroplasts. Plant Physiol. 1990, 94, 214–220. [Google Scholar] [CrossRef] [PubMed]
- Wada, H.; Schmidt, H.; Heinz, E.; Murata, N. In vitro ferredoxin-dependent desaturation of fatty acids in cyanobacterial thylakoid membranes. J. Bacteriol. 1993, 175, 544–547. [Google Scholar] [PubMed]
- Park, D.H.; Zeikus, J.G. Improved fuel cell and electrode designs for producing electricity from microbial degradation. Biotechnol. Bioeng. 2003, 81, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Schroder, U.; Niessen, J.; Scholz, F. A generation of microbial fuel cells with current outputs boosted by more than one order of magnitude. Angew. Chem. 2003, 42, 2880–2883. [Google Scholar] [CrossRef] [PubMed]
- Feng, C.; Ma, L.; Li, F.; Mai, H.; Lang, X.; Fan, S. A polypyrrole/anthraquinone-2,6-disulphonic disodium salt (PPy/AQDS)-modified anode to improve performance of microbial fuel cells. Biosens. Bioelectron. 2010, 25, 1516–1520. [Google Scholar] [CrossRef] [PubMed]
- Niessen, J.; Schroder, U.; Harnisch, F.; Scholz, F. Gaining electricity from in situ oxidation of hydrogen produced by fermentative cellulose degradation. Lett. Appl. Microbiol. 2005, 41, 286–290. [Google Scholar] [CrossRef] [PubMed]
- Rosenbaum, M.; Schroder, U.; Scholz, F. Utilizing the green alga Chlamydomonas reinhardtii for microbial electricity generation: A living solar cell. Appl. Microbiol. Biotechnol. 2005, 68, 753–756. [Google Scholar] [CrossRef] [PubMed]
- Pham, C.A.; Jung, S.J.; Phung, N.T.; Lee, J.; Chang, I.S.; Kim, B.H.; Yi, H.; Chun, J. A novel electrochemically active and Fe(III)-Reducing bacterium phylogenetically related to aeromonas hydrophila, isolated from a microbial fuel cell. FEMS Microbiol. Lett. 2003, 223, 129–134. [Google Scholar] [CrossRef]
- Gorman, D.S.; Levine, R.P. Cytochrome f and plastocyanin: Their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardii. Proc. Natl. Acad. Sci. USA 1965, 54, 1665–1669. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.M.; Zeeman, S.C. Quantification of starch in plant tissues. Nat. Protoc. 2006, 1, 1342–1345. [Google Scholar] [CrossRef] [PubMed]
- Wawrik, B.; Harriman, B.H. Rapid, colorimetric quantification of lipid from algal cultures. J. Microbiol. Methods 2010, 80, 262–266. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, L.-F.; Lin, J.-Y.; Pan, K.-Y.; Huang, C.-K.; Chu, Y.-K. Overexpressing Ferredoxins in Chlamydomonas reinhardtii Increase Starch and Oil Yields and Enhance Electric Power Production in a Photo Microbial Fuel Cell. Int. J. Mol. Sci. 2015, 16, 19308-19325. https://doi.org/10.3390/ijms160819308
Huang L-F, Lin J-Y, Pan K-Y, Huang C-K, Chu Y-K. Overexpressing Ferredoxins in Chlamydomonas reinhardtii Increase Starch and Oil Yields and Enhance Electric Power Production in a Photo Microbial Fuel Cell. International Journal of Molecular Sciences. 2015; 16(8):19308-19325. https://doi.org/10.3390/ijms160819308
Chicago/Turabian StyleHuang, Li-Fen, Ji-Yu Lin, Kui-You Pan, Chun-Kai Huang, and Ying-Kai Chu. 2015. "Overexpressing Ferredoxins in Chlamydomonas reinhardtii Increase Starch and Oil Yields and Enhance Electric Power Production in a Photo Microbial Fuel Cell" International Journal of Molecular Sciences 16, no. 8: 19308-19325. https://doi.org/10.3390/ijms160819308





