Histone Posttranslational Modifications of CD4+ T Cell in Autoimmune Diseases
Abstract
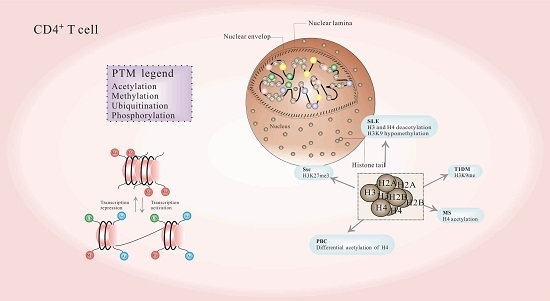
:1. Introduction
1.1. Acetylation
1.2. Methylation
1.3. Ubiquitination
1.4. Phosphorylation
2. Posttranslational Modification of Histones in CD4+ T Cells
2.1. Histone PTMs in Th1/Th2 Cell Development
2.2. Histone PTMs in Th17 Cell Development
2.3. Histone PTMs in Treg Cell Development
2.4. Histone PTMs in Other Effector T Cells
3. Histone PTMs and Autoimmune Diseases
3.1. Systemic Lupus Erythematosus (SLE)
3.2. Systemic Sclerosis (SSc)
3.3. Multiple Sclerosis (MS)
3.4. Primary Biliary Cirrhosis (PBC)
3.5. Type 1 Diabetes (T1D)
4. Conclusions
Acknowledgments
Conflicts of Interest
References
- Suganuma, T.; Workman, J.L. Crosstalk among histone modifications. Cell 2008, 135, 604–607. [Google Scholar] [CrossRef] [PubMed]
- Strahl, B.D.; Allis, C.D. The language of covalent histone modifications. Nature 2000, 403, 41–45. [Google Scholar] [CrossRef] [PubMed]
- Jenuwein, T.; Allis, C.D. Translating the histone code. Science 2001, 293, 1074–1080. [Google Scholar] [CrossRef] [PubMed]
- Richmond, T.J.; Davey, C.A. The structure of DNA in the nucleosome core. Nature 2003, 423, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Luger, K.; Richmond, T.J. The histone tails of the nucleosome. Curr. Opin. Genet. Dev. 1998, 8, 140–146. [Google Scholar] [CrossRef]
- Kornberg, R.D.; Lorch, Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell 1999, 98, 285–294. [Google Scholar] [CrossRef]
- Arents, G.; Moudrianakis, E.N. The histone fold: A ubiquitous architectural motif utilized in DNA compaction and protein dimerization. Proc. Natl. Acad. Sci. USA 1995, 92, 11170–11174. [Google Scholar] [CrossRef] [PubMed]
- Luger, K.; Mäder, A.W.; Richmond, R.K.; Sargent, D.F.; Richmond, T.J. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 1997, 389, 251–260. [Google Scholar] [PubMed]
- Tan, M.; Luo, H.; Lee, S.; Jin, F.; Yang, J.S.; Montellier, E.; Buchou, T.; Cheng, Z.; Rousseaux, S.; Rajagopal, N. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell 2011, 146, 1016–1028. [Google Scholar] [CrossRef] [PubMed]
- Kouzarides, T. Chromatin modifications and their function. Cell 2007, 128, 693–705. [Google Scholar] [CrossRef] [PubMed]
- Cheung, P.; Allis, C.D.; Sassone-Corsi, P. Signaling to chromatin through histone modifications. Cell 2000, 103, 263–271. [Google Scholar] [CrossRef]
- Wei, Y.; Yu, L.; Bowen, J.; Gorovsky, M.A.; Allis, C.D. Phosphorylation of histone H3 is required for proper chromosome condensation and segregation. Cell 1999, 97, 99–109. [Google Scholar] [CrossRef]
- Baarends, W.M.; Hoogerbrugge, J.W.; Roest, H.P.; Ooms, M.; Vreeburg, J.; Hoeijmakers, J.H.; Grootegoed, J.A. Histone ubiquitination and chromatin remodeling in mouse spermatogenesis. Dev. Biol. 1999, 207, 322–333. [Google Scholar] [CrossRef] [PubMed]
- El-Osta, A.; Wolffe, A.P. DNA methylation and histone deacetylation in the control of gene expression: Basic biochemistry to human development and disease. Gene Expr. 2000, 9, 63–75. [Google Scholar] [CrossRef] [PubMed]
- Turner, B.M. Histone acetylation and an epigenetic code. Bioessays 2000, 22, 836–845. [Google Scholar] [CrossRef]
- Glozak, M.A.; Sengupta, N.; Zhang, X.; Seto, E. Acetylation and deacetylation of non-histone proteins. Gene 2005, 363, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Shahbazian, M.D.; Grunstein, M. Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 2007, 76, 75–100. [Google Scholar] [CrossRef] [PubMed]
- Kristjuhan, A.; Walker, J.; Suka, N.; Grunstein, M.; Roberts, D.; Cairns, B.R.; Svejstrup, J.Q. Transcriptional inhibition of genes with severe histone h3 hypoacetylation in the coding region. Mol. Cell 2002, 10, 925–933. [Google Scholar] [CrossRef]
- Parthun, M.R. Hat1: The emerging cellular roles of a type B histone acetyltransferase. Oncogene 2007, 26, 5319–5328. [Google Scholar] [CrossRef] [PubMed]
- Grunstein, M. Histone acetylation in chromatin structure and transcription. Nature 1997, 389, 349–352. [Google Scholar] [CrossRef] [PubMed]
- Kuo, M.H.; Allis, C.D. Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays 1998, 20, 615–626. [Google Scholar] [CrossRef]
- Gallinari, P.; Di Marco, S.; Jones, P.; Pallaoro, M.; Steinkuhler, C. HDACs, histone deacetylation and gene transcription: From molecular biology to cancer therapeutics. Cell Res. 2007, 17, 195–211. [Google Scholar] [CrossRef] [PubMed]
- de Ruijter, A.J.; van Gennip, A.H.; Caron, H.N.; Kemp, S.; van Kuilenburg, A.B. Histone deacetylases (HDACs): Characterization of the classical HDAC family. Biochem. J. 2003, 370, 737–749. [Google Scholar] [CrossRef] [PubMed]
- Struhl, K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998, 12, 599–606. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zang, C.; Rosenfeld, J.A.; Schones, D.E.; Barski, A.; Cuddapah, S.; Cui, K.; Roh, T.Y.; Peng, W.; Zhang, M.Q.; et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat. Genet. 2008, 40, 897–903. [Google Scholar] [CrossRef] [PubMed]
- Creyghton, M.P.; Cheng, A.W.; Welstead, G.G.; Kooistra, T.; Carey, B.W.; Steine, E.J.; Hanna, J.; Lodato, M.A.; Frampton, G.M.; Sharp, P.A.; et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. USA 2010, 107, 21931–21936. [Google Scholar] [CrossRef] [PubMed]
- Bonasio, R.; Tu, S.; Reinberg, D. Molecular signals of epigenetic states. Science 2010, 330, 612–616. [Google Scholar] [CrossRef] [PubMed]
- Wood, A.; Shilatifard, A. Posttranslational modifications of histones by methylation. Adv. Protein Chem. 2004, 67, 201–222. [Google Scholar] [PubMed]
- Greer, J.M.; McCombe, P.A. The role of epigenetic mechanisms and processes in autoimmune disorders. Biologics 2012, 6, 307–327. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Kim, S.Y.; Artis, S.; Molfese, D.L.; Schumacher, A.; Sweatt, J.D.; Paylor, R.E.; Lubin, F.D. Histone methylation regulates memory formation. J. Neurosci. 2010, 30, 3589–3599. [Google Scholar] [CrossRef] [PubMed]
- Weake, V.M.; Workman, J.L. Histone ubiquitination: Triggering gene activity. Mol. Cell 2008, 29, 653–663. [Google Scholar] [CrossRef] [PubMed]
- Widom, J. Structure, dynamics, and function of chromatin in vitro. Annu. Rev. Biophys. Biomol. Struct. 1998, 27, 285–327. [Google Scholar] [CrossRef] [PubMed]
- Redman, K.L.; Rechsteiner, M. Extended reading frame of a ubiquitin gene encodes a stable, conserved, basic protein. J. Biol. Chem. 1988, 263, 4926–4931. [Google Scholar] [PubMed]
- Finley, D.; Chau, V. Ubiquitination. Annu. Rev. Cell Biol. 1991, 7, 25–69. [Google Scholar] [CrossRef] [PubMed]
- Jennissen, H.P. Ubiquitin and the enigma of intracellular protein degradation. Eur. J. Biochem. 1995, 231, 1–30. [Google Scholar] [PubMed]
- Mimnaugh, E.G.; Chen, H.Y.; Davie, J.R.; Celis, J.E.; Neckers, L. Rapid deubiquitination of nucleosomal histones in human tumor cells caused by proteasome inhibitors and stress response inducers: Effects on replication, transcription, translation, and the cellular stress response. Biochemistry 1997, 36, 14418–14429. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wang, L.; Erdjument-Bromage, H.; Vidal, M.; Tempst, P.; Jones, R.S.; Zhang, Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature 2004, 431, 873–878. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-S.; Shukla, A.; Schneider, J.; Swanson, S.K.; Washburn, M.P.; Florens, L.; Bhaumik, S.R.; Shilatifard, A. Histone crosstalk between H2B monoubiquitination and H3 methylation mediated by COMPASS. Cell 2007, 131, 1084–1096. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Guermah, M.; McGinty, R.K.; Lee, J.S.; Tang, Z.; Milne, T.A.; Shilatifard, A.; Muir, T.W.; Roeder, R.G. RAD6-Mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell 2009, 137, 459–471. [Google Scholar] [CrossRef] [PubMed]
- Ballal, N.; Kang, Y.; Olson, M.; Busch, H. Changes in nucleolar proteins and their phosphorylation patterns during liver regeneration. J. Biol. Chem. 1975, 250, 5921–5925. [Google Scholar] [PubMed]
- Berger, S.L. An embarrassment of niches: The many covalent modifications of histones in transcriptional regulation. Oncogene 2001, 20, 3007–3013. [Google Scholar] [CrossRef] [PubMed]
- Xhemalce, B.; Dawson, M.A.; Bannister, A.J. Histone modifications. Rev. Cell Biol. Mol. Med. 2011. [Google Scholar] [CrossRef]
- Oki, M.; Aihara, H.; Ito, T. Role of histone phosphorylation in chromatin dynamics and its implications in diseases. Subcell. Biochem. 2007, 41, 319–336. [Google Scholar] [PubMed]
- Wong, W.F.; Kohu, K.; Chiba, T.; Sato, T.; Satake, M. Interplay of transcription factors in T-cell differentiation and function: The role of Runx. Immunology 2011, 132, 157–164. [Google Scholar] [CrossRef] [PubMed]
- Fontenot, J.D.; Gavin, M.A.; Rudensky, A.Y. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat. Immunol. 2003, 4, 330–336. [Google Scholar] [CrossRef] [PubMed]
- Mullen, A.C.; Hutchins, A.S.; High, F.A.; Lee, H.W.; Sykes, K.J.; Chodosh, L.A.; Reiner, S.L. Hlx is induced by and genetically interacts with T-bet to promote heritable TH1 gene induction. Nat. Immunol. 2002, 3, 652–658. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Min, B.; Hu-Li, J.; Watson, C.J.; Grinberg, A.; Wang, Q.; Killeen, N.; Urban, J.F., Jr.; Guo, L.; Paul, W.E. Conditional deletion of Gata3 shows its essential function in TH1-TH2 responses. Nat. Immunol. 2004, 5, 1157–1165. [Google Scholar] [CrossRef] [PubMed]
- Pai, S.Y.; Truitt, M.L.; Ho, I.C. GATA-3 deficiency abrogates the development and maintenance of T helper type 2 cells. Proc. Natl. Acad. Sci. USA 2004, 101, 1993–1998. [Google Scholar] [CrossRef] [PubMed]
- Martins, G.A.; Hutchins, A.S.; Reiner, S.L. Transcriptional activators of helper T cell fate are required for establishment but not maintenance of signature cytokine expression. J. Immunol. 2005, 175, 5981–5895. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.R.; Kim, S.T.; Spilianakis, C.G.; Fields, P.E.; Flavell, R.A. T helper cell differentiation: Regulation by cis elements and epigenetics. Immunity 2006, 24, 369–379. [Google Scholar] [CrossRef] [PubMed]
- Avni, O.; Lee, D.; Macian, F.; Szabo, S.J.; Glimcher, L.H.; Rao, A. TH cell differentiation is accompanied by dynamic changes in histone acetylation of cytokine genes. Nat. Immunol. 2002, 3, 643–651. [Google Scholar] [CrossRef] [PubMed]
- Baguet, A.; Bix, M. Chromatin landscape dynamics of the IL4–IL13 locus during T helper 1 and 2 development. Proc. Natl. Acad. Sci. USA 2004, 101, 11410–11415. [Google Scholar] [CrossRef] [PubMed]
- Kondilis-Mangum, H.D.; Wade, P.A. Epigenetics and the adaptive immune response. Mol. Asp. Med. 2013, 34, 813–825. [Google Scholar] [CrossRef] [PubMed]
- Koyanagi, M.; Baguet, A.; Martens, J.; Margueron, R.; Jenuwein, T.; Bix, M. EZH2 and histone 3 trimethyl lysine 27 associated with Il4 and Il13 gene silencing in Th1 cells. J. Biol. Chem. 2005, 280, 31470–31477. [Google Scholar] [CrossRef] [PubMed]
- Messi, M.; Giacchetto, I.; Nagata, K.; Lanzavecchia, A.; Natoli, G.; Sallusto, F. Memory and flexibility of cytokine gene expression as separable properties of human TH1 and TH2 lymphocytes. Nat. Immunol. 2003, 4, 78–86. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, M.; Shinnakasu, R.; Nigo, Y.; Kimura, M.; Hasegawa, A.; Taniguchi, M.; Nakayama, T. Interleukin (IL)-4-independent maintenance of histone modification of the IL-4 gene loci in memory Th2 cells. J. Biol. Chem. 2004, 279, 39454–39464. [Google Scholar] [CrossRef] [PubMed]
- Onodera, A.; Yamashita, M.; Endo, Y.; Kuwahara, M.; Tofukuji, S.; Hosokawa, H.; Kanai, A.; Suzuki, Y.; Nakayama, T. STAT6-mediated displacement of polycomb by trithorax complex establishes long-term maintenance of GATA3 expression in T helper type 2 cells. J. Exp. Med. 2010, 207, 2493–2506. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, M.; Hirahara, K.; Shinnakasu, R.; Hosokawa, H.; Norikane, S.; Kimura, M.Y.; Hasegawa, A.; Nakayama, T. Crucial role of MLL for the maintenance of memory T helper type 2 cell responses. Immunity 2006, 24, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Wei, G.; Wei, L.; Zhu, J.; Zang, C.; Hu-Li, J.; Yao, Z.; Cui, K.; Kanno, Y.; Roh, T.Y.; Watford, W.T.; et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity 2009, 30, 155–167. [Google Scholar] [CrossRef] [PubMed]
- Allan, R.S.; Zueva, E.; Cammas, F.; Schreiber, H.A.; Masson, V.; Belz, G.T.; Roche, D.; Maison, C.; Quivy, J.P.; Almouzni, G.; et al. An epigenetic silencing pathway controlling T helper 2 cell lineage commitment. Nature 2012, 487, 249–253. [Google Scholar] [CrossRef] [PubMed]
- Tumes, D.J.; Onodera, A.; Suzuki, A.; Shinoda, K.; Endo, Y.; Iwamura, C.; Hosokawa, H.; Koseki, H.; Tokoyoda, K.; Suzuki, Y.; et al. The polycomb protein Ezh2 regulates differentiation and plasticity of CD4+ T helper type 1 and type 2 cells. Immunity 2013, 39, 819–832. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Davidson, T.S.; Wei, G.; Jankovic, D.; Cui, K.; Schones, D.E.; Guo, L.; Zhao, K.; Shevach, E.M.; Paul, W.E. Down-regulation of Gfi-1 expression by TGF-β is important for differentiation of Th17 and CD103I inducible regulatory T cells. J. Exp. Med. 2009, 206, 329–341. [Google Scholar] [CrossRef] [PubMed]
- Akimzhanov, A.M.; Yang, X.O.; Dong, C. Chromatin remodeling of interleukin-17 (IL-17)-IL-17F cytokine gene locus during inflammatory helper T cell differentiation. J. Biol. Chem. 2007, 282, 5969–5972. [Google Scholar] [CrossRef] [PubMed]
- Mele, D.A.; Salmeron, A.; Ghosh, S.; Huang, H.R.; Bryant, B.M.; Lora, J.M. BET bromodomain inhibition suppresses TH17-mediated pathology. J. Exp. Med. 2013, 210, 2181–2190. [Google Scholar] [CrossRef] [PubMed]
- Van Loosdregt, J.; Coffer, P.J. Post-translational modification networks regulating FOXP3 function. Trends Immunol. 2014, 35, 368–378. [Google Scholar] [CrossRef] [PubMed]
- Samstein, R.M.; Arvey, A.; Josefowicz, S.Z.; Peng, X.; Reynolds, A.; Sandstrom, R.; Neph, S.; Sabo, P.; Kim, J.M.; Liao, W.; et al. Foxp3 exploits a pre-existent enhancer landscape for regulatory T cell lineage specification. Cell 2012, 151, 153–166. [Google Scholar] [CrossRef] [PubMed]
- Kitagawa, Y.; Ohkura, N.; Sakaguchi, S. Epigenetic control of thymic Treg-cell development. Eur. J. Immunol. 2015, 45, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Morikawa, H.; Ohkura, N.; Vandenbon, A.; Itoh, M.; Nagao-Sato, S.; Kawaji, H.; Lassmann, T.; Carninci, P.; Hayashizaki, Y.; Forrest, A.R.; et al. Differential roles of epigenetic changes and Foxp3 expression in regulatory T cell-specific transcriptional regulation. Proc. Natl. Acad. Sci. USA 2014, 111, 5289–5294. [Google Scholar] [CrossRef] [PubMed]
- Schreiber, L.; Pietzsch, B.; Floess, S.; Farah, C.; Jansch, L.; Schmitz, I.; Huehn, J. The Treg-specific demethylated region stabilizes Foxp3 expression independently of NF-κB signaling. PLoS ONE 2014, 9, e88318. [Google Scholar] [CrossRef] [PubMed]
- Waight, J.D.; Takai, S.; Marelli, B.; Qin, G.; Hance, K.W.; Zhang, D.; Tighe, R.; Lan, Y.; Lo, K.M.; Sabzevari, H.; et al. Cutting Edge: Epigenetic Regulation of Foxp3 Defines a Stable Population of CD4+ Regulatory T Cells in Tumors from Mice and Humans. J. Immunol. 2015, 194, 878–882. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, L.; Han, R.; Beier, U.H.; Akimova, T.; Bhatti, T.; Xiao, H.; Cole, P.A.; Brindle, P.K.; Hancock, W.W. Two histone/protein acetyltransferases, CBP and p300, are indispensable for Foxp3+ T-regulatory cell development and function. Mol. Cell. Biol. 2014, 34, 3993–4007. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, L.; Predina, J.; Han, R.; Beier, U.H.; Wang, L.C.; Kapoor, V.; Bhatti, T.R.; Akimova, T.; Singhal, S.; et al. Inhibition of p300 impairs Foxp3+ T regulatory cell function and promotes antitumor immunity. Nat. Med. 2013, 19, 1173–1177. [Google Scholar] [CrossRef] [PubMed]
- Smith, P.M.; Howitt, M.R.; Panikov, N.; Michaud, M.; Gallini, C.A.; Bohlooly, Y.M.; Glickman, J.N.; Garrett, W.S. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 2013, 341, 569–573. [Google Scholar] [CrossRef] [PubMed]
- Arpaia, N.; Campbell, C.; Fan, X.; Dikiy, S.; van der Veeken, J.; de Roos, P.; Liu, H.; Cross, J.R.; Pfeffer, K.; Coffer, P.J.; et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 2013, 504, 451–455. [Google Scholar] [CrossRef] [PubMed]
- Tone, Y.; Furuuchi, K.; Kojima, Y.; Tykocinski, M.L.; Greene, M.I.; Tone, M. Smad3 and NFAT cooperate to induce Foxp3 expression through its enhancer. Nat. Immunol. 2008, 9, 194–202. [Google Scholar] [CrossRef] [PubMed]
- Vaeth, M.; Schliesser, U.; Muller, G.; Reissig, S.; Satoh, K.; Tuettenberg, A.; Jonuleit, H.; Waisman, A.; Muller, M.R.; Serfling, E.; et al. Dependence on nuclear factor of activated T-cells (NFAT) levels discriminates conventional T cells from Foxp3+ regulatory T cells. Proc. Natl. Acad. Sci. USA 2012, 109, 16258–16563. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.; Jiao, J.; Wang, L.; O’Brien, S.; Newick, K.; Wang, L.C.; Falkensammer, E.; Liu, Y.; Han, R.; Kapoor, V.; et al. HDAC5 controls the functions of Foxp3+ T-regulatory and CD8+ T cells. Int. J. Cancer 2016, 138, 2477–2486. [Google Scholar] [CrossRef] [PubMed]
- Crotty, S. Follicular helper CD4 T cells (TFH). Annu. Rev. Immunol. 2011, 29, 621–663. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.S.; Yang, J.A.; Crotty, S. Dynamic regulation of Bcl6 in follicular helper CD4 T (Tfh) cells. Curr. Opin. Immunol. 2013, 25, 366–372. [Google Scholar] [CrossRef] [PubMed]
- Johnston, R.J.; Poholek, A.C.; DiToro, D.; Yusuf, I.; Eto, D.; Barnett, B.; Dent, A.L.; Craft, J.; Crotty, S. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science 2009, 325, 1006–1110. [Google Scholar] [CrossRef] [PubMed]
- Lu, K.T.; Kanno, Y.; Cannons, J.L.; Handon, R.; Bible, P.; Elkahloun, A.G.; Anderson, S.M.; Wei, L.; Sun, H.; O’Shea, J.J.; et al. Functional and epigenetic studies reveal multistep differentiation and plasticity of in vitro-generated and in vivo-derived follicular T helper cells. Immunity 2011, 35, 622–632. [Google Scholar] [CrossRef] [PubMed]
- Ramming, A.; Druzd, D.; Leipe, J.; Schulze-Koops, H.; Skapenko, A. Maturation-related histone modifications in the PU.1 promoter regulate Th9-cell development. Blood 2012, 119, 4665–4674. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.; Pan, D.; Lee, Y.H.; Martinez, G.J.; Feng, X.H.; Dong, C. Cutting edge: Smad2 and Smad4 regulate TGF-β-mediated Il9 gene expression via EZH2 displacement. J. Immunol. 2013, 191, 4908–4912. [Google Scholar] [CrossRef] [PubMed]
- Hu, N.; Qiu, X.; Luo, Y.; Yuan, J.; Li, Y.; Lei, W.; Zhang, G.; Zhou, Y.; Su, Y.; Lu, Q. Abnormal histone modification patterns in lupus CD4+ T cells. J. Rheumatol. 2008, 35, 804–810. [Google Scholar] [PubMed]
- Hu, N.; Long, H.; Zhao, M.; Yin, H.; Lu, Q. Aberrant expression pattern of histone acetylation modifiers and mitigation of lupus by SIRT1-siRNA in MRL/lpr mice. Scand. J. Rheumatol. 2009, 38, 464–471. [Google Scholar] [CrossRef] [PubMed]
- Long, H.; Huang, W.; Yin, H.; Zhao, S.; Zhao, M.; Lu, Q. Abnormal expression pattern of histone demethylases in CD4+ T cells of MRL/lpr lupus-like mice. Lupus 2009, 18, 1327–1328. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Sun, Y.; Gao, F.; Wu, X.; Tang, J.; Yin, H.; Luo, Y.; Richardson, B.; Lu, Q. Epigenetics and SLE: RFX1 downregulation causes CD11a and CD70 overexpression by altering epigenetic modifications in lupus CD4+ T cells. J. Autoimmun. 2010, 35, 58–69. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Wu, X.; Zhang, Q.; Luo, S.; Liang, G.; Su, Y.; Tan, Y.; Lu, Q. RFX1 regulates CD70 and CD11a expression in lupus T cells by recruiting the histone methyltransferase SUV39H1. Arthritis Res. Ther. 2010, 12. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Qiu, X.; Luo, Y.; Yuan, J.; Li, Y.; Zhong, Q.; Zhao, M.; Lu, Q. Histone modifications and methyl-CpG-binding domain protein levels at the TNFSF7 (CD70) promoter in SLE CD4+ T cells. Lupus 2011, 20, 1365–1371. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Liao, J.; Zhao, M.; Liang, G.; Wu, X.; Zhang, P.; Ding, S.; Luo, S.; Lu, Q. Inhibited expression of hematopoietic progenitor kinase 1 associated with loss of jumonji domain containing 3 promoter binding contributes to autoimmunity in systemic lupus erythematosus. J. Autoimmun. 2011, 37, 180–189. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liao, J.; Zhao, M.; Wu, H.; Yung, S.; Chan, T.M.; Yoshimura, A.; Lu, Q. Increased expression of TLR2 in CD4+ T cells from SLE patients enhances immune reactivity and promotes IL-17 expression through histone modifications. Eur. J. Immunol. 2015, 45, 2683–2693. [Google Scholar] [CrossRef] [PubMed]
- Apostolidis, S.A.; Rauen, T.; Hedrich, C.M.; Tsokos, G.C.; Crispin, J.C. Protein phosphatase 2A enables expression of interleukin 17 (IL-17) through chromatin remodeling. J. Biol. Chem. 2013, 288, 26775–26784. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Xiao, Y.; Shi, Y.; Luo, Y.; Li, Y.; Zhao, M.; Lu, Q.; Xiao, R. Overexpression of JMJD3 may contribute to demethylation of H3K27me3 in CD4+ T cells from patients with systemic sclerosis. Clin. Immunol. 2015, 161, 396–399. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Feng, Y.; Kang, J.; Liu, C.; Li, Z.; Li, D.; Cao, W.; Qiu, J.; Guo, Z.; Bi, E.; et al. Critical regulation of CD4+ T cell survival and autoimmunity by β-arrestin 1. Nat. Immunol. 2007, 8, 817–824. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, M.M. Primary biliary cirrhosis. N. Engl. J. Med. 1996, 335, 1570–1580. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Huang, Y.; Liu, Y.; Sun, Y.; Zhou, Y.; Gu, M.; Chen, Y.; Xia, R.; Chen, S.; Deng, A.; et al. β-Arrestin 1 modulates functions of autoimmune T cells from primary biliary cirrhosis patients. J. Clin. Immunol. 2011, 31, 346–355. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Chang, C.; Zhou, Z. Molecular mechanisms in autoimmune type 1 diabetes: A critical review. Clin. Rev. Allergy Immunol. 2014, 47, 174–192. [Google Scholar] [CrossRef] [PubMed]
- Bettini, M.L.; Pan, F.; Bettini, M.; Finkelstein, D.; Rehg, J.E.; Floess, S.; Bell, B.D.; Ziegler, S.F.; Huehn, J.; Pardoll, D.M.; et al. Loss of epigenetic modification driven by the Foxp3 transcription factor leads to regulatory T cell insufficiency. Immunity 2012, 36, 717–730. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.Y.; Xu, J.F. Reduced Histone H3 Acetylation in CD4+ T Lymphocytes: Potential Mechanism of Latent Autoimmune Diabetes in Adults. Dis. Markers 2015, 2015. [Google Scholar] [CrossRef] [PubMed]



© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Z.; Yin, H.; Lau, C.S.; Lu, Q. Histone Posttranslational Modifications of CD4+ T Cell in Autoimmune Diseases. Int. J. Mol. Sci. 2016, 17, 1547. https://doi.org/10.3390/ijms17101547
Wang Z, Yin H, Lau CS, Lu Q. Histone Posttranslational Modifications of CD4+ T Cell in Autoimmune Diseases. International Journal of Molecular Sciences. 2016; 17(10):1547. https://doi.org/10.3390/ijms17101547
Chicago/Turabian StyleWang, Zijun, Heng Yin, Chak Sing Lau, and Qianjin Lu. 2016. "Histone Posttranslational Modifications of CD4+ T Cell in Autoimmune Diseases" International Journal of Molecular Sciences 17, no. 10: 1547. https://doi.org/10.3390/ijms17101547
APA StyleWang, Z., Yin, H., Lau, C. S., & Lu, Q. (2016). Histone Posttranslational Modifications of CD4+ T Cell in Autoimmune Diseases. International Journal of Molecular Sciences, 17(10), 1547. https://doi.org/10.3390/ijms17101547