Abstract
Testis-specific lactate dehydrogenase (LDH-C4) is one of the lactate dehydrogenase (LDH) isozymes that catalyze the terminal reaction of pyruvate to lactate in the glycolytic pathway. LDH-C4 in mammals was previously thought to be expressed only in spermatozoa and testis and not in other tissues. Plateau pika (Ochotona curzoniae) belongs to the genus Ochotona of the Ochotonidea family. It is a hypoxia-tolerant species living in remote mountain areas at altitudes of 3000–5000 m above sea level on the Qinghai-Tibet Plateau. Surprisingly, Ldh-c is expressed not only in its testis and sperm, but also in somatic tissues of plateau pika. To shed light on the function of LDH-C4 in somatic cells, Ldh-a, Ldh-b, and Ldh-c of plateau pika were subcloned into bacterial expression vectors. The pure enzymes of Lactate Dehydrogenase A4 (LDH-A4), Lactate Dehydrogenase B4 (LDH-B4), and LDH-C4 were prepared by a series of expression and purification processes, and the three enzymes were identified by the method of sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and native polyacrylamide gel electrophoresis (PAGE). The enzymatic kinetics properties of these enzymes were studied by Lineweaver-Burk double-reciprocal plots. The results showed the Michaelis constant (Km) of LDH-C4 for pyruvate and lactate was 0.052 and 4.934 mmol/L, respectively, with an approximate 90 times higher affinity of LDH-C4 for pyruvate than for lactate. At relatively high concentrations of lactate, the inhibition constant (Ki) of the LDH isoenzymes varied: LDH-A4 (Ki = 26.900 mmol/L), LDH-B4 (Ki = 23.800 mmol/L), and LDH-C4 (Ki = 65.500 mmol/L). These data suggest that inhibition of lactate by LDH-A4 and LDH-B4 were stronger than LDH-C4. In light of the enzymatic kinetics properties, we suggest that the plateau pika can reduce reliance on oxygen supply and enhance its adaptation to the hypoxic environments due to increased anaerobic glycolysis by LDH-C4.
1. Introduction
The Qinghai-Tibet Plateau, with an average elevation of over 3000 m above sea level, is the highest and also largest plateau on earth and is characterized by extremely harsh environmental conditions such as hypoxia, strong ultraviolet radiation, and cold temperature which expectedly have profound effects on human and animal survival. Over long-term adaption and evolution, both plateau-native humans and animals have developed a series of strategies to adapt to the plateau environment. Also, inhabitants have developed some unique features to avoid disadvantageous environmental aspects and effectively obtain material or energy from their survival environment as to ensure their normal growth and breeding.
Plateau pika (Ochotona curzoniae), which is a member of the genus Ochotona of the Ochotonidae family, is a small, non-hibernating rodent that lives in remote mountain areas at an elevation of 3000–5000 m on the Qinghai-Tibet Plateau. The pika plays an important role in biodiversity of the ecosystem on the plateau and is regarded as a key species since ancient times [1,2]. Fossil samples of pika that are nearly 37 million years old were found by archaeologistson the north edge of the Qinghai-Tibetan plateau [3]. During long-term evolution, the pikas evolved a series of physiological adaptations that allow them to thrive in the harsh environment and become a highly advanced hypoxia-tolerant mammal. Specifically, the pika obtained oxygen effectively from the hypoxic environment by larger superficial pulmonary alveoli and higher capillary density [4], thin walled pulmonary arterioles and blunted hypoxic pulmonary vasoconstriction (HPV) [5], an increase in erythrocyte count [6], reduction in the mean corpuscular volume [7], changes in hemoglobin (Hb) [8], and 2,3-diphosphoglycerate concentrations [5], and an increase in the oxygen affinity to Hb [8]; Secondly, a pika has a strong cardiac pumping function due to its larger heart and smaller weights of right-to-left ventricular plus septum [9]; Thirdly, a pika has a high oxygen utilization ratio by increasing the densities of capillary and mitochondrial [10], and myoglobin concentration in tissues [6,9]. In addition to these physiological mechanisms, a pika reduces dependence on oxygen by increasing anaerobic glycolysis in its skeletal muscle [11] and gluconeogenesis in liver [12]. The molecular mechanisms of these adaptations in pika have occurred due to a series of changes such as genetic evolution, the expression of tissue-specific proteins, and changes related to high altitude, including many functional cytokines as vascular endothelial growth factor (VEGF) [13,14], hemoglobin [3], HIF-1α [15,16], LDH-C4 [17], pyruvate carboxylase [12], myoglobin [9], inducible nitric oxide synthase (iNOS) [18], leptin [19,20] and cytochromec oxidase [21].
It is well known that LDH family enzymes catalyze the inter-conversion of pyruvate and lactate with the concomitant oxidation/reduction of the reduced form of nicotinamide adenine dinucleotide hydrogen (NADH) to nicotinamide adenine dinucleotide (NAD+) in the reactions [22]. Different forms of Lactate dehydrogenase (LDH) isozymes are comprised of expression products of three typical genes: Ldh-a, Ldh-b, and Ldh-c, which encode LDHA, LDHB and LDHC subunits, respectively [23,24]. LDH consists of A and B subunits that assemble into homo- or hetero- tetramers. These A and B subunits are distributed in various combinations in the body reflecting various energy metabolic requirements of different tissues, and are consistent with the catalytic properties of each isozyme [25,26]. However, the homo-tetramer LDH-C4 was previously detected only in testis and spermatozoa of mammals [27,28,29,30]. Surprisingly, our previous studies identified the expression of Ldh-c not only in testis and sperm, but also in somatic cells of plateau pika [17].
LDH-C4 has high thermostability [17], and its homolog from different species showed activity against longer carbon chain with α-hydroxy and α-keto acids than those of pyruvate and lactate [31,32,33,34,35,36,37,38,39,40,41], indicating that LDH-C4 has unique structural and functional properties [31,40,41,42]. In the present study, we compared the enzymatic kinetic characteristics among LDH-A4, LDH-B4, and LDH-C4 of plateau pika with the aim of exploring the pika’s adaptation mechanism to the hypoxic environment of the Qinghai-Tibet Plateau.
2. Results
2.1. Plasmid Construction and Recombinant Protein Expression
To study the enzyme kinetics of LDH-A4, LDH-B4, and LDH-C4 of plateau pika, the expression plasmids pCold-SUMO-Ldh-a, pET-30a-Ldh-b, and pET-30a-Ldh-c, respectively encoding the full-length pika LDHA, LDHB, and LDHC were constructed. In order to verify that the vectors contained the sequences of Ldh-a, Ldh-b, and Ldh-c and their correct expression, Polymerase Chain Reaction (PCR) of the plasmids and sodium dodecyl sulfate polyacrylamide gel electropheresis (SDS-PAGE) analysis of the expressed proteins were performed. Figure 1A–C show the agarose gel electrophoresis results of PCR products of Ldh-a, Ldh-b, and Ldh-c, respectively. Lane 1 in Figure 1A–C displayed clear and bright bands at approximately 1000 bp, matching the same number of base pairs of Ldh-a (999 bp), Ldh-b (1005 bp), and Ldh-c (999 bp) as our previous study [17]. By sequencing and alignment, the sequences of Ldh-a, Ldh-b and Ldh-c in the final plasmid constructs were determined to be 999 bp, 1005 bp and 999 bp in length and identical to those in the Genbank (HQ704676, HQ704677 and HQ704678, respectively).
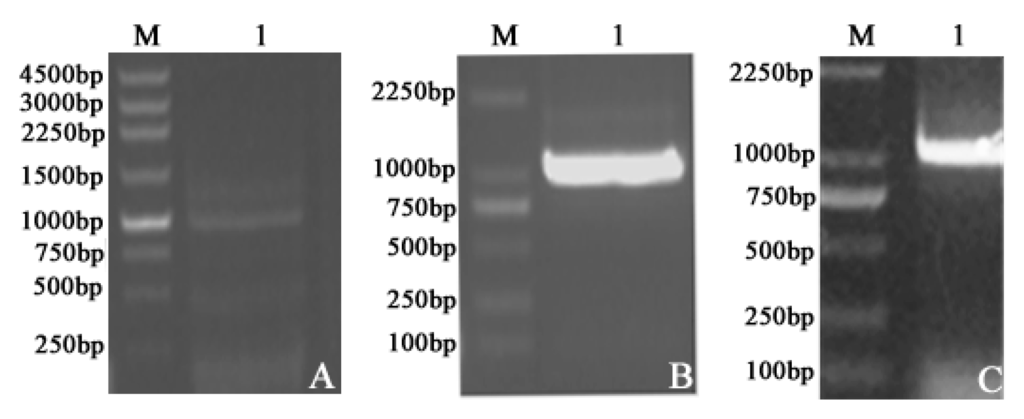
Figure 1.
Amplification of plateau pika Ldh-a, Ldh-b and Ldh-c. Lane 1: PCR products of (A) Ldh-a; (B) Ldh-b; and (C) Ldh-c. M: DNA marker. The PCR fragments of Ldh-a, Ldh-b, and Ldh-c were 999 bp, 1005 bp and 999 bp. The sequence of transcripts was identical to that in the GenBank for Ldh-a, Ldh-b, and Ldh-c, suggesting that the expression plasmids pCold-SUMO-Ldh-a, pET-30a-Ldh-b, and pET-30a-Ldh-c encoding the full-length pika LDHA, LDHB and LDHC, respectively, were successfully constructed.
The SDS-PAGE results of cell-free extracts (shown in Figure 2A–C) displayed the protein in the supernatant and precipitate fractions with dark bands around 40–50 kD. The recombinant LDH isoenzymes were expressed with His-tag (LDHA with His-tag and SUMO-tag) in the C-terminus, which was favorable to purification. These results demonstrated similar molecular weights to those of LDHA, LDHB, and LDHC, as we previously reported [17], indicating that expression plasmids pCold-SUMO-Ldh-a, pET-30a-Ldh-b and pET-30a-Ldh-c were successfully prepared, and expressed the proteins of interest correctly.
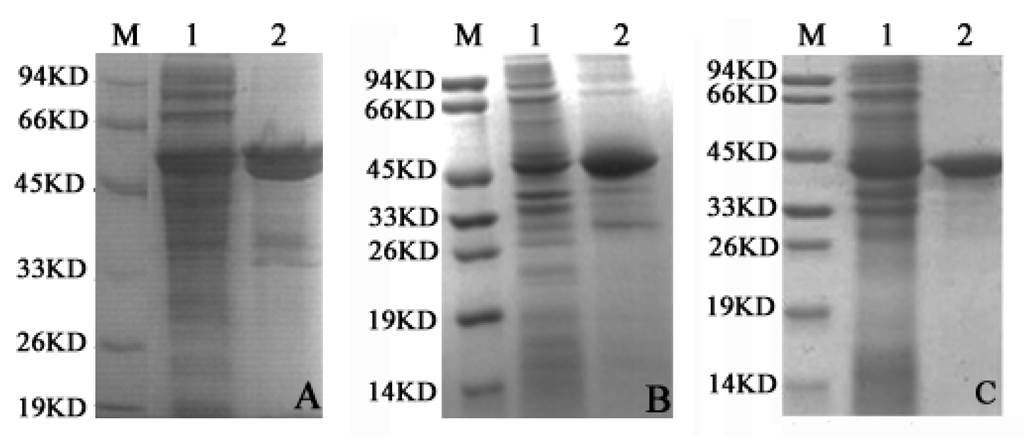
Figure 2.
Analysis of the cell-free extracts recombinant protein by SDS-PAGE. (A) LDHA; (B) LDHB; and (C) LDHC. For (A–C) Lane 1 and Lane 2 represent the total protein of supernatant and precipitation, respectively, in the E. coli BL21 (DE3) cells; M: molecular weight standards. SDS-PAGE analysis indicates LDHA, LDHB and LDHC expression in supernatant and precipitate fractions.
2.2. LDH-A4, LDH-B4 and LDH-C4 Purification and Activity Measurement
The native polyacrylamide gel electropheresis (native PAGE) results shown in Figure 3P1 indicate that LDH-A4 was observed in 100, 250 and 500 mmol/L imidazole buffer elutions with LDH activity, and LDH-B4 and LDH-C4 were observed in 250 and 500 mmol/L imidazole buffers elutions with LDH activity. LDH-A4, LDH-B4 and LDH-C4 bound so completely to Ni-NTA that no active protein was assayed in the remainder of the purified supernatant. As shown in Figure 3P2, the purified LDH-A4, LDH-B4, and LDH-C4, were represented with only one band as observed by SDS-PAGE, and the molecular weights were consistent with LDHA, LDHB, and LDHC, respectively. Figure 3P3 presents the LDH activity of the purified LDH-A4, LDH-B4, and LDH-C4 as determined by native PAGE. These results indicate that LDH-A4, LDH-B4, and LDH-C4 of plateau pika were successfully purified. As shown in Figure 4, the respective enzyme activity (n = 5) of LDH-A4, LDH-B4, and LDH-C4 was 2241 ± 9.3 U/g protein, 3918 ± 37.9 U/g protein, and 10,741 ± 80.9 U/g protein, and that of LDH-C4 was significantly higher than that of LDH-A4 and LDH-B4 (p < 0.01).
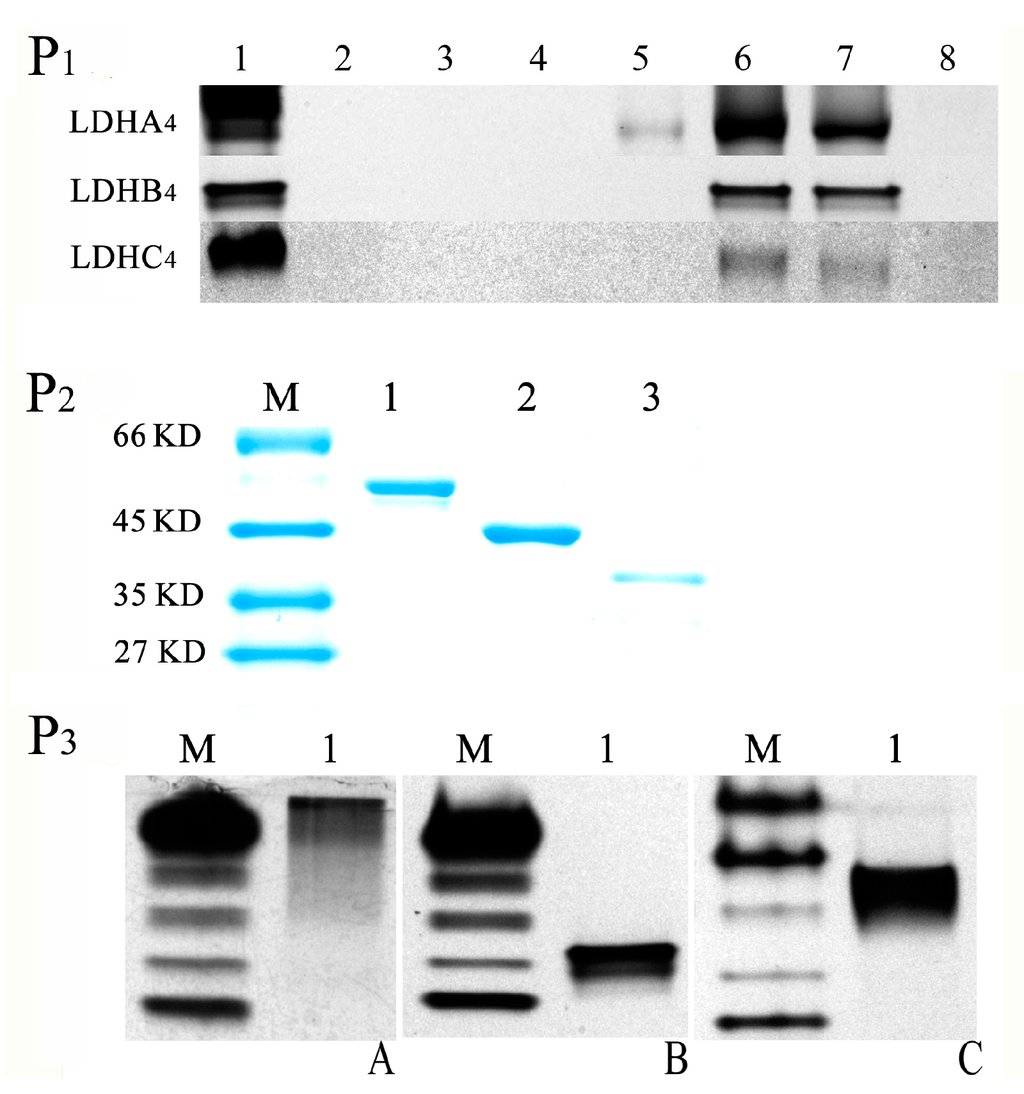
Figure 3.
Purification and identification of LDH-A4, LDH-B4, and LDH-C4. (P1) Native PAGE of recombinant protein washed by imidazole at different concentrations. Lane 1: the supernatant before purification, Lane 2–7: collected elutions from Ni-NTA resins eluted with 10, 20, 50, 100, 250 and 500 mmol/L imidazole buffers, respectively. Lane 8: the remainder in the purified supernatant; (P2) SDS-PAGE analysis of the purified LDH-A4, LDH-B4, and LDH-C4. Lane 1–3 represented theLDH-A4, LDH-B4, and LDH-C4, respectively; M: marker; (P3) Identification of purified LDH-A4, LDH-B4, and LDH-C4. Lane 1 represents (A) LDH-A4; (B) LDH-B4, and (C) LDH-C4 by Native polyacrylamide gel electrophoresis (PAGE), respectively. M: marker was prepared by mixed kidney and skeletal muscle tissues of plateau pika.
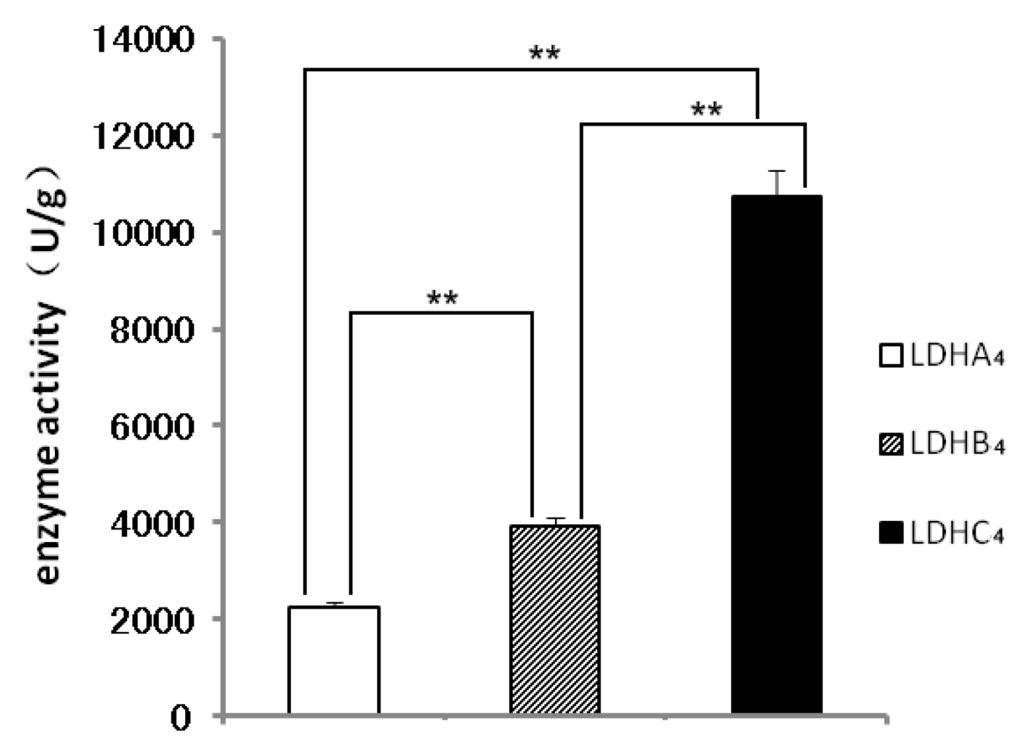
Figure 4.
The enzyme activities of LDH-A4, LDH-B4 and LDH-C4. The enzyme activity of LDH-A4, LDH-B4, and LDH-C4 was 2241 ± 9.3 U/g protein, 3918 ± 37.9 U/g protein, 10741 ± 80.9 U/g protein, each sample size was 5, ** p < 0.01. The enzyme activity of LDH-C4 was significantly higher that of LDH-A4 and LDH-B4 (p < 0.01).
2.3. Enzyme Kinetics Properties of LDH-A4, LDH-B4, and LDH-C4
The Km values for LDH-A4, LDH-B4 and LDH-C4 were determined from Lineweaver-Burk double-reciprocal plots. The activities of the LDH isoenzymes in forward reaction (pyruvate + NADH + H+ → lactate + NAD+) increased with increasing concentration of the substrate pyruvate, as shown in Figure 5. The kinetic parameters of the forward reaction for LDH-A4, LDH-B4, and LDH-C4, calculated from the Lineweaver-Burk double-reciprocal plots of reciprocal pyruvate concentration vs. reciprocal velocity, had Km values of 0.260, 0.172, and 0.052 mmol/L, respectively. The activities of LDH isoenzymes in reverse reaction (lactate + NAD+ → pyruvate + NADH + H+) also increased with increasing concentration of the substrate lactate, as shown in Figure 6. The kinetic parameters of the reverse reaction for LDH-A4, LDH-B4, and LDH-C4, calculated from the Lineweaver-Burk double-reciprocal plots of reciprocal lactate concentration vs. reciprocal velocity, had Km values of 19.968, 8.980 and 4.934 mmol/L, respectively.
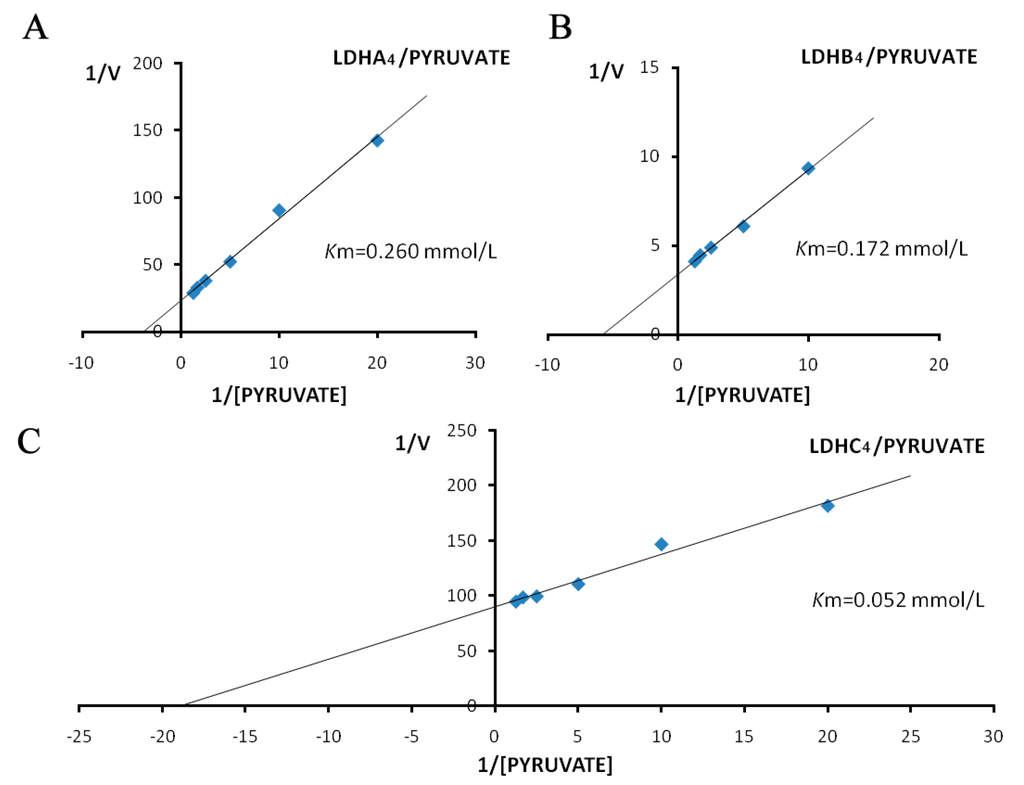
Figure 5.
Double reciprocal plots by pyruvate as substrate of plateau pika LDH isozymes. Double reciprocal plots of (A) LDH-A4; (B) LDH-B4; and (C) LDH-C4, respectively. Reciprocal velocity (V) was calculated with the reciprocal Δ340 nm/min, plots were preformed by calculating the reciprocal reaction velocity vs. reciprocal pyruvate concentration at a constant concentration of NADH. The concentrations of pyruvate used were 0.05, 0.1, 0.2, 0.4, 0.6 and 0.8 mmol/L, respectively. NADH concentration was maintained at 0.15 mmol/L. The resulting Km of LDH-A4, LDH-B4, and LDH-C4 for pyruvate were 0.260, 0.172 and 0.052 mmol/L, respectively.
The double reciprocal plots of initial velocities at different pyruvate concentrations and the inhibitory effect of lactate at different concentrations on pika LDH isozymes are shown in Figure 7. In all cases, the competitive inhibition was observed. Initial velocities were determined in the presence and absence of lactate of different concentrations. The Ki values calculated from the Lineweaver-Burk double-reciprocal plots of reciprocal pyruvate concentration vs. reciprocal velocity. Reciprocals of initial velocity with substrate concentrations below the above-mentioned concentrations gave linear plots with these substrates. The results showed that the Ki of lactate for LDH-C4 (Ki = 65.500 mmol/L) was almost three times higher than that for LDH-A4 (Ki = 26.900 mmol/L) and LDH-B4 (Ki = 23.800 mmol/L). The inhibitory effects of lactate at different concentrations on the enzyme reaction velocity of LDH isozymes are shown in Figure 8. Under the experimental conditions employed in the present study, lactate at higher concentrations exhibited a strong inhibition of LDH-A4 and LDH-B4 activity, but only slightly inhibited LDH-C4 activity. The results indicated that LDH-C4 was less inhibited by lactate, which was likely beneficial in catalyzing the conversion of pyruvate to lactate even at high concentrations of lactate.
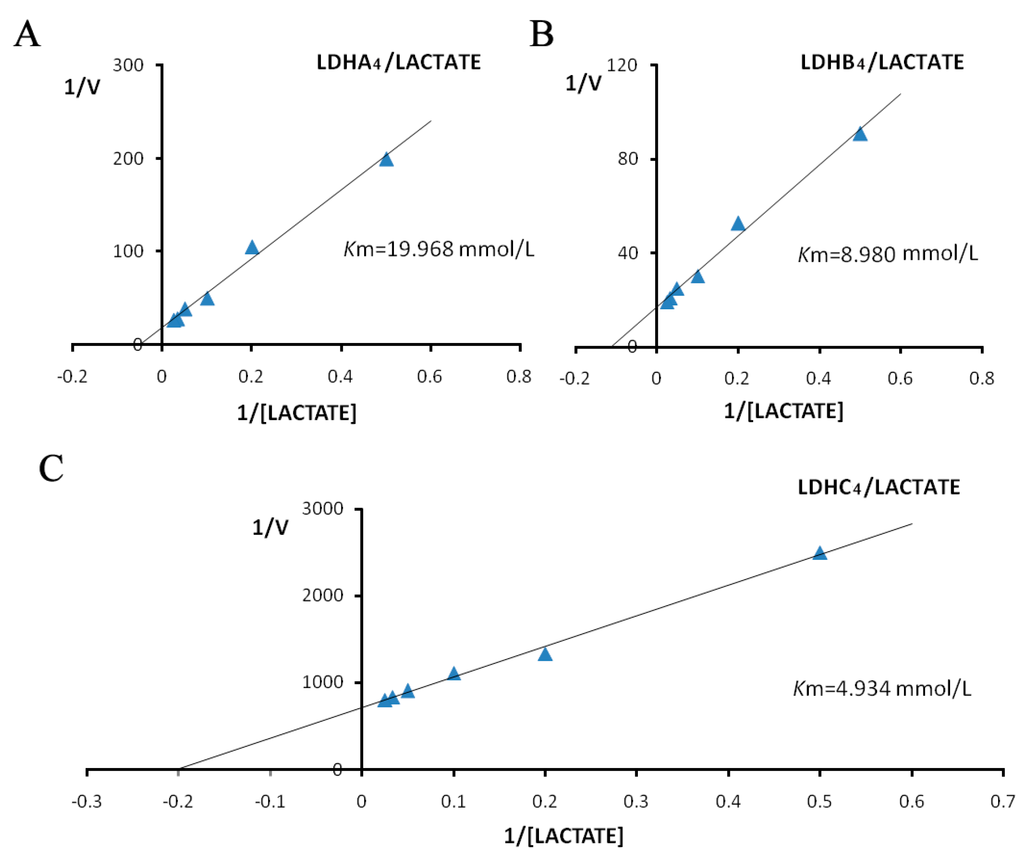
Figure 6.
Double reciprocal plots by lactate as substrate of plateau pika LDH isozymes. Double reciprocal plots of (A) LDH-A4; (B) LDH-B4; and (C) LDH-C4, respectively. Reciprocal velocity (V) was calculated with the reciprocal Δ340 nm/min, plots were preformed by calculating the reciprocal reaction velocity vs. reciprocal pyruvate concentration at a constant concentration of NAD+. The concentrations of lactate used were 2, 5, 10, 20, 30 and 40 mmol/L. NAD+ concentration was maintained at 0.5 mmol/L. The resulting Km of LDH-A4, LDH-B4, and LDH-C4 for lactate were 19.968, 8.980, 4.934 mmol/L, respectively.
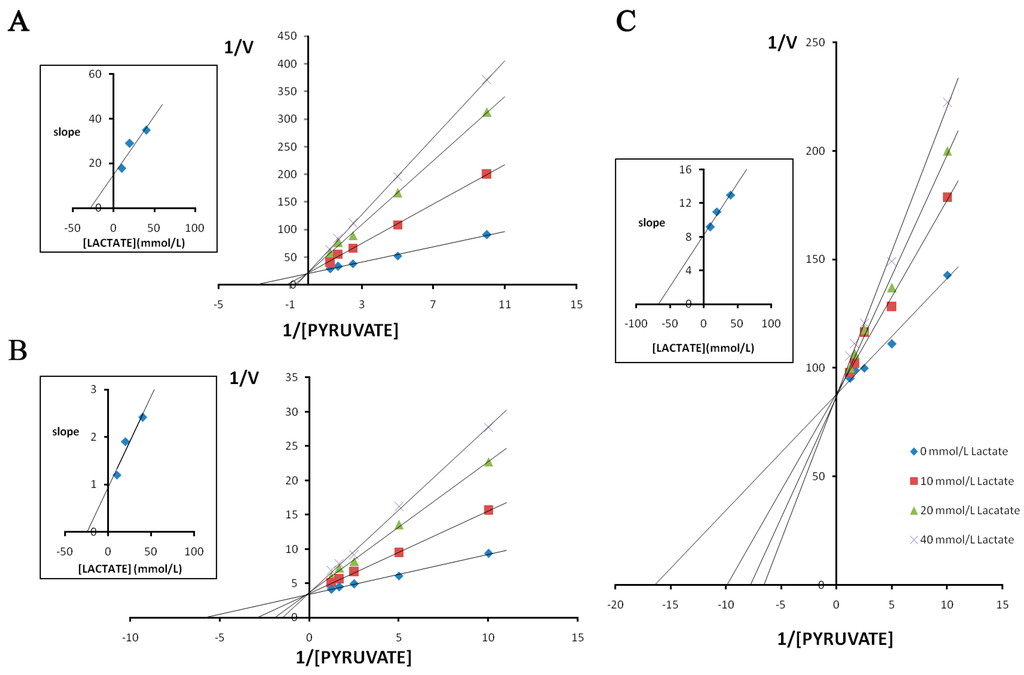
Figure 7.
Effect of pyruvate on the inhibitory activity of lactate on plateau pika LDH isozymes. Double reciprocal plots of (A) LDH-A4; (B) LDH-B4; and (C) LDH-C4, respectively. Reciprocal velocity (V) was calculated with the reciprocal Δ340 nm/min, plots were preformed by calculating the reciprocal value of reaction velocity vs. reciprocal value of pyruvate concentration at a constant concentration of NADH. The concentrations of pyruvate used were 0.1 0.2, 0.4, 0.6 and 0.8 mmol/L. NADH concentration was maintained at 0.15 mmol/L. Upper left: determination of Ki from plots of slope values against lactate concentrations. The resulting Ki for pika LDH-A4, LDH-B4, LDH-C4 by lactate were 25.9, 22.2 and 66.5 mmol/L, respectively.
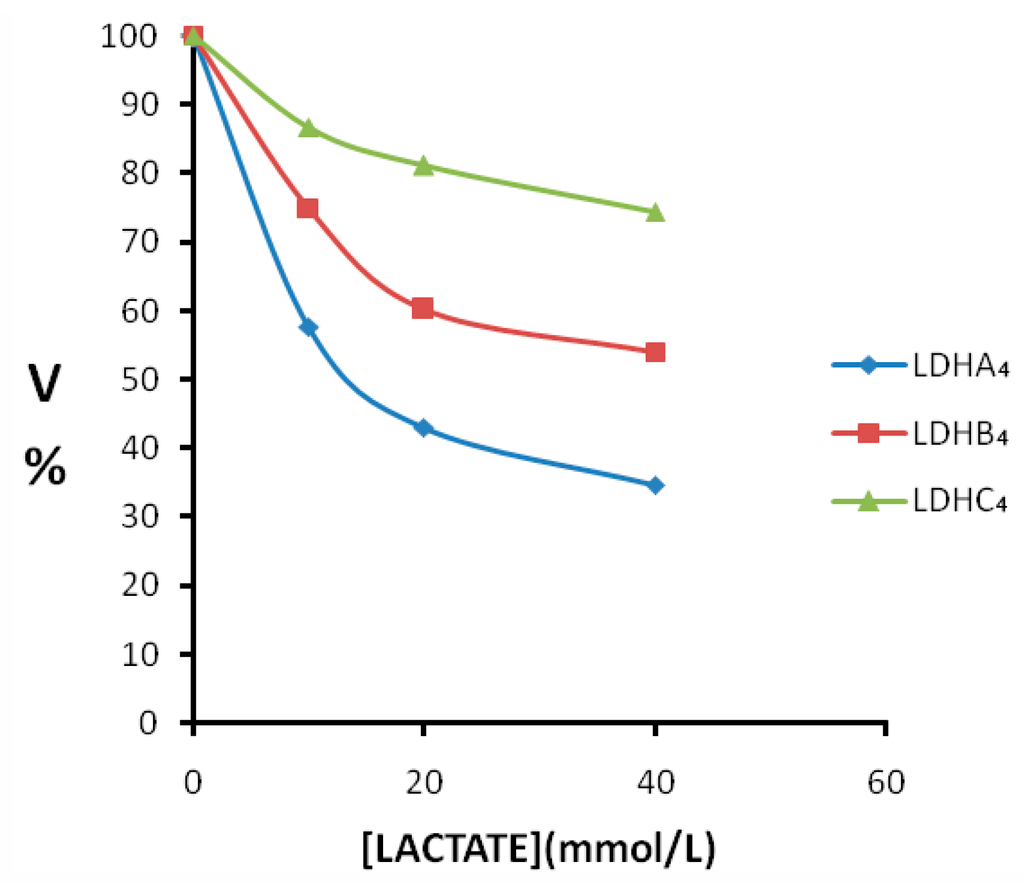
Figure 8.
Effect of increasing concentration of lactate on plateau pika LDH isozymes. Pyruvate was used as substrate and lactate as an inhibitor. Velocities were calculated taking the maximum activity without the inhibitor lactate as 100%. Lactate was added at the concentration of 10, 20 and 40 mmol/L. The enzyme reaction velocity of LDH-C4 was less affected than that of LDH-A4 and LDH-B4 when the lactate concentrations were at 0–40 mmol/L.
3. Discussion
LDH is the terminal enzyme of glycolysis with the role of reducing pyruvate to lactate with limiting oxygen [43]. LDH-C4 catalyzes the terminal reaction in the glycolytic pathway and displays a unique structure and functional properties [43,44].
Ultimately, the biochemical properties that distinguish LDH-C4 from the other LDH isoforms may be attributed to its high glycolytic rate. Studies have shown that since the same chemical reaction is being catalyzed, in spite of different kinetic constants, the equilibrium constant (Keq) is the same for all LDH isoenzymes, as stated in the Haldane equation [45,46]. The complex Haldane kinetics mechanism may be reduced to the Michaelis-Menten mechanism more tractably under special conditions of substrate-enzyme interactions [46]. Also, the basic Michaelis-Menten description can be modified in a logistical manner to fit the reaction parameters with in vitro conditions for the enzyme-catalyzed reactions [47]. In the sense it is even hyperbolic to logistic kinetics that kinetic constants may be considered as being the same, however with the amendment they would respectively correspond to various fractions from maximum velocity of outtake [45,47]. Based on this theory, the LDH isoenzyme pattern in itself may not change the equilibrium of the LDH reaction or influence the tissue lactate concentration [45]. However, the situation could be quite different during transition in energy metabolism where especially the glycolytic flux can undergo great rapid changes. During fast metabolic transitions involving significant pyruvate concentrations changes, primarily resulting from major changes glycolytic rate, the LDH isoenzyme pattern may be of great importance in the compound metabolic response to the altered energy metabolism [45].Therefore, we presumed that ATP was produced rapidly through anaerobic glycolysis by LDH-C4 in plateau pika, which stirs up the metabolic response to the changed energy metabolism, enhancing its adaptation to the hypoxic environment.
Previous studies have investigated in detail the enzymatic kinetics properties of LDH-C4 in other species [31,48,49]. The biochemical properties distinguishing LDH-C4 from the other LDH isoforms may be conducible to the high glycolytic rate. Compared with LDH-A4, pika LDH-C4 has a high Km for lactate (~2.0 mmol/L) and a low Km for pyruvate (~0.030 mmol/L) [31,48,49,50]. This finding suggests that LDH-C4 has a 60-fold higher affinity for pyruvate than that for lactate. The finding also implies that pyruvate conversion to lactate may occur even at high concentrations of extracellular or endogenous lactate [31,48,49,50]. This theory has been supported by an experiment that addition of excess lactate in the environment (50-fold excess in relation to pyruvate) did not affect adenosine triphosphate (ATP) production in capacitating spermatozoa [51]. In the present study, to investigate the enzyme kinetics of LDH-A4, LDH-B4, and LDH-C4 of plateau pika, the expression plasmids pCold-SUMO-Ldh-a, pET-30a-Ldh-b, and pET-30a-Ldh-c, encoding the full-length of pika LDHA, LDHB, and LDHC, were constructed and expressed. SDS-PAGE was used to determine the molecular weight and confirm the recombinant proteins. Figure 2A–C demonstrates that their molecular weights were about 50 kD, 40–45 kD and 40–45 kD for LDHA, LDHB, and LDHC, respectively. According to our previous paper [17], the open reading frames (ORF) of pika Ldh-a, Ldh-b and Ldh-c (accession numbers HQ704676, HQ704677, and HQ704678 in GenBank) were 999 bp, 1005 bp and 999 bp, respectively, encoding 332, 334 and 332 amino acids proteins, with their molecular weights of 36,557.5, 36,464.3 and 36,052.9 Da. Thus the molecular weight of the recombinant LDHA, LDHB, and LDHC in the current study (Figure 2A–C, Figure 3P2) was higher than the value calculated from their amino acid sequences deduced from the encoding cDNA. The addition of the His-tag resulted in a 5–10 kD deviation in the molecular weight of fusion proteins as shown in SDS-PAGE, and also indicated in other studies [52,53,54]. In addition, fusion proteins with the SUMO-tag added 12 kD to its molecular weight [55].
The recombinant expression shuttles (pET-30a-Ldh-b, pET-30a-Ldh-c, pCold-SUMO-Ldh-a) were transformed into E. coli BL21 cells and the enzymatic proteins (LDH-A4, LDH-B4, and LDH-C4) with functional activity were purified from protein fluid extracts. Our results indicate that the enzyme activity of the pika LDH-C4 was significantly higher than that of A4 and B4, that LDH-C4 was less sensitive to lactate inhibition than A4 and B4 (p < 0.01), and the Km values of LDH-C4 for pyruvate and lactate were also higher than that of A4 and B4. Compared with LDH-A4 and LDH-B4, LDH-C4 had a low Km for pyruvate (~0.052 mmol/L) and a high Km for lactate (~4.934 mmol/L); and the affinity of LDH-C4 for pyruvate is 90-fold higher than that for lactate. These properties of pika LDH-C4 were beneficial to catalyze the conversion of pyruvate to lactate even at high concentration of lactate.
Although the conversion mediated by LDH does not generate ATP, the reaction depends on NADH as a cofactor that accompanies oxidation to NAD+; the concentration of NAD+ is a rate-limiting factor in glycolysis and necessary for continued glycolysis [56]. Previous studies demonstrated that knockout of Ldh-c or inhibition of LDH-C4 in sperm led to rapid decline in sperm ATP levels [56,57], a decrease in progressive motility, and a failure to develop hyper-activated motility. It was revealed that all consumed13C-labeled pyruvate added in sperm culture medium was converted to lactate rather than oxidized in the tricarboxylic acid (TCA) cycle in metabolic tracing experiments; in the presence of exogenous pyruvate, the ATP concentration was increased by more than 50% [51]. When carbonyl cyanide m-chlorophenylhydrazone (CCCP) and NaCN was applied to inhibit the oxidative phosphorylation in mitochondria, the amount of ATP was kept at the equivalent level to that without CCCP and the vigorous motility of sperm was maintained [51,58]. These results suggest that LDH-C4 is essential in sperm glycolysis which has an important role in providing abundant ATP for sperm motility [56,58,59], and it is related to LDH-C4 enzymatic kinetics properties.
Plateau pika has a strong adaptability to the hypoxic plateau environment of the Qinghai-Tibet Plateau, given the sperm-specific lactate dehydrogenase (Ldh-c) gene is also expressed in their somatic cells. In light of the enzymatic kinetics properties, we propose that the pika could reduce dependence on oxygen and enhance the adaptation to the hypoxic environments due to increased anaerobic glycolysis by LDH-C4.
4. Materials and Methods
4.1. Reagents and Animal Procedures
All reagents were from Sangon (Sangon, Shanghai, China) unless otherwise noted. Plateau pikas were live-trapped from Haibei Alpine Meadow Ecosystem Research Station at an altitude of 3800 m in Qinghai Province, China. All animals were first anesthetized with sodium pentobarbital (5%), and then sacrificed by cervical dislocation immediately before dissection. Skeletal muscle, kidney, and testis were rapidly removed and frozen in liquid nitrogen for long-term storage. All procedures involved in the handling and care of animals were approved by the China Zoological Society according to the China Practice for the Care and Use of Laboratory Animals (permit number: GB 14923-2010).
4.2. Expression Plasmids Construction
In our previous study [17], Ldh-a, Ldh-b, and Ldh-c of plateau pika testis were cloned and deposited in GenBank with the accession numbers HQ704676, HQ704677, and HQ704678, respectively. Three lengths of BamHI/XhoI fragments (999 bp, 1005 bp, 999 bp representing Ldh-a, Ldh-b, and Ldh-c coding sequences, respectively) were PCR amplified from plateau pika testis cDNA using Premix Ex Taq Version Kit (Takara, Kyoto, Japan). The PCR primers for Ldh-a were 5′-CGGAATTCATGGCAGCTCTCAAGGATCAG-3′ (sense) and 5′-CCGCTCGAGGAACTGCAGCTCCTTCTGGAT-3′ (antisense), for Ldh-b were 5′-CGGAATTCATGGCAACCCTGAAGGAAAAACTCAT (sense) and 5′-CCGCTCGAGCAGGTCCTTCAGGTCCTTCTGGA-3′ (antisense), and for Ldh-c were 5′-CGGGATCCATGTCGACAGTCAAGGAGC-3′ (sense) and 5′-CCGCTCGAGAAACACCAGGTCCTTCTGGAC-3′ (antisense), respectively. PCR conditions were 5 min at 95 °C, 30 cycles of 45 s at 95 °C, 45 s at 65 °C, 1 min at 72 °C, and a final elongation step at 72 °C for 10 min. PCR products were determined with agarose gel electrophoresis. Subsequently, the BamHI/XhoI fragment, Ldh-a was inserted into pCold-SUMO expression vector (BPI, Beijing, China); the other two fragments, Ldh-b and Ldh-c were inserted into pET-30a (+) expression vector (Novagen, Darmstadt, Germany). The final plasmid constructs of pCold-SUMO-Ldh-a, pET-30a-Ldh-b, and pET-30a-Ldh-c had been sequenced in BGI (Beijing Genomics Institute, Beijing, China).The recombinant expression shuttles (pCold-SUMO-Ldh-a, pET-30a-Ldh-b, and pET-30a-Ldh-c) with six His-tags in the C-terminus were transformed into Escherichia coli (E. coli) BL21 (DE3) cells.
4.3. Protein Expression, Purification, Analysis and Enzyme Activity Measurement
E. coli BL21 (DE3) cells were cultured in media of Lysogeny broth (LB). E. coli clones with plasmids (pCold-SUMO-Ldh-a, pET-30a-Ldh-b, and pET-30a-Ldh-c) were inoculated into a liter of medium with 100 μg/mL ampicillin or 50 μg/mL kanamycin and cultured at 37 °C. When A600 value arrived at 0.6, isopropyl-β-d-thiogalactopyranoside (IPTG) reagent was added till the final concentration of 1 mmol/L, followed by further culturing at 25 °C for 10 h to induce the expression of Ldh-a, Ldh-b, and Ldh-c. To determine LDHA, LDHB and LDHC expression in the supernatant and precipitate of E. coli cells, bacterial cells were collected by centrifugation and resuspended in 20 mmol/L phosphate buffer saline (PBS) (pH 8.0), and disrupted by ultrasonication (36 × 5 s pulses with 5 s intervals). The supernatant was then collected; the precipitate was resuspended and washed in 20 mmol/L PBS (pH 8.0) with 8 mol/L urea. SDS-PAGE was performed to assay the collected supernatant and treated precipitate with a steady current voltage electrophoresis instrument (Beijing Liuyi Instrument Factory, Beijing, China) as follows: 6 μL samples were loaded on a 12% (w/v) separating gel with a 5% (w/v) stacking gel in Tris-glycine buffer (pH = 8.3). The electric voltage was 80 V in the stacking gel and 120 V in the separating gel. The gel was stained in with Coomassie staining solution (10% methyl alcohol, 10% acetic acid, 0.2% R-250) for 4 h, and finally stored in the destaining solution (10% methyl alcohol, 10% acetic acid).
To obtain the active enzyme protein in the supernatant of E. coli cells, bacterial cells were collected with following procedures: 4000 r/min centrifugation (15 min), resuspension in 20 mmol/L PBS (pH = 7.0), and disruption by repeated freeze-thaw cycles (in liquid nitrogen and 37 °C water bath) for 10 times. In freeze-thaw cycles, Triton X-100 (final concentration of 1%), lysozyme (final concentration of 1%) and DNA enzyme (final concentration of 0.2%) were added. The lysates were centrifuged at 15,000 r/min under a temperature of 4 °C for 10 min, and the obtained supernatant was used for purification. Ni-NTA resin (QIAGEN, Hilden, Germany) was used to purify the recombinant LDHs. The resins were washed twice with increasing concentrations of imidazole at 10, 20, 50, 100, 250 and 500 mmol/L in PBS gradient (50 mmol/L NaH2PO4, 300 mmol/L NaCl, pH = 8.0). Each fraction was collected using individual collection tubes and detected by native PAGE. Elutions with LDH activity were totally collected. Enzyme liquids were obtained from elutions after removing imidazole by ultrafiltration (Ultrafiltration Tubes, 50 kD, 15 mL, Merck Millipore, Billerica, MA, USA). On average, a final amount of 3 mg LDH isoenzymes were purified from every liter of bacterial culture following this approach. LDH isoenzymes stocks were finally stored in PBS (pH = 8.0) at 4 °C until following steps. The LDH isoenzymes were prepared simultaneously under the same conditions [60].
SDS-PAGE was used to detect the protein purification and native PAGE was used to test and verify the enzyme activity. Native PAGE experiment was also performed with a voltage electrophoresis apparatus (Beijing Liuyi Instrument Factory, Beijing, China) with the following conditions: 8% (w/v) separating gel with a 4% (w/v) stacking gel (non-denaturing polyacrylamide gel) in Tris-glycine (pH = 8.3), and 6 μL samples were loaded. LDH bands were stained at 37 °C in a specific staining reagent (a mixture of 0.5 mol/L phosphate buffer (pH = 7.5), 10 mL of 1 mg/mL nitrobenzenethiocyanate chloride (NBT), 4 mL of 5 mg/mL NAD+, 2.5 mL of 1 mol/L sodium lactate, 2.5 mL of 0.1 mol/L NaCl and 1 mL of 1 mg/mL phenazinemethosulfate (PMS)) for half an hour in the dark. The stained gels were then rinsed with ddH2O and stored in the storage liquid (10% glycerol and 7% acetic acid). The reference marker for native PAGE was made with a mixture of pika kidney and skeletal muscle, tissues were homogenized on ice as a 1:4 (w/v) dilution in 0.9% physiological saline, and centrifuged at 4000 r/min at 4 °C for 10 min, and finally the supernatant was collected.
Protein concentration was determined by Bradford assay (Bradford protein assay kit, Nanjing Jiancheng Biotech Inc., Nanjing, China). Enzyme activity was measured by Bergmeyer assay (Lactate dehydrogenase kit, Nanjing Jiancheng Biotech Inc., Nanjing, China).
4.4. LDH-A4, LDH-B4 and LDH-C4 Kinetics Properties
The activity of LDH was measured using UV spectrophotometer (Unicon 2800, Shanghai, China) by recording the change of 340 nm absorbance produced by NADH oxidation. Assays were kept at consistent temperature of 37 °C. Calculation of Km value for pyruvate and lactate was resulted from Lineweaver-Burk plots. Ki value of lactate was determined from Km and Vmax, which was obtained with or without inhibitor added to reaction buffer via various concentrations of pyruvate at a constant inhibitor concentration, followed by plotting the slope (Km/Vmax value) against inhibitor concentrations. The mixed reagent of reaction to from pyruvate lactate contained 0.15 mmol/L NADH, 50 mmol/L PBS (pH8.0) and the sodium pyruvate gradients as substrate were performed at 0.05, 0.1, 0.2, 0.4, 0.6 and 0.8 mmol/L for LDH-A4, LDH-B4, and LDH-C4, and the enzyme persisted at a constant inhibitor concentration. The reaction from lactate to pyruvate contained 0.5 mmol/L NAD+, 50 mmol/LPBS, (pH 8.0); sodium lactate gradients as substrate were performed at 2, 5, 10, 20, 30 and 40 mmol/L for LDH-A4, LDH-B4, and LDH-C4. The concentration gradients of sodium lactate as inhibitor were added as 10, 20, and 40 mmol/L, respectively; and the substrates, coenzymes, and other buffers followed the method of the reaction from pyruvate to lactate as outlined above. A ΔE340 value of 0.06–0.07 per minute was provided in the experiment. The enzyme preparation was diluted with PBS (pH = 8.0) and assayed in a 1 cm light path. Before starting the reaction by addition of substrate, the coenzyme was incubated with the buffer used in the assay for 10 min at 37 °C. In total, 3 mL volumes of reagents were added in the process [61]. All chemicals were analytically pure from Sangon (Sangon, Shanghai, China). All measurements in the study were repeated 5 times for the average values.
4.5. Statistics
The data are presented as mean ± standard deviation (SD). Statistical analysis was performed by one-way analysis of variance (ANOVA) followed by Duncan’s test using SPSS 22.0 (SPSS Inc., Chicago, IL, USA). Values of p < 0.01 was considered highly significant and p < 0.05 was considered statistically significant.
5. Conclusions
In light of the enzymatic kinetics properties, we suggest that the pika could reduce dependence on oxygen and enhance adaptation to the hypoxic environment due to its increasing anaerobic glycolysis to produce ATP rapidly by LDH-C4, which is often the role of LDH-A4 in most mammal species.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. 31260512, No. 30960054, and No. 31040011), the Key Project of Chinese Ministry of Education (209132) and the Natural Science Foundation of Qinghai Province, China (No. 2012-Z-905 and No. 2014-ZJ-714).
Author Contributions
Dengbang Wei planned the experiments; Yang Wang and Lian Wei performed the experiments; Yang Wang, Xiao Li, Linna Wei and Lina Xu analyzed the data; Yang Wang and Dengbang Wei wrote the paper.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Smith, A.T.; Foggin, J.M. The plateau pika (Ochotona curzoniae) is a keystone species for biodiversity on the Tibetan plateau. Anim. Conserv. 1999, 2, 235–240. [Google Scholar] [CrossRef]
- Lai, C.H.; Smith, A.T. Keystone status of plateau pikas (Ochotona curzoniae): Effect of control on biodiversity of native birds. Biodivers. Conserv. 2003, 12, 1901–1912. [Google Scholar] [CrossRef]
- Yang, Y.Z.; Cao, Y.; Jin, G.E.; Bai, Z.Z.; Yun, M.L.; Ge, R.L. Molecular cloning and characterization of hemoglobin α and β chains from plateau pika (Ochotona curzoniae) living at high altitude. Gene 2007, 403, 118–124. [Google Scholar]
- Wang, X.J.; Wei, D.B.; Wei, L.; Qi, X.Z.; Zhu, S.H. Characteristics of pulmonary acinus structure in the plateau zokor (Myospalax baileyi) and plateau pika (Ochotona curzoniae). Acta Zool. Sin. 2008, 54, 531–539. [Google Scholar]
- Ge, R.L.; Kubo, K.; Kobayashi, T.; Sekiguchi, M.; Honda, T. Blunted hypoxic pulmonary vasoconstrictive response in the rodent Ochotona curzoniae (pika) at high altitude. Am. J. Physiol. Heart Circ. Physiol. 1998, 274, H1792–H1799. [Google Scholar]
- Wang, X.J.; Wei, D.B.; Wei, L.; Zhang, J.M.; Yu, H.Y. Physiological character of erythrocyte adapting to hypoxia in plateau zokor and plateau pika. Sichuan J. Zool. 2008, 6, 038. (In Chinese) [Google Scholar]
- Ye, R.R.; Cao, Y.F.; Bai, Q.H. Blood indices of plateau pika and relationship with hypoxia adaptation. Acta Anim. Sci. Sin. 1994, 2, 114–119. [Google Scholar]
- He, J.; Xu, C.; Meng, X.; Li, H.; Wang, Y. Comparative analysis in transport and intake of oxygen between pikas (Ochotona curzoniae) and rats. J. Prev. Med. Chin. People Lib. 1994, 12, 431–435. (In Chinese) [Google Scholar]
- Qi, X.Z.; Wang, X.J.; Zhu, S.H.; Rao, X.F.; Wei, L.; Wei, D.B. Hypoxic adaptation of the hearts of plateau zokor (Myospalax baileyi) and plateau pika (Ochotona curzoniae). Acta Physiol. Sin. 2008, 60, 348–354. (In Chinese) [Google Scholar]
- Wei, D.B.; Wei, L.; Zhang, J.M.; Yu, H.Y. Blood-gas properties of plateau zokor (Myospalax baileyi). Comp. Biochem. Phys. A 2006, 145, 372–375. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.H.; Qi, X.Z.; Wang, X.J.; Rao, X.F.; Wei, L.; Wei, D.B. Difference in oxygen uptake in skeletal muscles between plateau zokor (Myospalax baileyi) and plateau pika (Ochotona curzoniac). Acta Physiol. Sin. 2009, 61, 373–378. (In Chinese) [Google Scholar]
- Sun, S.Z.; Wei, L.; Wei, D.B.; Wang, D.W.; Ma, B.Y. Differences of glycolysis in skeletal muscle and lactate metabolism in liver between plateau zokor (Myospalax baileyi) and plateau pika (Ochotona curzoniae). Acta Physiol. Sin. 2013, 65, 276–284. [Google Scholar]
- Li, H.G.; Guo, S.C.; Ren, Y.M.; Wang, D.P.; Yu, H.H.; Li, W.J.; Zhao, X.Q.; Chang, Z.J. VEGF189 expression is highly related to adaptation of the plateau pika (Ochotona curzoniae) inhabiting high altitudes. HighAlt. Med. Biol. 2013, 14, 395–404. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.N.; Zhu, R.J.; Wang, D.W.; Wei, L.; Wei, D.B. Gene coding and mRNA expression of vascular endothelial growth factor as well as microvessel density in brain of plateau zokor: Comparison with other rodents. Acta Physiol. Sin. 2011, 63, 155–163. (In Chinese) [Google Scholar]
- Li, H.G.; Ren, Y.M.; Guo, S.C.; Cheng, L.; Wang, D.P.; Yang, J.; Chang, Z.J.; Zhao, X.Q. The protein level of hypoxia-inducible factor-1α is increased in the plateau pika (Ochotona curzoniae) inhabiting high altitudes. J. Exp. Zool. A Ecol. Genet. Physiol. 2009, 311, 134–141. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.B.; Ning, H.X.; Zhu, S.S.; Sun, P.; Xu, S.X.; Chang, Z.J.; Zhao, X.Q. Cloning of hypoxia-inducible factor-1α cDNA from a high hypoxia tolerant mammal—plateau pika (Ochotona curzoniae). Biochem. Biophys. Res. Commun. 2004, 316, 565–572. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.W.; Wei, L.; Wei, D.B.; Rao, X.F.; Qi, X.Z.; Wang, X.J.; Ma, B.Y. Testis-specific lactate dehydrogenase is expressed in somatic tissues of plateau pikas. FEBS Open Biol. 2013, 3, 118–123. [Google Scholar] [CrossRef] [PubMed]
- Pichon, A.; Zhenzhong, B.; Favret, F.; Jin, G.; Shufeng, H.; Marchant, D.; Richalet, J.P.; Ge, R.L. Long-term ventilatory adaptation and ventilatory response to hypoxia in plateau pika (Ochotona curzoniae): Role of nNOS and dopamine. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2009, 297, R978–R987. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Wang, Z.L.; Zhao, X.Q.; Xu, B.H.; Ren, Y.H.; Tian, H.F. Natural selection and adaptive evolution of leptin in the ochotona family driven by the cold environmental stress. PLoS ONE 2008, 3, e1472. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhao, X.Q.; Guo, S.C.; Li, H.G.; Qi, D.L.; Wang, D.P.; Cao, J.H. Leptin cDNA cloning and its mRNA expression in plateau pikas (Ochotona curzoniae) from different altitudes on Qinghai-Tibet Plateau. Biochem. Biophys. Res. Commun. 2006, 345, 1405–1413. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.J.; Gao, W.X.; Gao, Y.Q.; Tang, S.; Huang, Q.Y.; Tan, X.L.; Chen, J.; Huang, T.S. Mitochondrial genome analysis of Ochotona curzoniae and implication of cytochrome c oxidase in hypoxic adaptation. Mitochondrion 2008, 8, 352–357. [Google Scholar] [CrossRef] [PubMed]
- Everse, J.; Kaplan, N.O. Lactate dehydrogenases: Structure and function. Adv. Enzymol. Relat. Areas Mol. Biol. 1973, 37, 61–133. [Google Scholar] [PubMed]
- Li, S. Lactate dehydrogenase isoenzymes A (muscle), B (heart) and C (testis) of mammals and the genes coding for these enzymes. Biochem. Soc. Trans. 1989, 17, 304. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; O’brien, D.; Hou, E.; Versola, J.; Rockett, D.; Eddy, E. Differential activity and synthesis of lactate dehydrogenase isozymes A (muscle), B (heart), and C (testis) in mouse spermatogenic cells. Biol. Reprod. 1989, 40, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Cahn, R.D.; Zwilling, E.; Kaplan, N.O.; Levine, L. Nature and development of lactic dehydrogenases. Science 1962, 136, 962–969. [Google Scholar] [CrossRef] [PubMed]
- Fine, I.; Kaplan, N.; Kuftinec, D. Developmental changes of mammalian lactic dehydrogenases. Biochemistry 1963, 2, 116–121. [Google Scholar] [CrossRef]
- Goldberg, E. Reproductive implications of LDH-C4 and other testis-specific isozymes. Exp. Clin. Immunogenet. 1984, 2, 120–124. [Google Scholar]
- Coonrod, S.; Vitale, A.; Duan, C.; Bristol-Gould, S.; Herr, J.; Goldberg, E. Testis-Specific lactate dehydrogenase (LDH-C4; Ldh3) in murine oocytes and preimplantation embryos. J. Androl. 2006, 27, 502–509. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, E. Lactate dehydrogenase-X from mouse testes and spermatozoa. Methods Enzymol. 1975, 41, 318. [Google Scholar] [PubMed]
- Goldberg, E. Lactate dehydrogenases and malate dehydrogenases in sperm: Studied by polyacrylamide gel electrophoresis. Ann. N. Y. Acad. Sci. 1964, 121, 560–570. [Google Scholar] [CrossRef] [PubMed]
- Coronel, C.E.; Burgos, C.; de Gerez Burgos, N.M.; Rovai, L.E.; Blanco, A. Catalytic properties of the sperm-specific lactate dehydrogenase (LDH X or C4) from different species. J. Exp. Zool. 1983, 225, 379–385. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.; Yañez, R.; Brown, D.M.; Dickey, A.; Parks, M.E.; McKee, R.W. Isolation and properties of lactate dehydrogenase isozyme X from Swiss mice. Biochim. Biophys. Acta 1971, 146, 454–460. [Google Scholar] [CrossRef]
- Allen, J. Multiple forms of lactic dehydrogenase in tissues of the mouse: Their specificity, cellular localization, and response to altered physiological conditions. Ann. N. Y. Acad. Sci. 1961, 94, 937–951. [Google Scholar] [CrossRef] [PubMed]
- Zinkham, W.H.; Blanco, A.; Clowry, L.J. An unusual isozyme of lactate dehydrogenase in mature testes: Localization, ontogeny, and kinetic properties. Ann. N. Y. Acad. Sci. 1964, 121, 571–588. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, J.; Withycombe, W.A. Organ specificity and lactate-dehydrogenase activity. Some properties of human spermatozoal lactate dehydrogenase. Biochem. J. 1965, 97, 663–668. [Google Scholar] [CrossRef] [PubMed]
- Blanco, A.; Zinkham, W.H.; Kupchyk, L. Genetic control and ontogeny of lactate dehydrogenase in pigeon testes. J. Exp. Zool. 1964, 156, 137–152. [Google Scholar] [CrossRef] [PubMed]
- Battellino, L.J.; Jaime, F.R.; Blanco, A. Kinetic properties of rabbit testicular lactate dehydrogenase isozyme. J. Biol. Chem. 1968, 243, 5185–5192. [Google Scholar] [PubMed]
- Schatz, L.; Segal, H.L. Reduction of α-ketoglutarate by homogeneous lactic dehydrogenase X of testicular tissue. J. Biol. Chem. 1969, 244, 4393–4397. [Google Scholar] [PubMed]
- Battellino, L.J.; Blanco, A. Catalytic properties of the lactate dehydrogenase isozyme “X” from mouse testis. J. Exp. Zool. 1970, 174, 173–186. [Google Scholar] [CrossRef] [PubMed]
- Hawtrey, C.O.; Goldberg, E. Some kinetic aspects of sperm specific lactate dehydrogenase in mice. J. Exp. Zool. 1970, 174, 451–461. [Google Scholar] [CrossRef] [PubMed]
- Fleisher, G.A.; Kolb, E.; Larner, J. Isolation and characterization of bovine lactate dehydrogenase X. Biochemistry 1970, 9, 4372–4380. [Google Scholar] [CrossRef]
- Rodríguez-Páez, L.; Chena-Taboada, M.A.; Cabrera-Hernández, A.; Cordero-Martínez, J.; Wong, C. Oxamic acid analogues as LDH-C4-specific competitive inhibitors. J. Enzym. Inhib. Med. Chem. 2011, 26, 579–586. [Google Scholar] [CrossRef] [PubMed]
- Blanco, A.; Zinkham, W.H. Lactate dehydrogenases in human testes. Science 1963, 139, 601–602. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, E. Lactic and malic dehydrogenases in human spermatozoa. Science 1963, 139, 602–603. [Google Scholar] [CrossRef] [PubMed]
- Quistorff, B.; Grunnet, N. The isoenzyme pattern of LDH does not play a physiological role; except perhaps during fast transitions in energy metabolism. Aging 2011, 3, 457–460. [Google Scholar] [PubMed]
- Putz, M.V. On the Reducible Character of Haldane-Radić Enzyme Kinetics to Conventional and Logistic Michaelis-Menten Models. Molecules 2011, 16, 3128–3145. [Google Scholar] [CrossRef] [PubMed]
- Putz, M.V.; Ana-Maria, L.; Vasile, O. Full analytic progress curves of enzymic reactions in vitro. Int. J. Mol. Sci. 2006, 7, 469–484. [Google Scholar] [CrossRef]
- Clausen, J.; Ovlisen, B. Lactate dehydrogenase isoenzymes of human semen. Biochem. J. 1965, 97, 513–517. [Google Scholar] [CrossRef] [PubMed]
- LeVan, K.M.; Goldberg, E. Properties of human testis-specific lactate dehydrogenase expressed from Escherichia coli. Biochem. J. 1991, 273, 587–592. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.; Rodrı́guez-Páez, L.; Nogueda, B.; Pérez, A.; Baeza, I. Selective inhibition of the sperm-specific lactate dehydrogenase isozyme-C4 by N-isopropyl oxamate. Biochim. Biophys. Acta 1997, 1343, 16–22. [Google Scholar] [CrossRef]
- Hereng, T.; Elgstøen, K.; Cederkvist, F.; Eide, L.; Jahnsen, T.; Skålhegg, B.; Rosendal, K. Exogenous pyruvate accelerates glycolysis and promotes capacitation in human spermatozoa. Hum. Reprod. 2011. [Google Scholar] [CrossRef] [PubMed]
- Harwood. Basic DNA and RNA Protocols. In Methods in Molecular Biology, 1st ed.; NJ Humana Press: Totowa, NJ, USA, 1994; Volume 58, pp. 491–510. [Google Scholar]
- Niu, X.; Guiltinan, M.J. DNA binding specificity of the wheat bZIP protein EmBP-1. Nucleic Acids Res. 1994, 22, 4969–4978. [Google Scholar] [CrossRef] [PubMed]
- Tang, W. The cause of deviation made in determining the molecular weight of His-tag fusion proteins by SDS-PAGE. Acta Phytophysiol. Sin. 2000, 26, 64–68. (In Chinese) [Google Scholar]
- Berta, M.A.; Mazure, N.; Hattab, M.; Pouysségur, J.; Brahimi-Horn, M.C. SUMOylation of hypoxia-inducible factor-1α reduces its transcriptional activity. Biochem. Biophys. Res. Commun. 2007, 360, 646–652. [Google Scholar] [CrossRef] [PubMed]
- Odet, F.; Gabel, S.A.; Williams, J.; London, R.E.; Goldberg, E.; Eddy, E.M. Lactate dehydrogenase C and energy metabolism in mouse sperm. Biol. Reprod. 2011, 85, 556–564. [Google Scholar] [CrossRef] [PubMed]
- Odet, F.; Duan, C.; Willis, W.D.; Goulding, E.H.; Kung, A.; Eddy, E.M.; Goldberg, E. Expression of the gene for mouse lactate dehydrogenase C (Ldhc) is required for male fertility. Biol. Reprod. 2008, 79, 26–34. [Google Scholar] [CrossRef] [PubMed]
- Mukai, C.; Okuno, M. Glycolysis plays a major role for adenosine triphosphate supplementation in mouse sperm flagellar movement. Biol. Reprod. 2004, 71, 540–547. [Google Scholar] [CrossRef] [PubMed]
- Williams, A.C.; Ford, W.C.L. The role of glucose in supporting motility and capacitation in human spermatozoa. J. Androl. 2001, 22, 680–695. [Google Scholar] [PubMed]
- Zhang, J.Y.; Jin, Z.; Sun, T.; Jiang, Y.; Han, Q.Q.; Song, Y.Z.; Chen, Q.; Xia, X.S. Prokaryotic expression, purification, and polyclonal antibody production of a truncated recombinant rabies virus L protein. Iran J. Biotechnol. 2015, 13, e1022. [Google Scholar]
- Bergmeyer, H.U. Methods of Enzymatic Analysis; Academic Press: New York, NY, USA, 2012; Volume 3, pp. 1196–1201. [Google Scholar]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).