Negative Glucocorticoid Response-Like Element from the First Intron of the Chicken Growth Hormone Gene Represses Gene Expression in the Rat Pituitary Tumor Cell Line
Abstract
:1. Introduction
2. Results
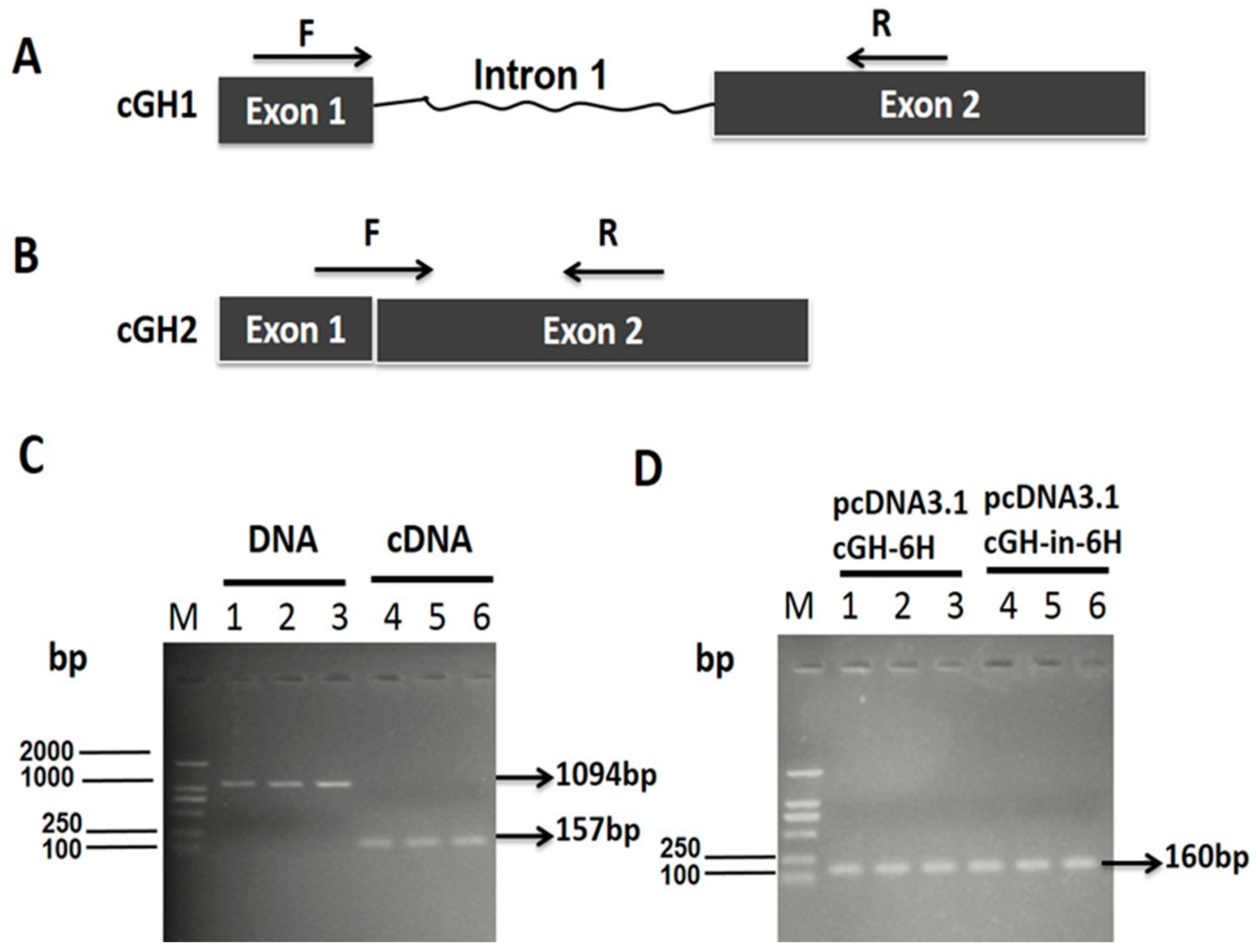
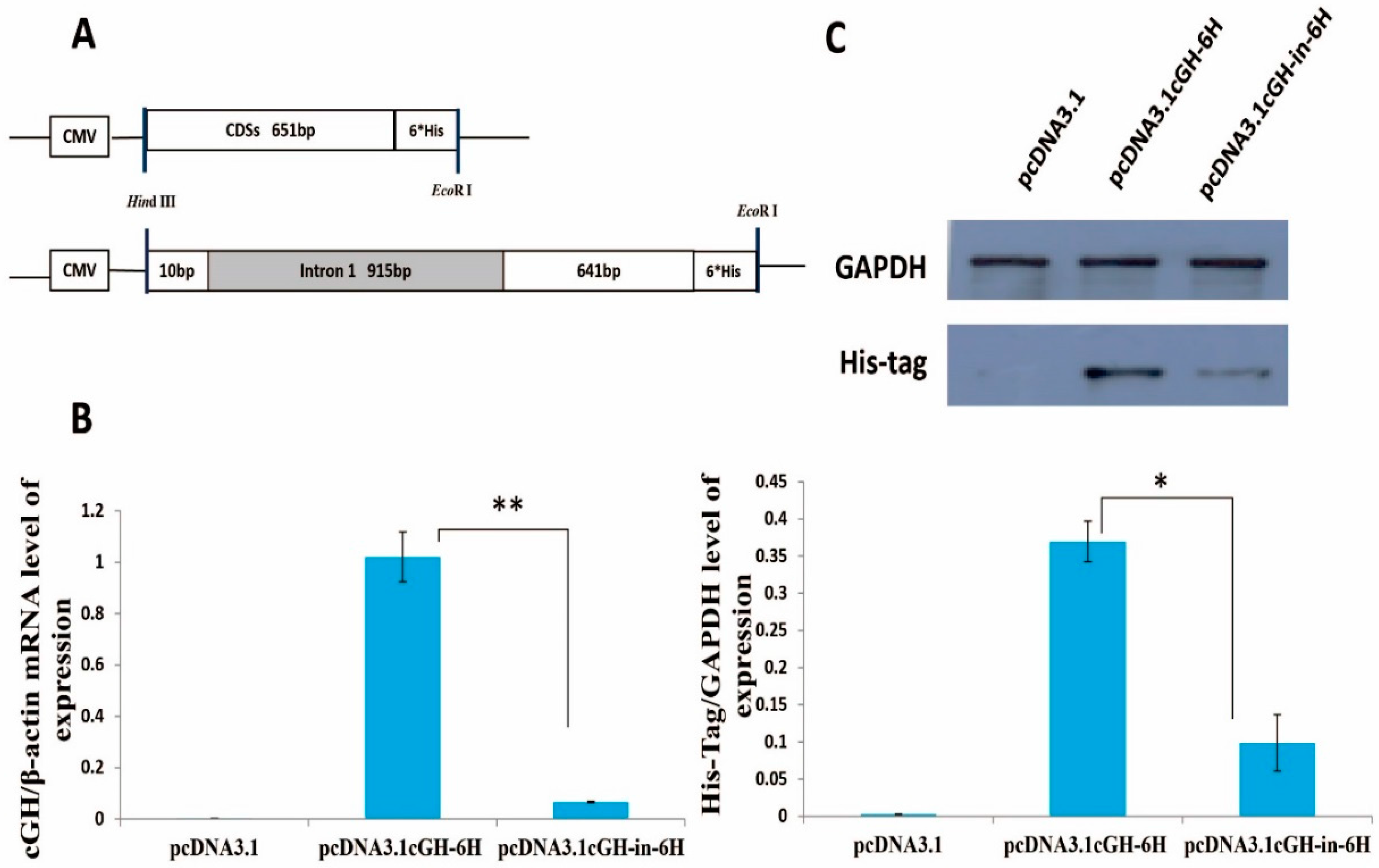
2.1. Effect of First Intron on Chicken Growth Hormone (cGH) Complete Coding Sequences (CDSs) Expression In Vitro
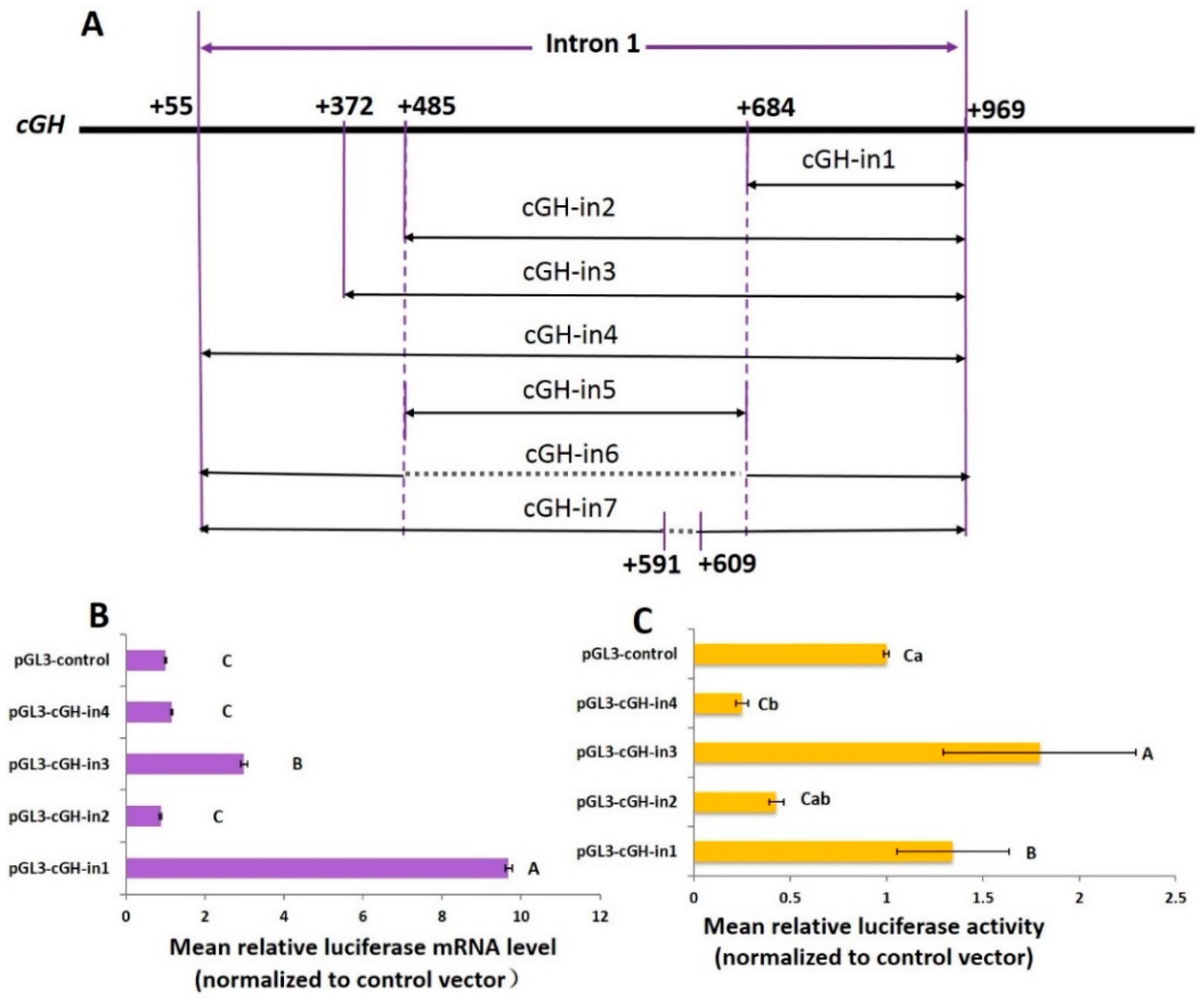
2.2. Effect of Intron 1 Fragments on Reporter Gene Expression In Vitro
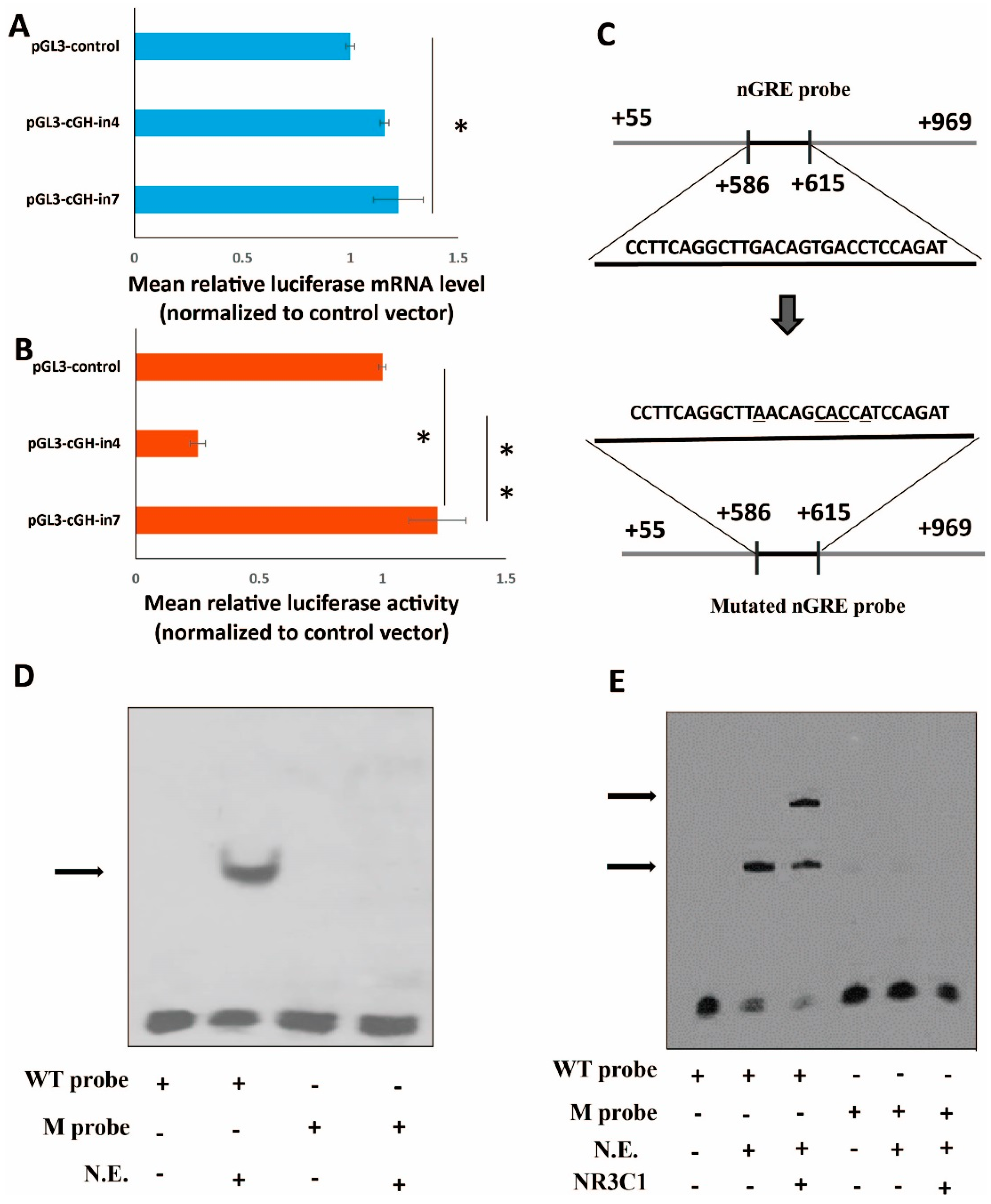
2.3. Transcriptional Factor Binding Sites (TFBSs) in Intron 1 of the cGH Gene
2.4. Role of the Intronic Negative Glucocorticoid Response Element (nGRE) in Reporter Gene Expression In Vitro
2.5. Effect of the Length of GH Intron 1 Fragments on Reporter Gene Expression In Vitro
3. Discussion
4. Materials and Methods
4.1. Construction of Plasmids
4.2. Cell Cultures
4.3. Cell Transfection
4.4. RNA Isolation, RT-PCR and Real-Time Quantitative PCR (RT-qPCR)
4.5. Dual-Luciferase Reporter Assay
4.6. Immunoblotting
4.7. Electrophoretic Mobility Shift Assay (EMSA)
4.8. Statistical Analysis
4.9. Ethics Standards
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Martínez, F.P.; Cruz, R.; Lu, F.; Plasschaert, R.; Deng, Z.; Rivera-Molina, Y.A.; Bartolomei, M.S.; Lieberman, P.M.; Tang, Q. CTCF binding to the first intron of the major immediate early (MIE) gene of human cytomegalovirus (HCMV) negatively regulates MIE gene expression and HCMV replication. J. Virol. 2014, 88, 7389–7401. [Google Scholar] [PubMed]
- Sudhakar, C.; Vaibhava, V.; Swarup, G. IRF-1-binding site in the first intron mediates interferon-γ-induced optineurin promoter activation. Biochem. Biophys. Res. Commun. 2013, 437, 179–184. [Google Scholar] [PubMed]
- Rockman, M.V.; Wray, G.A. Abundant raw material for cis-regulatory evolution in humans. Mol. Biol. Evol. 2002, 19, 1991–2004. [Google Scholar] [CrossRef] [PubMed]
- Camp, S.; de Jaco, A.; Zhang, L.; Marquez, M.; de La Torre, B.; Taylor, P. Acetylcholinesterase expression in muscle is specifically controlled by a promoter-selective enhancesome in the first intron. J. Neurosci. 2008, 28, 2459–2470. [Google Scholar] [CrossRef] [PubMed]
- Ratajewski, M.; de Boussac, H.; Sachrajda, I.; Bacquet, C.; Kovács, T.; Váradi, A.; Pulaski, L.; Arányi, T. ABCC6 expression is regulated by CCAAT/enhancer-binding protein activating a primate-specific sequence located in the first intron of the gene. J. Investig. Dermatol. 2012, 132, 2709–2717. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Cullen, B.R. Analysis of the stimulatory effect of splicing on mRNA production and utilization in mammalian cells. RNA 2003, 9, 618–630. [Google Scholar] [CrossRef] [PubMed]
- Buchman, A.R.; Berg, P. Comparison of intron-dependent and intron-independent gene expression. Mol. Cell. Biol. 1988, 8, 4395–4405. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Wu, Y.; Lan, Z.; Liu, Y.; Zhang, Y. Molecular analysis of the first intron in the bovine myostatin gene. Mol. Biol. Rep. 2011, 38, 4643–4649. [Google Scholar] [CrossRef] [PubMed]
- Charron, M.; Chern, J.Y.; Wright, W.W. The cathepsin L first intron stimulates gene expression in rat sertoli cells. Biol. Reprod. 2007, 76, 813–824. [Google Scholar] [CrossRef] [PubMed]
- Minkovsky, A.; Barakat, T.S.; Sellami, N.; Chin, M.H.; Gunhanlar, N.; Gribnau, J.; Plath, K. The pluripotency factor-bound intron 1 of Xist is dispensable for X chromosome inactivation and reactivation in vitro and in vivo. Cell Rep. 2013, 3, 905–918. [Google Scholar] [CrossRef] [PubMed]
- Bass, J.I.F.; Tamburino, A.M.; Mori, A.; Beittel, N.; Weirauch, M.T.; Reece-Hoyes, J.S.; Walhout, A.J. Transcription factor binding to Caenorhabditis elegans first introns reveals lack of redundancy with gene promoters. Nucleic Acids Res. 2014. [Google Scholar] [CrossRef]
- Almuly, R.; Skopal, T.; Funkenstein, B. Regulatory regions in the promoter and first intron of Sparus aurata growth hormone gene: Repression of gene activity by a polymorphic minisatellite. Comp. Biochem. Physiol. Part D Genom. Proteom. 2008, 3, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Nobuyoshi, M.; Lin, X.H.; Takimoto, Y.; Deuel, T.F.; Wang, Z.Y. Transcription regulation of the PDGF A-chain gene by first intron elements. Biochem. Biophys. Res. Commun. 1997, 230, 569–572. [Google Scholar] [CrossRef] [PubMed]
- Treacy, M.N.; Ryan, F.; Martin, F. Functional glucocorticoid inducible enhancer activity in the 5′-flanking sequences of the rat growth hormone gene. J. Steroid Biochem. Mol. Biol. 1991, 38, 1–15. [Google Scholar] [CrossRef]
- Slater, E.P.; Rabenau, O.; Karin, M.; Baxter, J.D.; Beato, M. Glucocorticoid receptor binding and activation of a heterologous promoter by dexamethasone by the first intron of the human growth hormone gene. Mol. Cell. Biol. 1985, 5, 2984–2992. [Google Scholar] [CrossRef] [PubMed]
- Palmiter, R.D.; Sandgren, E.P.; Avarbock, M.R.; Allen, D.D.; Brinster, R.L. Heterologous introns can enhance expression of transgenes in mice. Proc. Natl. Acad. Sci. USA 1991, 88, 478–482. [Google Scholar] [CrossRef] [PubMed]
- Heuck-Knubel, K.; Proszkowiec-Weglarz, M.; Narayana, J.; Ellestad, L.E.; Prakobsaeng, N.; Porter, T.E. Identification of cis elements necessary for glucocorticoid induction of growth hormone gene expression in chicken embryonic pituitary cells. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2012, 302, R606–R619. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhong, F.; Qi, S. Modification of pGH cDNA using the first intron and adenovirus-mediated expression in CHO cells. Chin. Med. J. 2003, 116, 1267–1269. [Google Scholar] [PubMed]
- Bernardini, S.; Argenton, F.; Vianello, S.; Colombo, L.; Bortolussi, M. Regulatory regions in the promoter and third intron of the growth hormone gene in rainbow trout, Oncorhynchus mykiss walbaum. Gen. Comp. Endocrinol. 1999, 116, 261–271. [Google Scholar] [CrossRef] [PubMed]
- Sekkali, B.; Brim, H.; Muller, M.; Argenton, F.; Bortolussi, M.; Colombo, L.; Belayew, A.; Martial, J.A. Structure and functional analysis of a tilapia (Oreochromis mossambicus) growth hormone gene: activation and repression by pituitary transcription factor Pit-1. DNA Cell Biol. 1999, 18, 489–502. [Google Scholar] [CrossRef] [PubMed]
- Argenton, F.; Vianello, S.; Bernardini, S.; Lopreiato, R.; Colombo, L.; Bortolussi, M. Trout GH promoter analysis reveals a modular pattern of regulation consistent with the diversification of GH gene control and function in vertebrates. Mol. Cell. Endocrinol. 2002, 189, 11–23. [Google Scholar] [CrossRef]
- Argenton, F.; Bernardini, S.; Puttini, S.; Colombo, L.; Bortolusi, M. A TGACG motif mediates growth-hormone factor-1/pituitary-transcriptional-activator-1-dependent cAMP regulation of the rainbow trout growth-hormone promoter. Eur. J. Biochem. 1996, 238, 591–598. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.Y.; Chan, K.M.; Lin, C.M.; Chen, T.T. Characterization of rainbow trout (Oncorhynchus mykiss) growth hormone 1 gene and the promoter region of growth hormone 2 gene. Arch. Biochem. Biophys. 1997, 340, 359–368. [Google Scholar] [CrossRef] [PubMed]
- Sekkali, B.; Belayew, A.; Bortolussi, M.; Martial, J.A.; Muller, M. Pit-1 mediates cell-specific and cAMP-induced transcription of the tilapia GH gene. Mol. Cell. Endocrinol. 1999, 152, 111–123. [Google Scholar] [CrossRef]
- Ip, S.C.; Lau, J.S.; Au, W.L.; Leung, F.C. Characterization of the 5′-flanking transcriptional regulatory region of chicken growth hormone gene. Exp. Biol. Med. 2004, 229, 640–649. [Google Scholar]
- Liu, W.S.; Ma, J.E.; Li, W.X.; Zhang, J.G.; Wang, J.; Nie, Q.H.; Qiu, F.F.; Fang, M.X.; Zeng, F.; Wang, X.; et al. The long intron 1 of growth hormone gene from Reeves’ Turtle (Chinemys reevesii) correlates with negatively regulated GH expression in four cell lines. Int. J. Mol. Sci. 2016, 17, 543. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.F.; Niu, D.K. Evidence against the energetic cost hypothesis for the short introns in highly expressed genes. BMC Evol. Biol. 2008, 8, 154. [Google Scholar] [CrossRef] [PubMed]
- Martinez, R.; Estrada, M.P.; Berlanga, J.; Guillen, I.; Hernandez, O.; Cabrera, E.; Pimentel, R.; Morales, R.; Herrera, F.; Morales, A.; et al. Growth enhancement in transgenic tilapia by ectopic expression of tilapia growth hormone. Mol. Mar. Biol. Biotechnol. 1996, 5, 62–70. [Google Scholar] [PubMed]
- Benbassat, C.; Shoba, L.N.; Newman, M.; Adamo, M.L.; Frank, S.J.; Lowe, W.L., Jr. Growth hormone-mediated regulation of insulin-like growth factor I promoter activity in C6 glioma cells. Endocrinology 1999, 140, 3073–3081. [Google Scholar] [CrossRef] [PubMed]
- Hoeflich, A.; Weber, M.M.; Fisch, T.; Nedbal, S.; Fottner, C.; Elmlinger, M.W.; Wanke, R.; Wolf, E. Insulin-like growth factor binding protein 2 (IGFBP-2) separates hypertrophic and hyperplastic effects of growth hormone (GH)/IGF-I excess on adrenocortical cells in vivo. FASEB J. 2002, 16, 1721–1731. [Google Scholar] [CrossRef] [PubMed]
- Qin, W.; Pan, J.; Qin, Y.; Lee, D.N.; Bauman, W.A.; Cardozo, C. Identification of functional glucocorticoid response elements in the mouse FoxO1 promoter. Biochem. Biophys. Res. Commun. 2014, 450, 979–983. [Google Scholar] [CrossRef] [PubMed]
- Sakata, H.; Maeno, M. Nkx2.5 is involved in myeloid cell differentiation at anterior ventral blood islands in the Xenopus embryo. Dev. Growth Differ. 2014, 56, 544–554. [Google Scholar] [CrossRef] [PubMed]
- Kurabe, N.; Murakami, S.; Tashiro, F. SGF29 and Sry pathway in hepatocarcinogenesis. World J. Biol. Chem. 2015, 6, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.; Li, X.; Guo, B.; Ke, Q.; Dong, M.; Li, F. PAK5-mediated E47 phosphorylation promotes epithelial-mesenchymal transition and metastasis of colon cancer. Oncogene 2016, 35, 1943–1954. [Google Scholar] [CrossRef] [PubMed]
- Hu, F.; Wang, C.; Du, J.; Sun, W.; Yan, J.; Mi, D.; Zhang, J.; Qiao, Y.; Zhu, T.; Yang, S. δEF1 promotes breast cancer cell proliferation through down-regulating p21 expression. Biochim. Biophys. Acta Mol. Basis Dis. 2010, 1802, 301–312. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Shu, Y.; Liang, X.; Chen, E.C.; Yee, S.W.; Zur, A.A.; Li, S.; Xu, L.; Keshari, K.R.; Lin, M.J.; et al. OCT1 is a high-capacity thiamine transporter that regulates hepatic steatosis and is a target of metformin. Proc. Natl. Acad. Sci. USA 2014, 111, 9983–9988. [Google Scholar] [CrossRef] [PubMed]
- Irimia, M.; Roy, S.W. Origin of spliceosomal introns and alternative splicing. Cold Spring Harb. Perspect. Biol. 2014, 6, a016071. [Google Scholar] [CrossRef] [PubMed]
- Niemelä, E.H.; Frilander, M.J. Regulation of gene expression through inefficient splicing of U12-type introns. RNA Biol. 2014, 11, 1325–1329. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Dostert, A.; Heinzel, T. Negative glucocorticoid receptor response elements and their role in glucocorticoid action. Curr. Pharm. Des. 2004, 10, 2807–2816. [Google Scholar] [PubMed]

| Position | Name |
|---|---|
| +55/+372 | AP1FJ, AP1, AP4, AML1, CAP, CDXA, CEBPB, CEBP, CREL, GATA3, GATA2, GATA1, GR, LMO2COM, MZF1, OCT1, P53, SRY, SP1, TST1 |
| +372/+485 | CAP, CDXA, CETS1P54, DELTAEF1, MYOD, NKX25, SRY, TATA |
| +485/+684 | AP4, AML1, CAP, CETS1P54, CEBPB, DELTAEF1, E47, GRE, LMO2COM, MZF1, NKX25, SRY, USF |
| +684/+969 | AP4, CAP, CEBPB, E2F, E47, HNF3β, IK2, LMO2COM, MZF1, MYB, STAT, USF |
| Groups | Length (bp) | Mean mRNA Expression Level ± SEM | Mean Protein Expression Level ± SEM |
|---|---|---|---|
| pGL3-cGH-in4 | 915 | 1.159 ± 0.0201 | 0.252 ± 0.0313 |
| pGL3-cGH-in7 | 897 | 1.716 ± 0.1975 | 1.223 ± 0.3150 |
| pGL3-cGH-in6 | 715 | 1.638 ± 0.1396 | 1.054 ± 0.1044 |
| pGL3-cGH-in3 | 598 | 2.992 ± 0.0904 | 1.79 ± 0.4998 |
| pGL3-cGH-in2 | 485 | 0.878 ± 0.0238 | 0.429 ± 0.0386 |
| pGL3-cGH-in1 | 286 | 9.681 ± 0.0885 | 1.344 ± 0.2907 |
| pGL3-cGH-in5 | 200 | 1.649 ± 0.1000 | 0.632 ± 0.0640 |
| pGL3-control | 0 | 1.007 ± 0.0224 | 1.003 ± 0.0436 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, J.-E.; Lang, Q.-Q.; Qiu, F.-F.; Zhang, L.; Li, X.-G.; Luo, W.; Wang, J.; Wang, X.; Lin, X.-R.; Liu, W.-S.; et al. Negative Glucocorticoid Response-Like Element from the First Intron of the Chicken Growth Hormone Gene Represses Gene Expression in the Rat Pituitary Tumor Cell Line. Int. J. Mol. Sci. 2016, 17, 1863. https://doi.org/10.3390/ijms17111863
Ma J-E, Lang Q-Q, Qiu F-F, Zhang L, Li X-G, Luo W, Wang J, Wang X, Lin X-R, Liu W-S, et al. Negative Glucocorticoid Response-Like Element from the First Intron of the Chicken Growth Hormone Gene Represses Gene Expression in the Rat Pituitary Tumor Cell Line. International Journal of Molecular Sciences. 2016; 17(11):1863. https://doi.org/10.3390/ijms17111863
Chicago/Turabian StyleMa, Jing-E., Qian-Qian Lang, Feng-Fang Qiu, Li Zhang, Xiang-Guang Li, Wen Luo, Juan Wang, Xing Wang, Xi-Ran Lin, Wen-Sheng Liu, and et al. 2016. "Negative Glucocorticoid Response-Like Element from the First Intron of the Chicken Growth Hormone Gene Represses Gene Expression in the Rat Pituitary Tumor Cell Line" International Journal of Molecular Sciences 17, no. 11: 1863. https://doi.org/10.3390/ijms17111863







