MicroRNA, Pm-miR-2305, Participates in Nacre Formation by Targeting Pearlin in Pearl Oyster Pinctada martensii
Abstract
:1. Introduction
2. Results
2.1. Precursor Prediction of Pm-miR-2305

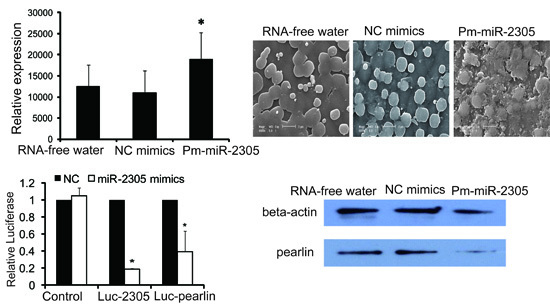
2.2. Expression Analysis of Pm-miR-2305
2.3. Functional Analysis of Pm-miR-2305 in Nacre Formation

2.4. Target Analysis of Pm-miR-2305

2.5. Target Verification between Pm-miR-2305 and Pearlin by Dual Luciferase Assay
2.6. Pearlin Antibody Preparation

2.7. Down-Regulation of Pearlin Protein after the Over-Expression of Pm-miR-2305

3. Discussion
4. Experimental Section
4.1. Experimental Samples and RNA Extraction
4.2. Precursor Prediction of Pm-miR-2305
4.3. Expression Analysis of Pm-miR-2305 and Pearlin
4.4. Over-Expression of Pm-miR-2305 in Vivo
4.5. Construction of Luciferase Reporter Plasmids
4.6. Target Analysis between Pm-miRNAs and Nacre Formation-Related Genes
4.7. Luciferase Assay
4.8. Protein Expression, Purification, and Antiserum Preparation
4.9. Western Blot
4.10. Statistical Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Yu, F.; Jiao, Y.; Zhu, Y.; Wang, Y.; Zhu, J.; Cui, X.; Liu, Y.; He, Y.; Park, E.Y.; Zhang, H.; et al. Microrna 34c gene down-regulation via DNA methylation promotes self-renewal and epithelial-mesenchymal transition in breast tumor-initiating cells. J. Biol. Chem. 2012, 287, 465–473. [Google Scholar] [CrossRef] [PubMed]
- Arfat, Y.; Xiao, W.Z.; Ahmad, M.; Zhao, F.; Li, D.J.; Sun, Y.L.; Hu, L.; Zhihao, C.; Zhang, G.; Shang, P.; et al. Role of micrornas in osteoblasts differentiation and bone disorders. Curr. Med. Chem. 2014, 22, 748–758. [Google Scholar] [CrossRef]
- Zhao, X.; Xu, D.; Li, Y.; Zhang, J.; Liu, T.; Ji, Y.; Wang, J.; Zhou, G.; Xie, X. Micrornas regulate bone metabolism. J. Bone Miner. Metab. 2014, 32, 221–231. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, M.; Saruwatari, K.; Kogure, T.; Yamamoto, Y.; Nishimura, T.; Kato, T.; Nagasawa, H. An acidic matrix protein, pif, is a key macromolecule for nacre formation. Science 2009, 325, 1388–1390. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.D.; Zhao, M.; Liu, W.G.; Guan, Y.Y.; Shi, Y.; Wang, Q.; Wu, S.Z.; He, M.X. Gigabase-scale transcriptome analysis on four species of pearl oysters. Mar. Biotechnol. 2013, 15, 253–264. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Yu, C.; Gu, Z.; Zhan, X.; Wang, Y.; Wang, A. Characterization of the pearl oyster (Pinctada martensii) mantle transcriptome unravels biomineralization genes. Mar. Biotechnol. 2013, 15, 175–187. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Kinoshita, S.; Nomura, N.; Riho, C.; Maeyama, K.; Nagai, K.; Watabe, S. The mining of pearl formation genes in pearl oyster Pinctada fucata by cDNA suppression subtractive hybridization. Mar. Biotechnol. 2011, 14, 177–188. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Zheng, Z.; Du, X.; Wang, Q.; Huang, R.; Deng, Y.; Shi, S.; Zhao, X. Identification and characterization of micrornas in pearl oyster Pinctada martensii by solexa deep sequencing. Mar. Biotechnol. 2014, 16, 54–62. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Wang, Q.; Jiao, Y.; Huang, R.; Deng, Y.; Wang, H.; Du, X. Identification of genes potentially related to biomineralization and immunity by transcriptome analysis of pearl sac in pearl oyster Pinctada martensii. Mar. Biotechnol. 2012, 14, 730–739. [Google Scholar] [CrossRef] [PubMed]
- Berezikov, E.; Guryev, V.; van de Belt, J.; Wienholds, E.; Plasterk, R.H.; Cuppen, E. Phylogenetic shadowing and computational identification of human microrna genes. Cell 2005, 120, 21–24. [Google Scholar] [CrossRef] [PubMed]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microrna targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Chen, L.; Zhao, M.; Tian, Q.; Zhang, W.H. Identification of drought-responsive micrornas in medicago truncatula by genome-wide high-throughput sequencing. BMC Genom. 2011, 12, 367. [Google Scholar] [CrossRef] [PubMed]
- Glazov, E.A.; Kongsuwan, K.; Assavalapsakul, W.; Horwood, P.F.; Mitter, N.; Mahony, T.J. Repertoire of bovine miRNA and miRNA-like small regulatory rnas expressed upon viral infection. PLoS ONE 2009, 4, e6349. [Google Scholar] [CrossRef] [PubMed]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef] [PubMed]
- Malumbres, M. MiRNAs versus oncogenes: The power of social networking. Mol. Syst. Biol. 2012, 8, 569. [Google Scholar] [CrossRef] [PubMed]
- Ren, Z.; Ambros, V.R. Caenorhabditis elegans micrornas of the let-7 family act in innate immune response circuits and confer robust developmental timing against pathogen stress. Proc. Natl. Acad. Sci. USA 2015, 112, E2366–E2375. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Tang, Y.; Cui, H.; Zhao, X.; Luo, X.; Pan, W.; Huang, X.; Shen, N. Let-7/miR-98 regulate Fas and Fas-mediated apoptosis. Genes Immun. 2011, 12, 149–154. [Google Scholar] [CrossRef] [PubMed]
- Boyerinas, B.; Park, S.M.; Hau, A.; Murmann, A.E.; Peter, M.E. The role of let-7 in cell differentiation and cancer. Endocr. Relat. Cancer 2010, 17, F19–F36. [Google Scholar] [CrossRef] [PubMed]
- Wan, Y.; Sun, G.; Wang, Z.; Guo, J.; Shi, L. MiR-125b promotes cell proliferation by directly targeting lin28 in glioblastoma stem cells with low expression levels of miR-125b. Neuroreport 2014, 25, 289–296. [Google Scholar] [CrossRef] [PubMed]
- Xia, H.F.; He, T.Z.; Liu, C.M.; Cui, Y.; Song, P.P.; Jin, X.H.; Ma, X. MiR-125b expression affects the proliferation and apoptosis of human glioma cells by targeting Bmf. Cell. Physiol. Biochem. 2009, 23, 347–358. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.H.; Zhang, Y.N.; Li, H.B.; Hu, C.Y.; Wang, N.; Cao, P.P.; Liao, B.; Lu, X.; Cui, Y.H.; Liu, Z. Over-expression of miR-125b, a novel regulator of innate immunity, in eosinophilic chronic rhinosinusitis with nasal polyps. Am. J. Respir. Crit. Care Med. 2012, 185, 140–151. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Deng, M.; He, H.; Zeng, D.; Zhang, W. MiR-125b regulates osteogenic differentiation of human bone marrow mesenchymal stem cells by targeting Smad4. J. Cent. South Univ. Med. Sci. 2013, 38, 341–346. [Google Scholar]
- Marin, F.; le Roy, N.; Marie, B. The formation and mineralization of mollusk shell. Front. Biosci. 2012, 4, 1099–1125. [Google Scholar] [CrossRef]
- Perovic, I.; Chang, E.P.; Lui, M.; Rao, A.; Colfen, H.; Evans, J.S. A nacre protein, n16.3, self-assembles to form protein oligomers that dimensionally limit and organize mineral deposits. Biochemistry 2014, 53, 2739–2748. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Liu, C.; Chen, L.; Sun, J.; Zhou, Y.; Li, Q.; Zheng, G.; Zhang, G.; Wang, H.; Xie, L.; et al. A new method to extract matrix proteins directly from the secretion of the mollusk mantle and the role of these proteins in shell biomineralization. Mar. Biotechnol. 2011, 13, 981–991. [Google Scholar] [CrossRef] [PubMed]
- Marie, B.; Joubert, C.; Tayale, A.; Zanella-Cleon, I.; Belliard, C.; Piquemal, D.; Cochennec-Laureau, N.; Marin, F.; Gueguen, Y.; Montagnani, C. Different secretory repertoires control the biomineralization processes of prism and nacre deposition of the pearl oyster shell. Proc. Natl. Acad. Sci. USA 2012, 109, 20986–20991. [Google Scholar] [CrossRef] [PubMed]
- Yan, F.; Jiao, Y.; Deng, Y.; Du, X.; Huang, R.; Wang, Q.; Chen, W. Tissue inhibitor of metalloproteinase gene from pearl oyster Pinctada martensii participates in nacre formation. Biochem. Biophys. Res. Commun. 2014, 450, 300–305. [Google Scholar] [CrossRef] [PubMed]
- Samata, T.; Hayashi, N.; Kono, M.; Hasegawa, K.; Horita, C.; Akera, S. A new matrix protein family related to the nacreous layer formation of Pinctada fucata. FEBS Lett. 1999, 462, 225–229. [Google Scholar] [CrossRef]
- Miyashita, T.; Takagi, R.; Okushima, M.; Nakano, S.; Miyamoto, H.; Nishikawa, E.; Matsushiro, A. Complementary DNA cloning and characterization of pearlin, a new class of matrix protein in the nacreous layer of oyster pearls. Mar. Biotechnol. 2000, 2, 409–418. [Google Scholar] [PubMed]
- Kapinas, K.; Delany, A.M. MicroRNA biogenesis and regulation of bone remodeling. Arthritis Res. Ther. 2011, 13, 220. [Google Scholar] [CrossRef] [PubMed]
- Eulalio, A.; Behm-Ansmant, I.; Izaurralde, E. P bodies: At the crossroads of post-transcriptional pathways. Nat. Rev. Mol. Cell Biol. 2007, 8, 9–22. [Google Scholar] [CrossRef] [PubMed]
- Giraldez, A.J.; Mishima, Y.; Rihel, J.; Grocock, R.J.; Van Dongen, S.; Inoue, K.; Enright, A.J.; Schier, A.F. Zebrafish miR-430 promotes deadenylation and clearance of maternal mrnas. Science 2006, 312, 75–79. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.L.; Huang, S.Q.; Guo, K.; Xiang, A.L.; Zhu, Y.Y.; Nie, L.; Yang, Z.M. Computational identification of novel micrornas and targets in Brassica napus. FEBS Lett. 2007, 581, 1464–1474. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Ridzon, D.A.; Broomer, A.J.; Zhou, Z.; Lee, D.H.; Nguyen, J.T.; Barbisin, M.; Xu, N.L.; Mahuvakar, V.R.; Andersen, M.R.; et al. Real-time quantification of micrornas by stem-loop RT-PCR. Nucleic Acids Res. 2005, 33, e179. [Google Scholar] [CrossRef] [PubMed]
- Kruger, J.; Rehmsmeier, M. Rnahybrid: MicroRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006, 34, W451–W454. [Google Scholar] [CrossRef] [PubMed]
- Cai, S.H.; Huang, Y.C.; Lu, Y.S.; Wu, Z.H.; Wang, B.; Tang, J.F.; Jian, J.C. Expression and immunogenicity analysis of accessory colonization factor a from vibrio alginolyticus strain hy9901. Fish Shellfish Immunol. 2013, 34, 454–462. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Yao, H.; Zhu, P.; Zhang, X.; Pan, Q.; Gong, C.; Huang, Y.; Hu, X.; Su, F.; Lieberman, J.; et al. Let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell 2007, 131, 1109–1123. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiao, Y.; Zheng, Z.; Tian, R.; Du, X.; Wang, Q.; Huang, R. MicroRNA, Pm-miR-2305, Participates in Nacre Formation by Targeting Pearlin in Pearl Oyster Pinctada martensii. Int. J. Mol. Sci. 2015, 16, 21442-21453. https://doi.org/10.3390/ijms160921442
Jiao Y, Zheng Z, Tian R, Du X, Wang Q, Huang R. MicroRNA, Pm-miR-2305, Participates in Nacre Formation by Targeting Pearlin in Pearl Oyster Pinctada martensii. International Journal of Molecular Sciences. 2015; 16(9):21442-21453. https://doi.org/10.3390/ijms160921442
Chicago/Turabian StyleJiao, Yu, Zhe Zheng, Rongrong Tian, Xiaodong Du, Qingheng Wang, and Ronglian Huang. 2015. "MicroRNA, Pm-miR-2305, Participates in Nacre Formation by Targeting Pearlin in Pearl Oyster Pinctada martensii" International Journal of Molecular Sciences 16, no. 9: 21442-21453. https://doi.org/10.3390/ijms160921442
APA StyleJiao, Y., Zheng, Z., Tian, R., Du, X., Wang, Q., & Huang, R. (2015). MicroRNA, Pm-miR-2305, Participates in Nacre Formation by Targeting Pearlin in Pearl Oyster Pinctada martensii. International Journal of Molecular Sciences, 16(9), 21442-21453. https://doi.org/10.3390/ijms160921442